Abstract
AIM
Genetic variants of the enzyme that metabolizes warfarin, cytochrome P-450 2C9 (CYP2C9) and of a key pharmacologic target of vitamin K antagonists, vitamin K epoxide reductase (VKORC1), contribute to differences in patients' responses to coumarin derivatives. The role of these variants in fluindione response is unknown. Our aim was to assess whether genetic factors contribute to the variability in the response to fluindione.
METHODS
Four hundred sixty-five patients with a venous thromboembolic event treated by fluindione for at least 3 months with a target international normalized ratio (INR) of 2.0 to 3.0 were studied. VKORC1, CYP2C9, CYP4F2 and EPHX1 genotypes were assessed. INR checks, fluindione doses and bleeding events were collected.
RESULTS
VKORC1 genotype had a significant impact on early anticoagulation (INR value ≥2 after the first two intakes) (P < 0.0001), on the time required to reach a first INR within the therapeutic range (P < 0.0001) and on the time to obtain a first INR value > 4 (P = 0.0002). The average daily dose of fluindione during the first period of stability was significantly associated with the VKORC1 genotype: 19.8 mg (±5.5) for VKORC1 CC, 14.7 mg (±6.2) for VKORC1 CT and 8.2 mg (±2.5) for VKORC1 TT (P < 0.0001). CYP2C9, CYP4F2 and EPHX1 genotypes did not significantly influence the response to fluindione.
CONCLUSIONS
VKORC1 genotype strongly affected anticoagulation induced by fluindione whereas CYP2C9, CYP4F2 and EPHX1 genotypes seemed less determining.
Keywords: CYP2C9, CYP4F2, EPHX1, fluindione, pharmacogenetics, VKORC1
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
CYP2C9 and VKORC1 genetic variants contribute to differences in patients' responses to anticoagulant coumarin derivatives. Patients carrying the VKORC1 1173TT genotype have a decreased time to the first INR within the therapeutic range and to the first INR >4, and also require lower warfarin maintenance doses. Patients carrying the *2 or *3 CYP2C9 allele have lower maintenance warfarin requirements than those carrying the wild-type allele. The role of CYP2C9 and VKORC1 genetic variants in fluindione response is unknown.
WHAT THIS STUDY ADDS
Our results showed that VKORC1 genotype had a significant impact on early anticoagulation (INR value ≥2 after the first two intakes) (P < 0.0001), on the time required to reach a first INR within the therapeutic range (P < 0.0001), on the time to obtain a first INR value >4 (P = 0.0002) and on the average daily dose of fluindione during the first period of stability (19.8 mg (±5.5) for VKORC1 CC, 14.7 mg (±6.2) for VKORC1 CT and 8.2 mg (±2.5) for VKORC1 TT, P < 0.0001). CYP2C9, CYP4F2 and EPHX1 genotypes did not significantly influence the response to fluindione. This report provides new information on the respective role of common genetic polymorphisms on anticoagulation induced by another class of anticoagulant drugs rather than coumarin derivatives.
Introduction
Until the use of next generation oral anticoagulants becomes more common, vitamin K antagonists (VKAs) are still the drugs prescribed for long term oral anticoagulation. VKA therapy has demonstrated efficacy to reduce the occurrence of thromboembolic events in various clinical settings. However, because of a narrow therapeutic index, treatment with VKA is difficult to manage and needs frequent biological monitoring to adjust the dose and avoid the risk of thromboembolic or bleeding events. Despite these safety measures, VKA therapy is frequently associated with serious adverse reactions leading to an important morbidity and mortality [1].
Warfarin, a coumarin derivative, is the most widely prescribed VKA around the world. Nevertheless, as a ‘French exception’, fluindione, an indanedione derivative, is the first VKA used in France, accounting for about 70% of oral anticoagulant prescription. Fluindione has been sparsely studied and no clear pharmacological advantage over warfarin supported this choice.
Genetic variants of the enzyme that metabolizes warfarin, cytochrome P-450 2C9 (CYP2C9) and the key pharmacologic target of warfarin, vitamin K epoxide reductase (VKORC1) contribute to differences in patients' responses to various warfarin doses [2–10]. More recently, two other genotype variants, EPHX1 and CYP4F2 that encode proteins involved in warfarin action and metabolism were found to be associated with warfarin response and to be additional predictive variables for the maintenance dose of warfarin [11–14]. No data exist on the influence of these polymorphisms on fluindione action.
Our objective was to assess whether genetic factors (VKORC1, CYP2C9, CYP4F2 and EPHX1) contribute to the variability of fluindione response in patients treated for a venous thromboembolic event.
Methods
Patients
All patients included in this analysis were cases of an ongoing hospital-based case control study (EDITH study) designed to evaluate interactions between inherited and acquired risk factors of venous thromboembolism [15]. They had an objectively confirmed venous thromboembolic event occurring between May 2000 and December 2004. This thrombotic event was considered as unprovoked, i.e. absence of surgery or plaster cast immobilization in the past 3 months, pregnancy or post partum in the past 3 months or active cancer.
The protocol was approved by our hospital's scientific and ethics board. A specific written consent for participation in the study and for the DNA analysis was obtained from all patients.
Only patients receiving fluindione for at least 3 months to treat this thrombotic event were selected for this report. The international normalized ratio (INR) was used to follow the anticoagulant treatment and adapt the fluindione doses. The target INR therapeutic range was 2.0 to 3.0. Patients with a different INR therapeutic range were excluded. Since the patients were not followed-up in an anticoagulant clinic, available quantitative and qualitative data for each patient were variable. We retained patients for whom we were able to collect enough data to assess one of our following outcomes.
Data collection
The following variables were recorded prospectively: age, gender, height, weight and smoking status defined as non-smoker, past smoker or current smoker. All other variables retained for this analysis were collected retrospectively from a review of medical charts and records by two trained abstractors and one of the authors (KL). The date of starting fluindione treatment and all concomitant medications, taken at the time of the first intake of fluindione, were recorded. Concomitant medications were classified according to their impact on CYP2C9 (inhibitors or inducers) using the drug interaction table defined by Flockhart [16]. INR was monitored by hospital practitioners during the hospital stay and by general or hospital practitioners after hospital discharge. During hospitalization, we collected all INR values included in the medical record and asked the hospital laboratory for other INR checks. We recorded fluindione daily dose used and adapted after each INR measurement. After discharge, we collected INR values found in the medical record from local laboratories or measured during a hospital visit. We also asked local laboratories for other INR checks. We recorded all available fluindione doses preceding INR measurement by checking medical records, information from local laboratories, from the patient's anticoagulant notebook and eventually by calling the patient or the general practitioner.
First doses of fluindione were not standardized and were done at the discretion of the primary care physician who initiated fluindione therapy. Adaptation of doses was also performed at the discretion of the primary care physician without standardized method. Patients were followed-up from the first date of fluindione use until the last INR value recorded in the database.
We also collected data on bleeding events. Bleeding was classified as major if it was intraocular, spinal/epidural, intracranial or retroperitoneal, if haemoglobin decreased by ≥2 g dl−1, if transfusion of ≥2 U of blood or significant medical or surgical intervention was required or if it resulted in death. Clinically relevant but not major haemorrhage was defined as macroscopic haematuria (spontaneous or persistent more than 24 h after an invasive procedure), or a significant epistaxis leading to local treatment or a cutaneous haematoma larger than 10 cm2. Minor bleedings were all other bleedings and were not recorded. The date of the haemorrhage, INR value and fluindione dosage at the time of the bleeding event were recorded, if available as well as comedications and treatments used for the bleeding event.
Genotyping
Blood was collected in 0.05 m EDTA for DNA analysis. Genomic DNA was extracted from peripheral leucocytes from EDTA-anticoagulated blood using a commercially available DNA isolation kit (Qiagen, S.A., Courtaboeuf, France). Genomic DNA was analyzed by investigators blinded to patient characteristics or outcomes.
For VKORC1, we chose the VKORC1 C1173 T SNP (rs9934438) to tag the major VKORC1 haplotype groups A and B used in the nomenclature proposed by Rieder et al. [4]. The C wild type allele of the C1173T VKORC1 genetic polymorphism corresponds to the group B VKORC1 haplotype and the T allele to the group A VKORC1 haplotype. This SNP is in complete linkage disequilibrium with at least four other SNPs which individually allow the identification of VKORC1 haplotype groups [4, 17, 18].
Genotyping for the CYP2C9*2 (rs1799853), *3 (rs1057910) and C1173T VKORC1 (rs9934438) allele variants was performed using the TaqMan allelic discrimination assay (ABI prism 7000, Applied Biosystems, Courtaboeuf, France) as previously described [19, 20].
For the G357A EPHX1 (rs 2292566, GenBank accession NT 167186.1), and C1347T CYP4F2 (rs2108622, GenBank accession NT011295.11) polymorphisms, SNPs primers and probes were designed using the Primer express software from Applied Biosystem (Forster City, CA, USA). For the C1347T CYP4F2 polymorphism, the wild type allele «C» 5′-ACAACCCAGCTGTGT-3′ and variant allele «T» 5′-ACAACCCAGCTATGT-3′ probes were labeled with VIC and FAM fluorescent marker respectively at their 5′ extremity. The forward 5′-GCCTCATCAGTGTTTTCGGAAC-3′ and reverse 5′-GGAATGGACAAAAACAGAGAGAGG-3′ primers were used for amplification.
For the G357A EPHX1 polymorphism, the wild type allele «G» 5′-CTTCAAGACTAAGATTGA-3′ and variant allele «A» 5′-CTTCAAGACTAAAATTGA-3′ probes were labelled with VIC and FAM fluorescent marker respectively at their 5′ extremity. The forward 5′-AGCAGGTGGAGATTCTCAACAGA-3′ and reverse 5′-AGAAGGCTGTTCTCATGACATACATC-3′ primers were used for PCR.
Each SNP genotyping procedure was performed in duplicate (separate experiments) for each patient. For any obtained discrepancy, samples were analyzed by DNA sequencing to confirm the genotype. Sequenced wild-type, homozygous and heterozygous patient samples were used as controls. All PCR reagents were purchased from Applied Biosystems.
Outcomes
In this report, we determined the influence of VKORC1, CYP2C9, CYP4F2 and EPHX1 genotypes on the following outcomes:
Early anticoagulation, defined as the INR value after the first two fluindione intakes (about 36 h after the first intake). We evaluated the association of each genotype variant and the risk of this first INR value ≥2.
Time to achieve a first INR in the therapeutic range (2 ≤ INR ≤ 3) during the first 6 months of follow-up.
Risk of over-anticoagulation, defined as INR value >4, and the time to achieve this INR value during the first 6 months of follow-up.
Chance and time to achieve a first period of stability. This period was calculated as the time until the first of two consecutive INR measurements within the therapeutic range (2 ≤ INR ≤ 3), with these INR measurements encompassing a period of at least 2 weeks, without modification of fluindione dose. For this analysis, we retained among all patients those with at least 4 weeks of follow-up and at least six INR values during this period.
Average daily dose of fluindione during the first period of stability. We also evaluated the contribution of the study recorded genetic and environmental factors on the fluindione dose requirements at the first period of stability.
Bleeding events occurring during the fluindione treatment period.
Statistical analyses
Descriptive statistics for the patient population were generated with the use of point estimates (means ± SD or frequencies) where appropriate.
For all genetic polymorphisms, chi square tests (or Fisher's exact tests when appropriate) were performed, with a significance level of α = 0.05, to ensure that the allele frequencies were within Hardy–Weinberg equilibrium.
For the genetic variables, we defined three groups according to the different allelic variants and coded genotypes as 0 (wild type), 1 (heterozygous), or 2 (homozygous) except for CYP2C9 SNPs coded as 0 (wild type), 1 (heterozygous for CYP2C9*2 or CYP2C9*3), 2 (homozygous for CYP2C9*2 or CYP2C9*3 or compound heterozygous for CYP2C9*2 and CYP2C9*3).
Univariate analysis for binary outcomes (first INR ≥ 2, INR > 4, chance to achieve a first period of stability and haemorrhage), we used a chi square test to compare frequencies between genotypes. The time to first INR within the therapeutic range and the time to the first INR > 4 were compared among genotype groups using log-rank tests. The average daily dose of fluindione during the first period of stability was compared among genotypes using the Kruskall Wallis test.
Multivariate analyses were performed for genotypes significantly associated with pre-defined outcomes in univariate analyses. Logistic regression analyses were performed to determine the odds ratio (OR) for having a first INR value ≥ 2, an INR > 4, a chance to achieve a first period of stability or a haemorrhage according to genotype variant group. The Cox proportional hazard model was used to estimate hazard ratios (HR) for the time to the first INR in the therapeutic range and the time to a first INR > 4 according to genotype variant group. All multivariate analyses were adjusted on potential confounding variables: age, gender, smoking status, body mass index, initial dose of fluindione and medications impacting on CYP2C9. To determine the relative contribution of genetic and environnemental factors on average daily dose of fluindione during the first period of stability, confounding variables, except initial dose of fluindione, and all genotypes were entered into a least square regression model which was then reduced using stepwise backward elimination with a significance threshold for staying in the model set at α = 0.10.
To maintain an overall two-sided significance level of 0.05 in the analyses of outcomes, the nominal significance level was adjusted according to Bonferroni's method. A two-tailed P value of less than 0002 was considered to indicate statistical significance [six outcomes, four polymorphisms: 0.05/(6 × 4) = 0.002].
All statistical analyses were carried out using SAS Version 9.1.
Results
Population characteristics
Of the 677 patients with idiopathic venous thromboembolism included in the Edith study from May 2000 to December 2004, 495 were treated by fluindione. Among those 495, 30 were excluded from this analysis because of the following reasons: not enough collected data to assess one of the pre-specified outcomes (n = 6), out of range target INR (n = 2), no blood sample, no DNA or DNA of poor quality (n = 22). Finally, 465 patients were included in this analysis. Their characteristics are presented in Table 1.
Table 1.
Baseline characteristics of the 465 patients treated with fluindione for a venous thromboembolic event
| Variables | n (%) or mean ± SD or median [range] |
|---|---|
| Gender | |
| Women | 261 (56) |
| Men | 204 (44) |
| Ethnic group | |
| Caucasian | 463 (99.6) |
| Other | 2 (0.4) |
| Age (years) | 69.3 ± 16.5 |
| Weight (kg) | 71.9 ± 15.3 |
| Body mass index (kg m–2) | 26.2 ± 4.6 |
| Smoking status | |
| Non smoker | 235 (50.5) |
| Past smoker | 144 (31.0) |
| Current smoker | 52 (11.2) |
| Unknown | 34 (7.3) |
| Follow-up (days) | 83.8 ± 148.2 |
| 33 [3–1930] | |
| Number of INR determinations | 10.2 ± 7.5 |
| 8 [2–60] | |
| Initial dosage of fluindione (mg) | 17.6 ± 4.1 |
| 20 [5–30] | |
| Concomitant medications | |
| Inhibitors of 2C9 | 75 (16.1) |
| Inducers of 2C9 | 0 |
Allelic frequencies
VKORC1 C1173T, CYP2C9 and EPHX1 genotypes were missing for two, three and one patients respectively. For VKORC1 C1173T, the frequency of T allele was 38%. For CYP2C9, the frequencies of 2C9*1, 2C9*2 and 2C9*3 were 82.3%, 12.2% and 5.5% respectively. For CYP4F2, the frequency of T allele was 29.4%. For EPHX1, the frequency of A allele was 11.9%. No significant deviation from Hardy-Weinberg equilibrium was observed for any polymorphism (P > 0.20).
Early anticoagulation: risk of the first measured INR ≥ 2
An INR value after the first two fluindione intakes (about 36 h after the start of fluindione) was available for 336 patients. The VKORC1 polymorphism had a significant impact on the risk of the first INR value ≥2 whereas CYP2C9, CYP4F2 and EPHX1 did not (Table 2). Using VKORC1 CC as the reference, the ORs estimating the risk of the first INR value ≥ 2 were 3.2 (95% CI 1.3, 7.8) for VKORC1 CT and 22.7 (95% CI 8.4, 62) for VKORC1 TT, after adjustment for age, gender, body mass index, smoking status, initial dose of fluindione and concomitant medications.
Table 2.
Impact of VKORC1, CYP2C9, CYP4F2 and EPHX1 polymorphisms on early anticoagulation (the INR value after the first two fluindione intakes)
| INR value after the first two intakes of fluindione (n = 336) | |||
|---|---|---|---|
| INR < 2 (n = 277) | INR≥2 (n = 59) | P value | |
| VKORC1 (n = 334) | n = 275 | n = 59 | |
| CC | 113 (94.2%) | 7 (5.8%) | <0.0001 |
| CT | 140 (84.3%) | 26 (15.7%) | |
| TT | 22 (45.8%) | 26 (54.2%) | |
| CYP2C9 (n = 334) | n = 276 | n = 58 | |
| *1/*1 | 191 (83.8%) | 37 (16.2%) | 0.72 |
| *1/*2 or *1/*3 | 73 (80.2%) | 18 (19.8%) | |
| *2/*2 or *3/*3 or *2/*3 | 12 (80.0%) | 3 (20.0%) | |
| CYP4F2 (n = 336) | n = 277 | n = 59 | |
| CC | 141 (82.9%) | 29 (17.1%) | 0.97 |
| CT | 109 (82.0%) | 24 (18.0%) | |
| TT | 27 (81.8%) | 6 (18.2%) | |
| EPHX1 (n = 335) | n = 276 | n = 59 | |
| GG | 218 (82.3%) | 47 (17.7%) | 0.82 |
| GA | 52 (83.9%) | 10 (16.1%) | |
| AA | 6 (75.0%) | 2 (25.0%) | |
Time to achieve a first INR in the therapeutic range (2 ≤ INR ≤ 3)
All patients (n = 465) were included in this subsection analysis. Figure 1A–D shows the time to achieve the first INR in the therapeutic range among various genotypes. The VKORC1 C1173T genotypes had a significant impact on the time required to reach a first INR within the therapeutic range (P < 0.0001). The mean times to reach a first INR within the therapeutic range were 7.9 days (±10.8) for VKORC1 CC, 7.0 days (±10.5) for VKORC1 CT and 5.1 days (±5.9) for VKORC1 TT. Among patients with VKORC1 TT, the time to achieve a first INR within the therapeutic range was shorter than for patients with VKORC1 CC after adjustment for confounding covariates (adjusted HR 2.4, 95% CI 1.7, 3.3; P < 0.0001). The same findings were found for patients with VKORC1 CT (adjusted HR 1.5, 95% CI 1.2, 1.9; P = 0.0001).
Figure 1.
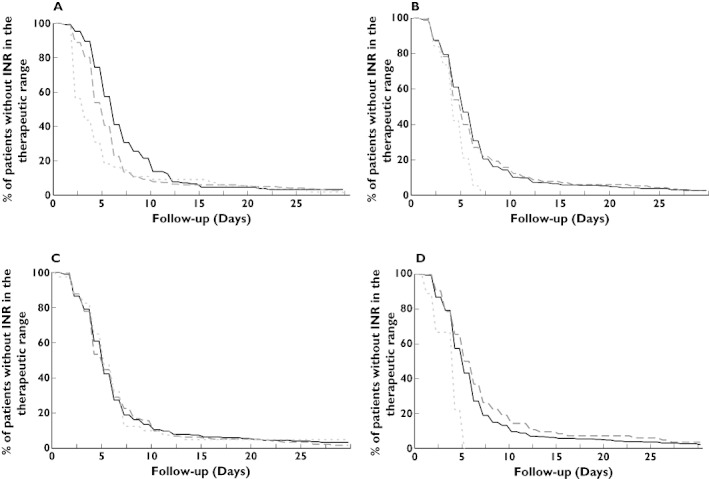
The graphs show the association between the time to first international normalized ratio (INR) within the therapeutic range for patients carrying genetic variants for VKORC1 (A), CYP2C9 (B), CYP4F2 (C) and EPHX1 (D). The results presented on the graphs are limited to the first 30-day period. P values correspond to the log-rank test results. (A) VKORC1 CC ( ); VKORC1 CT (
); VKORC1 CT ( ); VKORC1 TT (
); VKORC1 TT ( ); (B) 2C9 *1/*1 (
); (B) 2C9 *1/*1 ( ); 2C9 *1/*2 or *1/*3 (
); 2C9 *1/*2 or *1/*3 ( ); 2C9 *2/*2 or *2/*3 or *3/*3 (
); 2C9 *2/*2 or *2/*3 or *3/*3 ( ); (C) CYP4F2 CC (
); (C) CYP4F2 CC ( ); CYP4F2 CT (
); CYP4F2 CT ( ); CYP4F2 TT (
); CYP4F2 TT ( ); (D) EPHX1 GG (
); (D) EPHX1 GG ( ); EPHX1 GA (
); EPHX1 GA ( ); EPHX1 AA (
); EPHX1 AA ( )
)
CYP2C9, CYP4F2 and EPHX1 genotypes had no significant impact on the time that was required to reach a first INR within the therapeutic range (respectively P = 0.03, P = 0.92 and P = 0.003).
Risk of over-anticoagulation (INR value > 4) and time to achieve a first INR > 4 during the first 6 months of follow-up
All patients (n = 465) were included in this subsection analysis. None of the four polymorphisms had a significant impact on the risk of at least one INR value >4 during the first 6 months of follow-up (Table 3) even if a higher proportion of patients with VKORC1 CT and VKORC1 TT had a risk to be over-anticoagulated compared with patients with VKORC1 CC.
Table 3.
Impact of VKORC1, CYP2C9, CYP4F2 and EPHX1 polymorphisms on the risk of INR > 4 (at least one INR value > 4) during the first 6 months of follow-up
| INR≤4 (n = 218) | INR > 4 (n = 247) | P value | |
|---|---|---|---|
| VKORC1 (n = 463) | n = 218 | n = 245 | |
| CC | 95 (54.9%) | 78 (45.1%) | <0.03 |
| CT | 98 (42.8%) | 131 (57.2%) | |
| TT | 25 (41.0%) | 36 (59.0%) | |
| CYP2C9 (n = 462) | n = 217 | n = 245 | |
| *1/*1 | 153 (48.3%) | 164 (51.7%) | 0.68 |
| *1/*2 or *1/*3 | 55 (43.7%) | 71 (56.3%) | |
| *2/*2 or *3/*3 or *2/*3 | 9 (47.4%) | 10 (52.6%) | |
| CYP4F2 (n = 465) | n = 218 | n = 247 | |
| CC | 108 (46.2%) | 126 (53.8%) | 0.56 |
| CT | 87 (46.0%) | 102 (54.0%) | |
| TT | 23 (54.8%) | 19 (45.2%) | |
| EPHX1 (n = 464) | n = 217 | n = 247 | |
| GG | 163 (44.9%) | 200 (55.1%) | 0.21 |
| GA | 50 (54.9%) | 41 (45.1%) | |
| AA | 4 (40.0%) | 6 (60.0%) |
Figure 2A–D shows the time to achieve the first INR value >4 among various genotypes. The VKORC1 C1173T genotype had a significant impact on the time to obtain a first INR value >4 (P = 0.0002). On the contrary, CYP2C9, CYP4F2 and EPHX1 genotypes had no impact on the time to obtain a first INR value >4 (respectively P = 0.61, P = 0.49 and P = 0.23).
Figure 2.
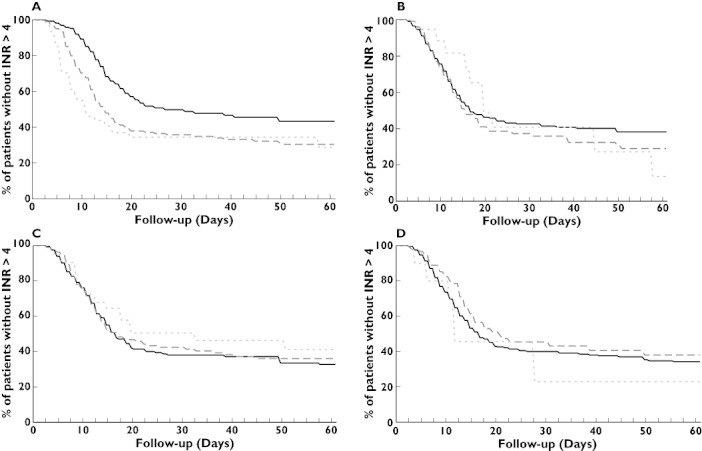
The graphs show the association between the time to first international normalized ratio (INR) of more than 4 for patients carrying genetic variants for VKORC1 (A), CYP2C9 (B), CYP4F2 (C) and EPHX1 (D). The results presented on the graphs are limited to the first 60-day period. P values correspond to the log-rank test results. (A) VKORC1 CC ( ); VKORC1 CT (
); VKORC1 CT ( ); VKORC1 TT (
); VKORC1 TT ( ); (B) 2C9 *1/*1 (
); (B) 2C9 *1/*1 ( ); 2C9 *1/*2 or *1/*3 (
); 2C9 *1/*2 or *1/*3 ( ); 2C9 *2/*2 or *2/*3 or *3/*3 (
); 2C9 *2/*2 or *2/*3 or *3/*3 ( ); (C) CYP4F2 CC (
); (C) CYP4F2 CC ( ); CYP4F2 CT (
); CYP4F2 CT ( ); CYP4F2 TT (
); CYP4F2 TT ( ); (D) EPHX1 GG (
); (D) EPHX1 GG ( ); EPHX1 GA (
); EPHX1 GA ( ); EPHX1 AA (
); EPHX1 AA ( )
)
The mean times to obtain a first INR value > 4 were 54.9 days (±173.2) for VKORC1 CC, 30.2 days (±54.3) for VKORC1 CT and 24.9 days (±47.6) for VKORC1 TT.
Among patients with VKORC1 TT, the time to achieve an INR > 4 during the first 6 months was shorter than for patients with VKORC1 CC after adjustment on confounding covariates (adjusted HR 2.2, 95% CI 1.5, 3.3; P = 0.0001). The same findings were found for patients with VKORC1 CT (adjusted HR 1.6, 95% CI 1.2, 2.2; P = 0.001).
Chance and time to achieve a first period of stability
For this analysis, among the 465 included patients, we retained those who had a follow-up of at least 4 weeks and at least six INR values during this period leading to a selection of 232 patients. Among those 232 patients, 131 (56.5%) achieved a first period of stability as previously defined. None of the four polymorphisms had a significant influence on the chance to achieve a first period of stability (results not shown). None of the four polymorphisms influenced the time required to reach a first period of stability (results not shown).
Average daily dose of fluindione during the first period of stability
The average daily dose of fluindione during the first period of stability was determined for the 131 patients who achieved a first period of stability. The average daily dose of fluindione was significantly associated with the VKORC1 genotype. The average daily doses of fluindione were 19.8 mg (±5.5) for VKORC1 CC, 14.7 mg (±6.2) for VKORC1 CT and 8.2 mg (±2.5) for VKORC1 TT (P < 0.0001).
CYP2C9, CYP4F2 and EPHX1 genotypes did not influence the maintenance dose of fluindione (P = 0.72, P = 0.72 and P = 0.16 respectively).
Among genetic and environmental factors reported in the study, VKORC1 CT, VKORC1 TT and age were the main factors playing a possible role in fluindione dose requirement at the first period of stability. The model predicted 39% of the variability of fluindione response. Among these factors, VKORC1 accounted for 34% of the variability and age for 5%.
Bleedings
Data concerning bleedings were available for 216 patients. Nineteen patients (8.6%) during follow-up had a significant bleeding: 7 (3.2%) were major haemorrhages and 12 (5.4%) were clinically significant but not major haemorrhages. Half of bleedings occurred within the first month of treatment [median 32 days (1–626)]. At the time of bleeding, the mean INR value was 4.82 (±3.14) and the mean fluindione dose was 16.3 mg (±8.3). None of the four polymorphisms influenced significantly the risk of haemorrhage (results not shown). Among the 19 patients with haemorrhage, 11 had at least one variant allele of VKORC1.
Discussion
To our knowledge, this is the first report evaluating the influence of genetic factors on anticoagulation by fluindione treatment, the most used oral anticoagulant in France.
The main result of this observational study was that VKORC1 C1173T polymorphism significantly affected anticoagulation induced by fluindione. Patients carrying the T allele were more ‘sensitive’ to fluindione treatment. Study subjects with one or two T alleles reached their first INR in the therapeutic range faster and also experienced an INR > 4 more frequently and sooner than study subjects with wild genotype. The maintenance dose of fluindione in patients with the VKORC1 TT genotype was less than half the dose required for patients with VKORC1 CC genotype. VKORC1 genetic variants accounted for 34% of dose variability. In contrast, CYP2C9, CYP4F2 and EPHX1 polymorphisms seemed less determining in the anticoagulation induced by fluindione.
Vitamin K epoxide reductase multiprotein complex (VKORC1) is the target of all vitamin K antagonists independent of their biochemical family (indanedione or coumarin derivatives). Therefore, one could expect that the observed results with fluindione concerning VKORC1 polymorphism were comparable with those observed with acenocoumarol and warfarin. Indeed, our findings are in agreement with previous studies showing genetic variation of VKORC1 as a major determinant of interindividual variability in sensitivity to warfarin mainly in early anticoagulation [2, 4, 9, 21]. Our results with fluindione were also in line with those of previous studies suggesting that subjects receiving warfarin with the A/A haplotype of VKORC1 (corresponding to our VKORC1 1173 TT genotype) had a decreased time to the first INR within the therapeutic range and to the first INR > 4 [9], and required lower warfarin maintenance doses [2–4, 6].
The microsomal epoxide hydrolase 1 encoded by EPHX1 has been proposed as a presumed subunit of the vitamin K epoxide reductase [22, 23]. The rs2292566 polymorphism of EPHX1 was significantly associated with the warfarin maintenance dose, accounting for 1.7% of the dose variability in a cohort of 300 elderly Caucasian inpatients with a mean age of 86 years [12]. In our study, we found that patients carrying two EPHX1 variant alleles of this same synonymous polymorphism (rs2292566) tended to reach an INR within the therapeutic range faster. Nevertheless, the global influence of EPHX1 genotype appeared minor compared with the impact of VKORC1 genotype on fluindione response.
Little is known about fluindione metabolism. If CYP2C9 is clearly involved in warfarin metabolism, its participation in fluindione metabolism is unknown. The *2 and *3 polymorphisms are associated with reduced metabolism of warfarin and individuals carrying the *2 or *3 CYP2C9 allele have lower maintenance warfarin requirements than those carrying the wild-type allele [3, 5, 6]. Our data showed that the CYP2C9 genotype had little impact on fluindione anticoagulation suggesting that the metabolism of fluindione is not mainly mediated by CYP2C9.
Recent studies have suggested that CYP4F2 might play a role in vitamin K1 metabolism and polymorphisms of CYP4F2 also play a role in warfarin and acenocoumarol pharmacogenetics [12–14, 24, 25]. The presence of the mutant 433 M allele (presented as the T allele in our study) could explain the requirement for higher doses of coumarin anticoagulants in 433 M allele carriers [24]. In our study, the maintenance dose of fluindione in patients with two mutant alleles was also higher but not statistically significant.
One strong point of our study is that all included patients had the same indication for fluindione use, i.e. venous thromboembolism and the same INR target. Another strong point was the relatively large number of patients available for each predefined outcome, adding strength to our results. However, our study has also some limitations. First, because INR and fluindione doses were recorded retrospectively, we may have missed some INR values or fluindione doses that finally could have changed some variables, for example our classification for stability. Second, we did not use a standardized procedure to manage the patients and we cannot exclude that the practitioner's experience had played a role in the anticoagulation response to fluindione, especially for the chance and time to achieve a first period of stability. On the other hand, despite the absence of a standardized adjustment of fluindione doses, our results were in good agreement with previous results published for coumarin derivatives. Third, our study was not powered to determine the influence of genetic polymorphisms on bleeding events, the most relevant tolerance outcome when evaluating anticoagulant therapy. However, in concordance with previous studies, more than 50% of bleedings were observed in patients carrying two or more mutant allele of VKORC1.
In conclusion, our study provides first results on the contribution of genetic factors to the variability in fluindione response in patients with venous thromboembolism. As for coumarin derivatives, we found that VKORC1 C1173T genetic polymorphism strongly affects the fluindione response. VKORC1 genotype influences all variables reflecting fluindione sensitivity and accounts for about one third of the dose variability. The impact of CYP2C9, CYP4F2 and EPHX1 genotypes appears less important for fluindione than for warfarin. The relevance of these findings for clinical practice remains to be determined. However, the arrival of new anticoagulants without such interindividual variability may moderate this issue.
Acknowledgments
The authors are indebted to all patients who participated in the study. They wish to thank all local laboratories contacted by phone who agreed to give us the requested INR values. They also thank all members of the Centre d'Investigation Clinique de Brest (CIC 05–02) for their precious work and Zarrin Alavi and Emmanuel Nowak for their pertinent advice.
The EDITH study was supported by grants from INSERM (Contrat de Recherche Stratégique 2001, CRES N° 4CR05G), and from Région Bretagne (Programme 1044–04013235 n°1440, Programme Hospitalier de Recherche Clinique 2000). This specific report was supported by grants from Assistance Publique Hôpitaux de Paris (Contrat d'innovation en recherche Clinique 2006) and specific grants from INSERM in 2006. The Centre Hospitalier Régional et Universitaire de Brest promoted the study. The funders of this study had no role in the design or conduct of the study, in the collection, analysis or interpretation of the data or in the preparation, review, or approval of the manuscript.
Competing Interests
There are no competing interests to declare.
REFERENCES
- 1.Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, Farrar K, Park BK, Breckenridge AM. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ. 2004;329:15–9. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Andrea G, D'Ambrosio RL, Di Perna P, Chetta M, Santacroce R, Brancaccio V, Grandone E, Margaglione M. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–9. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 3.Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, Wood P, Kesteven P, Daly AK, Kamali F. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–33. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 4.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, Rettie AE. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–93. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 5.Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysis. Genet Med. 2005;7:97–104. doi: 10.1097/01.gim.0000153664.65759.cf. [DOI] [PubMed] [Google Scholar]
- 6.Aquilante CL, Langaee TY, Lopez LM, Yarandi HN, Tromberg JS, Mohuczy D, Gaston KL, Waddell CD, Chirico MJ, Johnson JA. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther. 2006;79:291–302. doi: 10.1016/j.clpt.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Limdi NA, McGwin G, Goldstein JA, Beasley TM, Arnett DK, Adler BK, Baird MF, Acton RT. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008;83:312–21. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flockhart DA, O'Kane D, Williams MS, Watson MS, Flockhart DA, Gage B, Gandolfi R, King R, Lyon E, Nussbaum R, O'Kane D, Schulman K, Veenstra D, Williams MS, Watson MS. ACMG Working Group on Pharmacogenetic Testing of CYP2C9, VKORC1 Alleles for Warfarin Use. Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin. Genet Med. 2008;10:139–50. doi: 10.1097/GIM.0b013e318163c35f. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, Kim RB, Roden DM, Stein CM. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Schwarz UI, Ritchie MD, Roden DM, Stein CM, Kurnik D. Relative contribution of CYP2C9 and VKORC1 genotypes and early INR response to the prediction of warfarin sensitivity during initiation of therapy. Blood. 2009;113:3925–30. doi: 10.1182/blood-2008-09-176859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, Ritchie MD, Stein CM, Roden DM, Smith JD, Veenstra DL, Rettie AE, Rieder MJ. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–7. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pautas E, Moreau C, Gouin-Thibault I, Golmard JL, Mahé I, Legendre C, Taillandier-Hériche E, Durand-Gasselin B, Houllier AM, Verrier P, Beaune P, Loriot MA, Siguret V. Genetic factors (VKORC1, CYP2C9, EPHX1, and CYP4F2) are predictor variables for warfarin response in very elderly, frail inpatients. Clin Pharmacol Ther. 2010;87:57–64. doi: 10.1038/clpt.2009.178. [DOI] [PubMed] [Google Scholar]
- 13.Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, Hubbard J, Turpaz Y, Langaee TY, Eby C, King CR, Brower A, Schmelzer JR, Glurich I, Vidaillet HJ, Yale SH, Qi Zhang K, Berg RL, Burmester JK. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–12. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;(5):e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oger E, Lacut K, Le Gal G, Couturaud F, Guénet D, Abalain JH, Roguedas AM, Mottier D. Hyperhomocysteinemia and low B vitamin levels are independently associated with venous thromboembolism: results from the EDITH study: a hospital-based case-control study. J Thromb Haemost. 2006;4:793–9. doi: 10.1111/j.1538-7836.2006.01856.x. [DOI] [PubMed] [Google Scholar]
- 16.Flockhart DA. Drug interactions: cytochrome P450 drug interaction table. Indiana University School of Medicine. 2007. Available at http://medicine.iupui.edu/clinpharm/ddis/table.asp (last accessed 2 September 2010)
- 17.Bodin L, Verstuyft C, Tregouet DA, Robert A, Dubert L, Funck-Brentano C, Jaillon P, Beaune P, Laurent-Puig P, Becquemont L, Loriot MA. Cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase (VKORC1) genotypes as determinants of acenocoumarol sensitivity. Blood. 2005;106:135–40. doi: 10.1182/blood-2005-01-0341. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Zhang W, Zhang Y, Yang Y, Sun L, Hu S, Chen J, Zhang C, Zheng Y, Zhen Y, Sun K, Fu C, Yang T, Wang J, Sun J, Wu H, Glasgow WC, Hui R. VKORCI haplotypes are associated with arterial vascular diseases (stroke, coronary heart disease, and aortic dissection) Circulation. 2006;113:1615–21. doi: 10.1161/CIRCULATIONAHA.105.580167. [DOI] [PubMed] [Google Scholar]
- 19.Morin S, Bodin L, Loriot MA, Thijssen HH, Robert A, Strabach S, Verstuyft C, Tregouet DA, Dubert L, Laurent-Puig P, Funck-Brentano C, Jaillon P, Beaune PH, Becquemont L. Pharmacogenetics of acenocoumarol pharmacodynamics. Clin Pharmacol Ther. 2004;75:403–14. doi: 10.1016/j.clpt.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Larramendy-Gozalo C, Yang JQ, Verstuyft C, Bodin L, Dubert L, Zhang Y, Xu C, Fan L, Jaillon P, Becquemont L. Genetic polymorphism of vitamin K epoxide reductase (VKORC1) 1173C>T in a Chinese and a Caucasian population. Basic Clin Pharmacol Toxicol. 2006;98:611–3. doi: 10.1111/j.1742-7843.2006.pto_440.x. [DOI] [PubMed] [Google Scholar]
- 21.Wadelius M, Chen LY, Lindh JD, Eriksson N, Ghori MJ, Bumpstead S, Holm L, McGinnis R, Rane A, Deloukas P. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113:784–92. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wadelius M, Chen LY, Eriksson N, Bumpstead S, Ghori J, Wadelius C, Bentley D, McGinnis R, Deloukas P. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121:23–34. doi: 10.1007/s00439-006-0260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morisseau C, Hammock BD. Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu Rev Pharmacol Toxicol. 2005;45:311–33. doi: 10.1146/annurev.pharmtox.45.120403.095920. [DOI] [PubMed] [Google Scholar]
- 24.McDonald MG, Rieder MJ, Nakano M, Hsia CK, Rettie AE. CYP4F2 is a vitamin K1 oxidase: an explanation for altered warfarin dose in carriers of the V433m variant. Mol Pharmacol. 2009;75:1337–46. doi: 10.1124/mol.109.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pérez-Andreu V, Roldán V, Antón AI, García-Barberá N, Corral J, Vicente V, González-Conejero R. Pharmacogenetic relevance of CYP4F2 V433M polymorphism on acenocoumarol therapy. Blood. 2009;113:4977–9. doi: 10.1182/blood-2008-09-176222. [DOI] [PubMed] [Google Scholar]


