Abstract
Receptor tyrosine kinases (RTKs) control a multitude of biological processes and are therefore subjected to multiple levels of regulation. Negative feedback is one of the mechanisms that provide an effective means to control RTK-mediated signaling. Sef has recently been identified as a specific antagonist of fibroblast growth factor (FGF) signaling in zebrafish and subsequently in mouse and human. Sef encodes a putative type I transmembrane protein that antagonizes the Ras/mitogen-activated protein kinase pathway in all three species. Mouse Sef was also shown to inhibit the phosphatidylinositol 3-kinase pathway. We show here that an alternative splicing mechanism generates an isoform of human Sef, hSef-b, which unlike the previously reported Sef (hSef-a) is a cytosolic protein. Contrary to hSef-a, which is ubiquitously expressed, hSef-b transcripts display a restricted pattern of expression in human tissues. hSef-b inhibits FGF-induced cell proliferation and prevents the activation of mitogen-activated protein kinase without affecting the upstream component MAPK kinase. Furthermore, hSef-b does not antagonize FGF induction of the phosphatidylinositol 3-kinase pathway. In addition to the effects on FGF signaling, hSef-b inhibited cellular response to platelet-derived growth factor but not other RTK ligands. Therefore, alternative splicing of the hSef gene expands the Sef feedback inhibition repertoire of RTK signaling.
Growth factor signaling by receptor tyrosine kinases (RTKs) is essential for proper function of multicellular organisms and is conserved throughout evolution (1). Inappropriate signaling by RTKs has been implicated in the onset and progression of a variety of human diseases including cancer and genetic disorders, implying that the strength and duration of signaling must be tightly controlled (1–4). This provides a strong impetus to identify molecules that regulate RTK-mediated signaling and to study their mechanism of action.
Several mechanisms collectively known as “negative signaling” have been evolved to attenuate signaling by RTKs (5). One such mechanism involves ligand-induced antagonists of RTK signaling. The Sprouty and SPRED (Sprouty-related EVH1-domain-containing) proteins belong to this category and are regarded as general inhibitors of RTK signaling. They suppress the RTK-induced mitogen-activated protein kinase (MAPK) pathway (reviewed in refs. 5 and 6). Sef is a newly identified antagonist of fibroblast growth factor (FGF) signaling. Sef (for similar expression to FGF genes) encodes a putative type I transmembrane protein that is conserved across zebrafish, mouse, and human, but not invertebrates (7–9). Zebrafish Sef (zfSef) antagonizes FGF activity during embryogenesis by acting as a feedback-induced antagonist of the Ras/MAPK-mediated FGF signaling (7, 8). Subsequent studies showed that the mouse and human homologues of zfSef similarly inhibit FGF-induced activation of MAPK, and mouse Sef also inhibits FGF-induced activation of protein kinase B (pkB/Akt), a key protein in the phosphatidylinositol 3-kinase (PI3-kinase) pathway (10–13).
FGFs comprise a family of 22 structurally related polypeptide mitogens that control cell proliferation, differentiation, survival, and migration and play a key role in embryonic patterning (14–16). They signal via binding and activation of a family of cell-surface tyrosine kinase receptors designated FGF receptors 1–4 (FGFR1–FGFR4) (17–20). Activated receptors trigger several signal transduction cascades including the Ras/MAPK and the PI3-kinase pathway (15, 21). Depending on the cell type, FGF can also activate other MAPK pathways, such that leading to the activation of p38-MAPK (22, 23).
Here, we report the cloning of an isoform of human Sef (hSef-b) and show that it is a product of an alternative splicing mechanism. This isoform differs from previously reported Sef proteins in its biochemical properties, subcellular localization, and specificity.
Materials and Methods
Enzymes, Growth Factors, Reagents, and Chemicals. Restriction enzymes and Taq polymerases were obtained from New England Biolabs, Amersham Biosciences, and Roche Biochemicals. Purified recombinant FGF2 was produced as described (24–26). Bovine brain FGF1, recombinant human FGF4, epidermal growth factor, and platelet-derived growth factor (PDGF) were obtained from R & D Systems. [35S]Methionine (1,000 Ci/mmol) and [3H]thymidine (25 Ci/mmol) were obtained from Amersham Biosciences. Fibronectin, fetal and newborn calf serum, and media were from Biological Industries (Beit Haemek, Israel) or GIBCO. Fluoromount-GTM was from Southern Biotechnology Associates. BSA was from ICN. All other chemicals were from Sigma.
cDNA Cloning and Plasmid Construction. RT-PCR was used to amplify the entire coding region of hSef-a from human brain or fibroblast RNA and hSef-b from testes. First strand was synthesized by using a primer derived from the 3′ UTR of a partial hSef EST clone (AL133097, 5′-AGTGGCAATGCTTAGACTCTTTCGT-3′), and amplification of the coding region of each isoform was performed with nested primer and primer flanking the amino-terminal part unique to each isoform: testes EST clone BG721995, 5′-GCGTGCCAGACAGAGTGCTAGGCAT-3′; or EST clone BE75048, GAGGATCCTGACGGCCATGGCCCCGTGGCTGCAGCTC. After sequencing of several independent clones, the cDNA of hSef-a or hSef-b was cloned into pcDNA3.1, pTET splice, and pcDNA3.1/myc-His expression vectors (Invitrogen).
Analysis of the Expression Pattern of hSef Transcripts. Total RNA was extracted from human tissues and cell lines as described (19). Two micrograms of total RNA were used for first-strand synthesis with random hexamer primer. RT-PCR was performed with primers common to both hSef isoforms and primer sets specific to hSef-a or hSef-b isoforms.
Cell Culture and Transfection Methods. Human embryonic kidney (HEK) 293 and NIH 3T3 cells were grown in DMEM containing 10% FBS or newborn calf serum, respectively. Transient transfections in HEK 293 cells were performed with Lipofectamine Plus in Opti-MEM (Invitrogen). For coimmunoprecipitation, cells in 6-cm dishes were transfected with 3 μg each of FGFR1 and hSef-a plasmid. For hSef-b, we used 10-cm dishes, 4 μg of FGFR1, and 12 μg of hSef-b plasmid because hSef-b expression levels are significantly lower than those of hSef-a. Stable transfections in NIH 3T3 were performed with calcium phosphate (27). Tet-off NIH 3T3 cells (S2-6 cells, gift from David G. Schatz, Yale University, New Haven, CT; ref. 28) were cultured in histidine-deficient DMEM containing 0.5 mM l-histidinol, serum, and 1 μg/ml tetracycline (tet). hSef-b Tet-off NIH 3T3 cell lines were established by cotransfection of the S2-6 cells with pTet splice-hSef-b or an empty vector and pTK-Hyg (Clontech), followed by selection in complete medium plus 150 μg/ml hygromycin. Colonies of resistant cells were isolated 3 weeks after transfection.
Cell Growth and Apoptosis Assays. [3H]Thymidine incorporation assay was done in 96-well microtiter plates as described (24, 26). Confluent cultures were growth arrested in 0.3% serum for 24 h, and when indicated, tet was removed 24 h before stimulation with serum (10%) or growth factors. Apoptosis was examined with confocal microscopy by using the In Situ Cell Death detection kit (terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling; Roche Molecular Biochemicals) in control S2-6 or S2-6/hSef-b cells, which were grown for 48 h in the presence or absence of tet.
Protein Detection. In vitro translation of hSef-b was done with pcDNA3.1/Hygro-hSef-b vector by using the TnT Quick Coupled Transcription/Translation system in the presence of [35S]methionine, according to manufacturer instructions (Promega). Resulting products were analyzed by SDS/PAGE and visualized by phosphoimaging. Polyclonal antibodies against hSef were generated by injecting rabbits with a polypeptide containing the last 402 residues of hSef fused to the amino-terminal portion of bacteriophage T7 θ10 protein (29). Coimmunoprecipitation and immunoblotting were done essentially as described (30). For coimmunoprecipitation, cells were lysed 24 h after transfection in HTNG buffer (20 mM Hepes, pH 7.4/150 mM NaCl/10% glycerol/1% Triton X-100/1 mM EGTA/1 mM NaVO4/protease inhibitors). After incubation with antibody, immunocomplexes were captured on protein G Dynabeads (Dynal, Great Neck, NY) and washed with HTNG buffer. After SDS/PAGE and immunoblotting, bound antibodies were visualized by chemiluminescence. For immunofluorescence, transfected HEK 293 cells were fixed 48 h after transfection with 4% paraformaldehyde and incubated in PBS containing 1% BSA, 0.1% saponin, and antibodies as indicated in the text. Nuclear staining was done with 10 μM DRAQ5 (Biostatus, Leicestershire, U.K.). Confocal microscopy was performed with an MRC-1024 laser confocal microscope (Bio-Rad). Commercial antibodies that were used are anti-myc epitope (9E10), p38, MAPK kinase (MEK), Akt, cyclin-dependent kinase 4 (cdk4), and FGFR1 (H-76) rabbit polyclonal antibodies (Santa Cruz Biotechnology), phospho-p44/42 MAPK (Thr-202/Tyr-204), E10 mouse monoclonal antibodies (New England Biolabs), phospho-Akt (Ser-473), phospho-p38, peroxidase-conjugated goat anti-rabbit or anti-mouse IgG (Sigma), cyclin D1 and phospho-MEK (Cell Signaling Technology, Beverly, MA), FITC-conjugated goat anti-rabbit IgG (ICN), rhodamine-red-X-conjugated Affinipure goat anti-mouse IgG (Jackson ImmunoResearch), and extracellular signal-regulated kinase (ERK)2 rabbit polyclonal antibodies (gift from Y. Granot, Ben Gurion University, Beer-Sheva, Israel).
Results
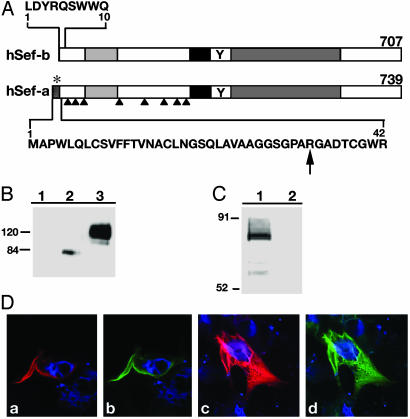
Cloning and Characterization of the hSef-b Isoform. A database search with the zfSef sequence revealed an EST (GenBank accession no. AL133097) containing the entire 3′ UTR of hSef and most of its coding region, except for the first 170 residues. The remainder of the coding region was obtained by searching the human genome database for upstream exons of hSef and based on homology with bovine Sef (GenBank accession no. BE750478). A cDNA fragment encoding the entire ORF of hSef was amplified from primary human fibroblast or human fetal brain RNA. Human Sef has been mapped to a single locus on chromosome 3p14.3. Additional database searches with the amino-terminal sequence of hSef revealed an EST clone from human testes that was 577 nt long and contained an ORF of 122 residues. This EST clone differed from the original hSef in its amino terminus and the upstream 5′ UTR sequences. Because the human genome contains a single Sef locus, these findings suggested the existence of alternatively spliced Sef isoforms. To examine this possibility, RT-PCR was performed with human testes RNA by using primers complementary to the 3′ UTR of hSef and complementary to the 5′ UTR of the testes EST. A single product of 2,200 nt was obtained, confirming the existence of alternate isoforms of hSef (designated hSef-a and hSef-b for the brain and testes isoforms, respectively). hSef-b contains an ORF of 707 aa residues, as compared with the 739 residues of hSef-a; the last 697 residues are identical in both isoforms. As a result of the alternative splicing, the first 42 residues in the ORF of hSef-a were replaced with 10 new residues in the ORF of hSef-b. Similar to hSef-a, the new isoform contained eight potential N-linked glycosylation sites, a potential transmembrane-spanning domain and tyrosine phosphorylation site, and Ig and IL-17 receptor-like domains. Unlike hSef-a, it lacked a signal for secretion (Fig. 1A). Interestingly, the unique hSef-b region has a CUG initiation codon. The next putative initiation codon (AUG) is located within the region that is identical in both isoforms. Translation from the CUG codon would result in a protein of 707 aa and predicted molecular mass of 78 kDa, whereas translation from the AUG codon would result in a protein of 595 residues and a predicted molecular mass of 65 kDa.
Fig. 1.
Schematic representation of hSef-a and hSef-b-(A) Residues unique to each Sef isoform are shown in bold letters. *, signal for secretion; arrows, potential N-linked glycosylation sites; black box, transmembrane domain; Y, putative tyrosine phosphorylation site; light and dark gray boxes, Ig-like domain and the IL-17R-like domain. (B) Identification of hSef products. HEK 293 cells were transiently transfected with control empty vector (lane 1) or Myc-tagged hSef-b or hSef-a vectors (lanes 2 and 3, respectively). Equal amounts of protein were subjected to gel electrophoresis, and hSef products were identified by immunoblotting with α-myc antibody. (C) In vitro translation of hSef-b. Lane 1, translation in the presence of hSef-b vector; lane 2, translation in the presence of an empty vector. The assay was performed as described in Materials and Methods. Products were analyzed by SDS/PAGE and visualized by phosphoimaging. (D) Subcellular localization of hSef isoforms. HEK 293 cells, transfected with myc-tagged hSef-a (a and b) or myc-tagged hSef-b (c and d) expression vectors, were fixed 48 h later and stained with α-myc (red; a and c) or α-hSef antibody (green; b and d). Nuclei were counterstained with bisbenzimide (blue).
To characterize the hSef-b product and elucidate whether the alternative initiation codon in hSef-b can serve as a translation start site, a cDNA fragment encoding the entire ORF of hSef-b was subcloned into a eukaryotic expression vector. A myc epitope tag was added at the carboxyl terminus, allowing the detection of the hSef-b product. Expression in HEK 293 cells revealed a single protein product with a molecular mass of ≈80 kDa (Fig. 1B). A major product with a similar molecular mass was obtained after in vitro translation (Fig. 1C). These findings strongly suggest that the CUG codon functions as a major translation start site in the hSef-b isoform.
The similarity between the apparent and predicted molecular mass of hSef-b suggested that this protein does not undergo any significant posttranslational modifications. Contrary to hSef-b, the hSef-a product, when expressed in HEK 293 cells, appeared as a broad band with an average molecular mass of 120 kDa, as compared with 82 kDa of the predicted protein (see Fig. 1B). Treatment of cells expressing each isoform with tunicamycin, an inhibitor of N-linked glycosylation, reduced the molecular mass of hSef-a but had no effect on the molecular mass of the hSef-b product (data not shown). Collectively, our findings indicate that hSef-a, but not hSef-b, is a glycoprotein synthesized in the classical secretory pathway and further suggest that the hSef-b product is an intracellular protein. Immunofluorescent staining of HEK 293 cells expressing each of the human Sef isoforms was used to investigate this possibility. Detection was performed with antibodies directed against the myc epitope tag fused to the hSef products or antibodies directed against the carboxyl terminus of hSef proteins. As shown in Fig. 1D, hSef-b product was cytosolic, whereas the hSef-a product was located at the cell surface as shown (8).
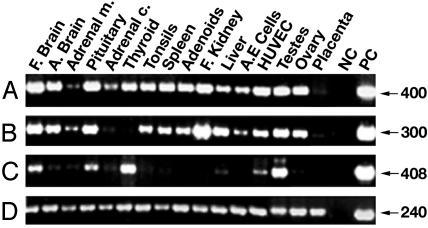
Tissue Type-Specific Expression of Human Sef Isoforms. The pattern of expression of hSef isoforms in a variety of human tissues and cell lines was examined by RT-PCR. Primers were designed to amplify a region common to both hSef transcripts or to specifically amplify each transcript. All 16 samples examined were positive for Sef transcripts when amplified with the primers from the common region of Sef isoforms (Fig. 2A). hSef-a transcript was differentially expressed in 15 samples (Fig. 2B). hSef-a transcript was highly expressed in both fetal and adult brain, pituitary, tonsils, spleen, adenoids, fetal kidney, liver, testes, and ovary, and moderate levels were detected in primary aortic endothelial cells, human umbilical vein endothelial cells, and adrenal medulla. Low levels of hSef-a were observed in adrenal cortex, barely detected in placenta, and absent in thyroid. In contrast, hSef-b transcript was highly expressed in thyroid and testes and moderately expressed in pituitary, fetal brain, and human umbilical vein endothelial cells; the remaining tissues were either negative or expressed barely detectable levels of the hSef-b transcript (Fig. 2C). These findings suggest that the expression of the human Sef isoforms is regulated at the level of splicing or mRNA stability.
Fig. 2.
Expression pattern of hSef isoforms. Expression was determined by RT-PCR by using total RNA from the indicated human tissues and human primary cells. Amplification was performed with primers derived from hSef common region (A) and with specific primers for hSef-a (B) or hSef-b (C). Amplification of GAPDH transcript (D) compares RNA levels in each sample. Adrenal m. and adrenal c. denote adrenal medulla and cortex, respectively. A.E. cells, primary aortic endothelial cells. HUVEC, human umbilical vein endothelial cells; F, fetal; A, adult; NC, negative control. Templates for positive controls (PC) are plasmids containing hSef-a (A and B), hSef-b (C), or GAPDH (D).
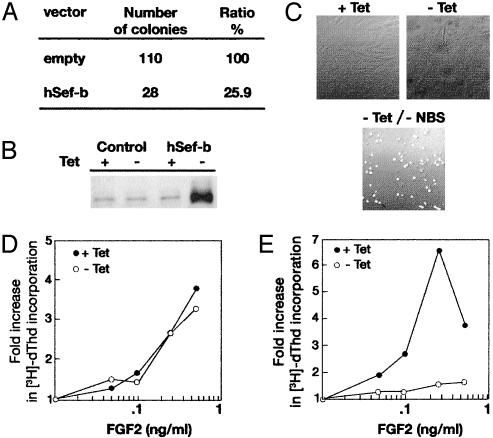
hSef-b Inhibits Proliferation of NIH 3T3 Cells. The different biochemical properties and subcellular localization of the two hSef isoforms raised the question whether hSef-b can inhibit FGF biological activity similar to hSef-a. NIH 3T3 cells were chosen to study the effect of hSef-b on biological responses to FGFs. These cells proliferate in response to various members of the FGF family and have been extensively used as a model to study oncogenesis, regulation of cell proliferation, and growth factor-mediated signaling. RT-PCR analysis revealed that they express the mouse-Sef gene (data not shown). First, we examined the general effect of hSef-b on growth of NIH 3T3 cells by colony assay. Cells transfected with hSef-b expression vector formed 75% fewer colonies when compared with cells transfected with control vector (Fig. 3A). To determine how hSef-b inhibits cell growth and to study its effect on FGF-mediated signaling, we established NIH 3T3 stable cell lines in which the expression of hSef-b is regulated by tet. Clones that did not express detectable levels of hSef-b in the presence of tet were chosen for further analysis. Maximal level of hSef-b protein was obtained 16 h after removal of tet (Fig. 3B and data not shown).
Fig. 3.
(A) hSef-b suppresses colony formation in NIH 3T3 cells. Cells were stably transfected with expression vector bearing hSef-b (pCDNA/hSef-b) or an empty vector (pCDNA). After 1 day, cells were diluted (1/25) and marker-selected for 2–3 weeks. Resistant clones were counted at the end of the selection process (five plates for each vector). The results are representative of three experiments. (B) Induced expression of hSef-b in the tet-off NIH 3T3 cells. Cells were grown in 10% serum in the presence and absence of tet. After 24 h, the cells were lysed, and hSef-b expression was analyzed by immunoblotting with hSef-specific antibodies. Control cultures denote parental cells transfected with an empty pTet-splice vector. (C) The effect of hSef-b on apoptosis. NIH 3T3/hSef-b cells were grown for 48 h in the presence or absence of tet or in the absence of tet and serum (–tet, –NBS). Cells were washed and fixed, and apoptosis was then evaluated by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling. (D and E) hSef-b inhibits the mitogenic activity of FGF2. Confluent cultures of control cells (D) or hSef-b-expressing cells (E) were serum starved and grown in the presence or absence of tet for 24 h. FGF2 was added at the above-indicated concentrations. [3H]Thymidine incorporation assay was performed as described (24, 26).
The inhibitory effect of hSef-b on colony formation could have resulted from either induction of apoptosis or inhibition of cell growth. Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling and [3H]thymidine incorporation assays were used to explore these possibilities. No significant level of apoptosis was observed in hSef-b-expressing cells grown for 48 h in the absence of tet, whereas apoptotic cells were readily detected in the same cells grown in serum-free medium (Fig. 3C). In contrast, the mitogenic activity of FGF2 was strongly inhibited in cells expressing hSef-b but not in cells transfected with control vector (compare Fig. 3 D and E). These findings indicate that hSef-b acts by restricting cell division and not by inducing apoptosis.
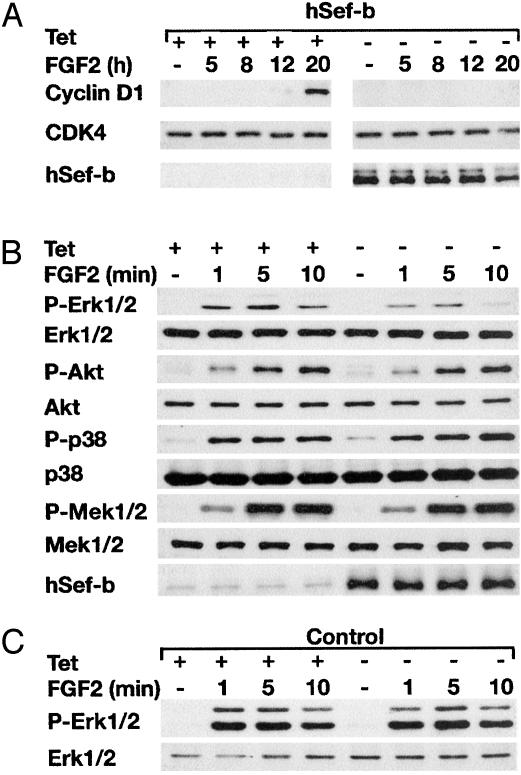
The hSef-b Isoform Prevents the Activation of ERK1/2 MAPK and Causes a Dramatic Reduction in Cyclin D1 Levels. Sef-inducible cell lines were used to explore the mechanism underlying hSef-b inhibition of cell division. Because D-type cyclins regulate S-phase entry (31), we first examined the effect of hSef-b on the levels of cyclin D1 protein in growth-arrested cells treated with FGF2. Cyclin D1 protein levels were evaluated at different time intervals post-growth factor stimulation. Cyclin D1 protein was readily observed in FGF2-treated NIH 3T3/hSef-b cells grown in the presence of tet, whereas it was not detected in FGF2-treated cells induced to express hSef-b (Fig. 4A). Because the levels of cdk4 do not fluctuate during the cell cycle, we used cdk4 as probe for protein levels in each time point (31). cdk4 levels were similar in all time points in cultures grown with and without tet (Fig. 4A). In addition, the levels of cyclin D1 remained unchanged in the control cells grown with or without tet (data not shown).
Fig. 4.
hSef-b reduces cyclin D1 levels and inhibits the Ras/MAPK pathway in FGF2-stimulated cells. Control and hSef-b inducible NIH 3T3 cells were serum starved for 24 h in the presence and absence of tet and then stimulated with FGF2 (20 ng/ml) for the indicated time periods. (A) The effect of hSef-b on the levels of cyclin D1 protein. The levels of cyclin D1 were evaluated in total cell lysates over 20 h of stimulation. Cyclin D1 and cdk proteins were analyzed by immunoblotting with anti-D1 monoclonal antibody or rabbit anti-cdk4. (B) The effect of hSef-b on FGF2-induced signaling pathways. Equal amounts of total cell lysates were analyzed by immunoblotting. The membranes were successively incubated with the indicated antibodies. (C) Activation of ERK1/2 MAPK in the control cultures. In cells transfected with control vector, there was no effect of tet removal on ERK1/2 activation. Each experiment was repeated at least twice and by using two independent clones of hSef-b inducible cells. P-ERK1/2, P-Akt, P-p38, and P-MEK1/2 are antibodies directed against the phosphorylated (P) form of the kinases.
The ERK1/2 MAPK, pkB/Akt, and p38 MAPK are known to regulate cyclin D1 levels by transcriptional and posttranslational mechanisms (31, 32). ERK1/2 MAPK enhance cyclin D1 expression and regulate the assembly of cyclin–cdk complex, whereas pkB/Akt regulates the turnover of cyclin D1 protein (31, 32). The p38 MAPK can inhibit cyclin D1 expression in a cell type-dependent manner. It has been generally associated with cellular response to stress, but several reports suggest its involvement in cellular responses to growth factors including FGF2 (22, 23). Therefore, we examined the effect of hSef-b on the activation of these three kinases by using antibodies that specifically recognize their activated form. Stimulation over time of control cells and hSef-b-expressing cells revealed that hSef-b inhibited the activation of ERK1/2 MAPK, whereas total ERK1/2 levels remained unaltered (Fig. 4 B and C). hSef-b had no effect on FGF2-induced activation of pkB/Akt or p38 MAPK (Fig. 4B). To localize the site of hSef-b action, we examined its effect on the activation of the dual specificity MEK1/2, which phosphorylates ERK1/2, in response to growth factor stimulation. Fig. 4B shows that hSef-b had no effect on MEK1/2 activation in FGF2-stimulated cells, suggesting that the MAPK signaling pathway is blocked at the level or downstream of MEK.
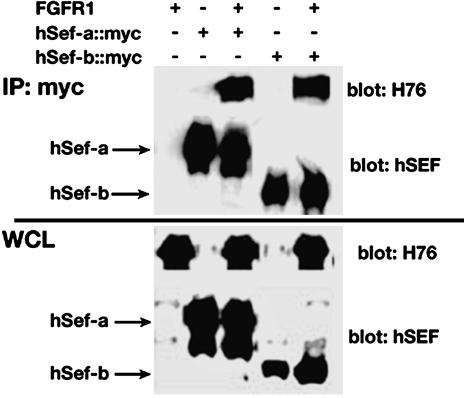
The hSef-b Protein Associates with FGFR1. The cell-surface Sef isoform associates with several members of the FGFR family in coimmunoprecipitation assays, and the interaction site was mapped to the intracellular domain (8, 10, 13). Because this domain is conserved in both hSef isoforms, we tested whether hSef-b can form a complex with FGFR1 also. To this end, HEK 293 cells were transiently transfected with myc-tagged Sef constructs with or without FGFR1. As shown in Fig. 5, FGFR1 coimmunoprecipitated with hSef-a in the absence of ligand stimulation, in agreement with published data (8, 10, 13). hSef-b, notwithstanding its lower expression levels compared to hSef-a, efficiently associated with FGFR1 but not with epidermal growth factor receptor (Fig. 5 and data not shown). These results lend further support to the importance of the carboxyl-terminal domain of Sef for the association with FGFRs.
Fig. 5.
Coimmunoprecipitation of Sef isoforms and FGFR1. HEK 293 cells were transfected with the indicated constructs, and whole-cell lysates were either immunoblotted with α-FGFR1 (H76) or α-hSef antibodies or immunoprecipitated with α-myc antibody and immunoblotted with H76 or α-hSef antibodies.
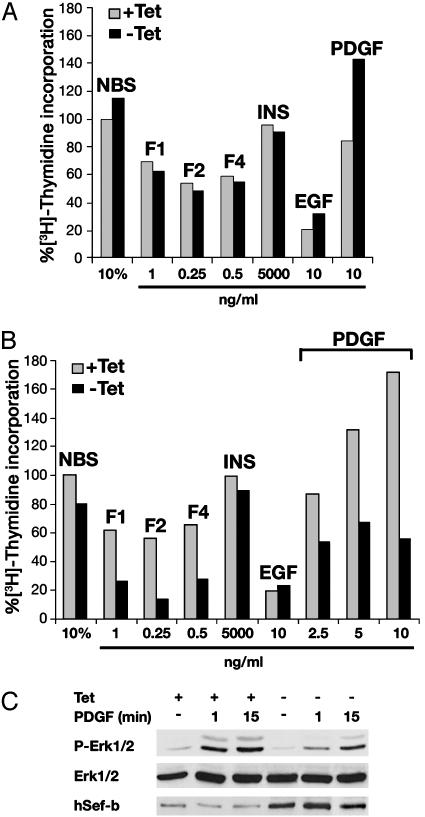
The Spectrum of the Inhibitory Activity of hSef-b. The subcellular localization of hSef-b may extend the repertoire of RTKs that it can inhibit. To explore this hypothesis, we examined the effect of hSef-b on mitogenic activity of additional members of the FGF family, as well as a subset of other RTK ligands, and serum. A dose-response curve was performed for each ligand, and representative results are shown in Fig. 6. In addition to FGF2, hSef-b inhibited the mitogenic activity of FGF1 and FGF4 (80% inhibition of FGF2 and 60% inhibition of FGF1 or FGF4) as well as the activity of PDGF (40, 57 and 68% inhibition at 2.5, 5, and 10 ng/ml ligand, respectively) (Fig. 6B). Inhibition of PDGF was accompanied with reduction in the activation of ERK1/2 MAPK (Fig. 6C). This is in contrast to the reported lack of an effect of Sef-a on PDGF signaling (10). In contrast with its effect on the mitogenic activity of FGF and PDGF, hSef-b had little or no effect on the activity of serum, insulin, or epidermal growth factor (Fig. 6B). The mitogenic activity of serum and each of the other growth factors was similar in control cultures grown in the presence or absence of tet (Fig. 6A).
Fig. 6.
The spectrum of inhibitory activity of hSef-b. Mitogenic assay in control cultures (A) or cells expressing hSef-b (B) was performed as described in the legend to Fig. 3. Fold increase (FI) in biological activity was calculated by dividing cpm values obtained in the presence of the indicated stimulators with those obtained in 0.2% serum alone. Percent [3H]thymidine incorporation is relative to FI obtained in cultures stimulated with 10% serum in the presence of tet that was set at 100%. The concentrations of FGFs, insulin, epidermal growth factor, and serum are those that gave rise to a maximal biological response. F, FGF; INS, insulin. (C) hSef-b inducible NIH 3T3 cells were serum starved for 24 h in the presence and absence of tet and then stimulated with PDGF (20 ng/ml) for the indicated time periods. Equal amounts of total cell lysates were analyzed by immunoblotting with anti-P-ERK1/2 and anti-ERK antibodies. These experiments were repeated at least three times and by using two independent hSef-b-expressing clones.
Discussion
Negative feedback mechanisms provide effective means to control growth factor-mediated signaling, either by restriction of the incoming signal itself or by induction of counter regulatory mechanisms affecting the propagation of the signal (33). In the present study, we describe the isolation and characterization of an isoform of hSef, a recently identified antagonist of FGF-induced signaling.
Our results indicate that different hSef isoforms are generated via an alternative splicing mechanism. One isoform, hSef-a, is similar to the previously reported Sef from zebrafish and mammals (7–9, 13). This isoform is thought to encode a type I transmembrane protein, and our immunofluorescent staining confirmed that hSef-a product is located at the cell surface. In hSef-b, the isoform described in the present study, the leader sequence and the next eight residues of hSef-a were replaced by 10 different residues. This isoform lacks a signal for secretion, and immunofluorescent staining revealed that it is a cytosolic protein. Furthermore, the hSef-b product does not undergo significant posttranslational modifications, a property that is typical for proteins that are translated in the cytosolic compartment. Collectively, our results show that alternative splicing differentially influences the subcellular localization of hSef isoforms.
Besides lacking a signal for secretion, the alternate sequence contained a CUG initiation codon instead of the conventional AUG initiation codon. Initiation from this CUG codon is consistent with the apparent molecular mass of both the in vitro- and in vivo-expressed hSef-b. The utilization of CUG as a translation start site may be responsible for the observed differences in the expression levels of hSef-a and hSef-b products, as non-AUG codons direct less efficient translation initiation (34, 35).
The hSef-b isoform exhibits a restricted pattern of expression compared with hSef-a. It is highly expressed in testes and thyroid and, to a much lesser extent, in tissues of neuronal origin and primary endothelial cells, whereas hSef-a transcript is expressed in all of the tissues and primary cells that were examined except for the thyroid. Interestingly, the expression profile of hSef-a parallels that of FGFRs (19, 20, 36–38), suggesting that this isoform regulates a wide array of biological processes where FGFs are implicated. The high levels of hSef-b in thyroid and testes could imply that this isoform regulates unique biological processes and may be more specific to cells of epithelial origin. With respect to FGFs, it may control signaling by specific receptor isoforms. Therefore, an important avenue of future research would be to determine how each Sef isoform affects signaling by the distinct FGFRs.
hSef-b inhibited mitogenic response of NIH 3T3 cells to several members of the FGF family but not to serum, insulin, or epidermal growth factor. Unlike the cell-surface Sef (10), hSef-b inhibited PDGF-induced mitogenic response, suggesting that intracellular machinery, common to signaling by FGF and PDGF, is affected. Consistent with this is the finding that hSef-b inhibited the Ras/MAPK pathway and reduced cyclin D1 levels. However, hSef-b does not globally affect RTK-induced signaling pathways because it had no effect on FGF induction of the PI3-kinase or the p38 MAPK pathway, which is consistent with hSef-b action downstream of Ras. Lack of an effect on the PI3-kinase pathway also correlates with the finding that hSef-b inhibited cell growth but did not lead to an increase in apoptosis. The inhibition of the MAPK pathway and maintenance of PI3-kinase pathway may allow cells expressing hSef-b to cease proliferation but remain viable on growth factor stimulation.
Our results restrict hSef-b activity to a narrow window at the level of, or downstream from, MEK. Similar to the hSef-a isoform (refs. 8, 10, 13 and present data), the hSef-b isoform can associate with FGFR1 in coimmunoprecipitation assays. Because, unlike hSef-a, the hSef-b isoform is cytosolic, its association with the receptor could function as a means to bring hSef-b in the vicinity of the components of the Ras/MAPK pathway. Although both isoforms interact with FGFR1, the outcome of this association is not identical because the cell-surface Sef isoform inhibits multiple FGF-signaling pathways (ref. 10 and data not shown). Because the entire hSef-b isoform is located inside the cells, its folding must be quite different from that of the hSef-a isoform and is likely to influence its mode of action. For example, the amino-terminal domain of hSef-b, which also contains the unique hSef-b residues, can interact with proteins in the signaling cascade, whereas in hSef-a, this domain is extracellular and is not required for inhibitory activity (8, 10, 13). Alternatively, it could function as an autoregulatory domain that prevents hSef-b from interacting with certain proteins in the signaling cascade.
In summary, we show that alternative splicing generates a human Sef isoform that differs from the previously reported isoform by biochemical properties, subcellular localization, and specificity. Thus, signaling inhibitors with different properties are encoded by a single Sef gene.
Acknowledgments
We thank Dr. Dan Cassel for critical review of the manuscript and Sharon Mink for technical assistance. This work was supported by Israel Ministry of Health Grant 5195 and Israel Cancer Research Fund Grant 200701 (to D.R.).
Abbreviations: RTK, receptor tyrosine kinase; FGF, fibroblast growth factor; PI3-kinase, phosphatidylinositol 3-kinase; FGFR, FGF receptor; MAPK, mitogen-activated protein kinase; MEK, MAPK kinase; ERK, extracellular signal-regulated kinase; PDGF, platelet-derived growth factor; tet, tetracycline; HEK, human embryonic kidney; cdk, cyclin-dependent kinase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY489047).
References
- 1.Schlessinger, J. (2000) Cell 103, 211–225. [DOI] [PubMed] [Google Scholar]
- 2.Pawson, T. & Saxton, T. M. (1999) Cell 97, 675–678. [DOI] [PubMed] [Google Scholar]
- 3.Simon, M. A. (2000) Cell 103, 13–15. [DOI] [PubMed] [Google Scholar]
- 4.Hunter, T. (2000) Cell 100, 113–127. [DOI] [PubMed] [Google Scholar]
- 5.Christofori, G. (2003) Nat. Cell Biol. 5, 377–379. [DOI] [PubMed] [Google Scholar]
- 6.Dikic, I. & Giordano, S. (2003) Curr. Opin. Cell Biol. 15, 128–135. [DOI] [PubMed] [Google Scholar]
- 7.Furthauer, M., Lin, W., Ang, S. L., Thisse, B. & Thisse, C. (2002) Nat. Cell Biol. 4, 170–174. [DOI] [PubMed] [Google Scholar]
- 8.Tsang, M., Friesel, R., Kudoh, T. & Dawid, I. B. (2002) Nat. Cell Biol. 4, 165–169. [DOI] [PubMed] [Google Scholar]
- 9.Lin, W., Furthauer, M., Thisse, B., Thisse, C., Jing, N. & Ang, S. L. (2002) Mech. Dev. 113, 163–168. [DOI] [PubMed] [Google Scholar]
- 10.Kovalenko, D., Yang, X., Nadeau, R. J., Harkins, L. K. & Friesel, R. (2003) J. Biol. Chem. 278, 14087–14091. [DOI] [PubMed] [Google Scholar]
- 11.Krasilnikov, M. A. (2000) Biochemistry 65, 59–67. [PubMed] [Google Scholar]
- 12.Scheid, M. P. & Woodgett, J. R. (2001) Nat. Rev. Mol. Cell Biol. 2, 760–768. [DOI] [PubMed] [Google Scholar]
- 13.Yang, R. B., Ng, C. K., Wasserman, S. M., Komuves, L. G., Gerritsen, M. E. & Topper, J. N. (2003) J. Biol. Chem. 278, 33232–33238. [DOI] [PubMed] [Google Scholar]
- 14.Klint, P. & Claesson-Welsh, L. (1999) Front. Biosci. 4, D165–D177. [DOI] [PubMed] [Google Scholar]
- 15.Powers, C. J., McLeskey, S. W. & Wellstein, A. (2000) Endocr. Relat. Cancer 7, 165–197. [DOI] [PubMed] [Google Scholar]
- 16.Ornitz, D. M. & Itoh, N. (2001) Genome Biol. 2, 3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKeehan, W. L., Wang, F. & Kan, M. (1998) Prog. Nucleic Acids Res. Mol. Biol. 59, 135–176. [DOI] [PubMed] [Google Scholar]
- 18.Miki, T., Fleming, T. P., Bottaro, D. P., Rubin, J. S., Ron, D. & Aaronson, S. A. (1991) Science 251, 72–75. [DOI] [PubMed] [Google Scholar]
- 19.Eisemann, A., Ahn, J. A., Graziani, G., Tronick, S. R. & Ron, D. (1991) Oncogene 6, 1195–1202. [PubMed] [Google Scholar]
- 20.Ron, D., Reich, R., Chedid, M., Lengel, C., Cohen, O. E., Chan, A. M., Neufeld, G., Miki, T. & Tronick, S. R. (1993) J. Biol. Chem. 268, 5388–5394. [PubMed] [Google Scholar]
- 21.Ong, S. H., Hadari, Y. R., Gotoh, N., Guy, G. R., Schlessinger, J. & Lax, I. (2001) Proc. Natl. Acad. Sci. USA 98, 6074–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maher, P. (1999) J. Biol. Chem. 274, 17491–17498. [DOI] [PubMed] [Google Scholar]
- 23.Boilly, B., Vercoutter-Edouart, A. S., Hondermarck, H., Nurcombe, V. & Le, B. X (2000) Cytokine Growth Factor Rev. 11, 295–302. [DOI] [PubMed] [Google Scholar]
- 24.Reich-Slotky, R., Shaoul, E., Berman, B., Graziani, G. & Ron, D. (1995) J. Biol. Chem. 270, 29813–29818. [DOI] [PubMed] [Google Scholar]
- 25.Ron, D., Bottaro, D. P., Finch, P. W., Morris, D., Rubin, J. S. & Aaronson, S. A. (1993) J. Biol. Chem. 268, 2984–2988. [PubMed] [Google Scholar]
- 26.Sher, I., Weizman, A., Lubinsky-Mink, S., Lang, T., Adir, N., Schomburg, D. & Ron, D. (1999) J. Biol. Chem. 274, 35016–35022. [DOI] [PubMed] [Google Scholar]
- 27.Ron, D., Tronick, S. R., Aaronson, S. A. & Eva, A. (1988) EMBO J. 7, 2465–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shockett, P., Difilippantonio, M., Hellman, N. & Schatz, D. G. (1995) Proc. Natl. Acad. Sci. USA 92, 6522–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studier, F. W., Rosenberg, A. H., Dunn, J. J. & Dubendorff, J. W. (1990) Methods Enzymol. 185, 60–89. [DOI] [PubMed] [Google Scholar]
- 30.Shaoul, E., Reich-Slotky, R., Berman, B. & Ron, D. (1995) Oncogene 10, 1553–1561. [PubMed] [Google Scholar]
- 31.Sherr, C. J. & Roberts, J. M. (1999) Genes Dev. 13, 1501–1512. [DOI] [PubMed] [Google Scholar]
- 32.Lavoie, J. N., L'Allemain, G., Brunet, A., Muller, R. & Pouyssegur, J. (1996) J. Biol. Chem. 271, 20608–20616. [DOI] [PubMed] [Google Scholar]
- 33.Niehrs, C. & Meinhardt, H. (2002) Nature 417, 35–36. [DOI] [PubMed] [Google Scholar]
- 34.Kozak, M. (1989) Mol. Cell. Biol. 9, 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak, M. (1991) J. Biol. Chem. 266, 19867–19870. [PubMed] [Google Scholar]
- 36.Miki, T., Bottaro, D. P., Fleming, T. P., Smith, C. L., Burgess, W. H., Chan, A. M. & Aaronson, S. A. (1992) Proc. Natl. Acad. Sci. USA 89, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finch, P. W., Cunha, G. R., Rubin, J. S., Wong, J. & Ron, D. (1995) Dev. Dyn. 203, 223–240. [DOI] [PubMed] [Google Scholar]
- 38.Yazaki, N., Hosoi, Y., Kawabata, K., Miyake, A., Minami, M., Satoh, M., Ohta, M., Kawasaki, T. & Itoh, N. (1994) J. Neurosci. Res. 37, 445–452. [DOI] [PubMed] [Google Scholar]