Abstract
Members of the Bcl-2 protein family that share only the Bcl-2 homology 3 (BH3) domain are known mostly as sentinels for apoptotic stimuli and initiators of apoptosis. One BH3-only protein, Bim, is the major physiological antagonist of the prosurvival proteins in B and T lymphocytes. It is required for hematopoietic homeostasis and to preclude autoimmunity. Here, we show that the BimEL isoform, which was predominant in T cells, existed in both phosphorylated and unphosphorylated forms. Whereas the unphosphorylated BimEL was sequestered to microtubules by means of a direct interaction with tubulin, the phosphorylated protein was released from microtubules. The freed BimEL was subjected to caspase cleavage at an early stage of apoptosis induced by stimuli that activate either the mitochondria- or death receptor-dependent apoptosis pathway. The N-terminally cleaved BimEL became hyperactive in inducing apoptosis because of its more efficient targeting of Bcl-2. Thus, unlike many other BH3-only proteins, BimEL can be activated downstream of the caspase cascade, leading to a positive feedback amplification of apoptotic signals.
Akey step in the mitochondria-dependent apoptotic pathway is the disruption of the mitochondria membrane. The integrity of the membrane is controlled primarily by a balance between the antagonistic actions of the proapoptotic and antiapoptotic members of the Bcl-2 family. The Bcl-2 homology 3 (BH3)-only family members constitute a key group of proapoptotic proteins that resemble Bcl-2 only in the BH3 domain. This domain is required for the interaction of these proteins with other Bcl-2 family members (1, 2). BH3-only proteins normally reside in other subcellular compartments or structures and translocate to the mitochondria in response to apoptotic stimuli. When at the mitochondria, they induce the conformational change and oligomerization of Bax and Bak, two Bcl-2 family members with BH1, BH2, and BH3 domains. The pore-forming capability of the oligomerized Bax and Bak results in the destabilization of the mitochondrial outer membrane and the subsequent release of the death molecules from the confines of the mitochondria (2). Antiapoptotic Bcl-2 family members, such as Bcl-2 and Bcl-xL, bind directly to the proapoptotic members, neutralize their activities, and abolish the apoptotic signaling. Given the pivotal role of the BH3-only proteins in sensing apoptotic stimuli and initiating apoptosis, their activities have to be kept in check to prevent inappropriate cell death (1).
Investigation into the diverse functions and regulations of the ≈10 known mammalian BH3-only proteins suggests that different proteins may initiate apoptosis in different cell types and/or transduce distinctive apoptotic stimuli in a given cell type (1, 3). Gene knock-out studies have revealed an essential role of one such protein, Bim, in shaping the development of the immune system and transducing the apoptotic signals caused by cytokine deprivation, calcium ion flux, and microtubule perturbation, but not other insults, in lymphocytes (4–7). Bim also facilitates HIV-1 Tat-induced apoptosis in T cells (8), which contributes in part to the progressive T cell depletion associated with AIDS.
Bim is the major physiological antagonist of the prosurvival proteins, at least in B and T lymphocytes (5). It is essential for the development of T cells (5) and apoptosis of activated T cells (7). Here, we report a previously undescribed mechanism by which Bim controls the apoptotic signaling in T cells. Three main isoforms of Bim (BimEL, BimL, and BimS) exist because of alternative splicing (9). The expressions of these isoforms vary in different cell types and tissues (10), with BimEL being the predominant one in T cells. Our data indicate that BimEL was sequestered to microtubules by means of a direct interaction with tubulin. Phosphorylated BimEL (pBimEL) was released from microtubules and cleaved by caspases at an early stage of apoptosis, induced by stimuli that activate either the mitochondria- or the death receptor-dependent apoptotic pathway. The N-terminally cleaved BimEL demonstrated a higher affinity for Bcl-2 and a markedly enhanced apoptotic activity. The activation of BimEL downstream of the caspase cascade may provide a positive feedback amplification of apoptotic signals.
Methods
Cell Death Analyses. Apoptosis was induced in Jurkat T cells by 50 ng/ml tumor necrosis factor α (TNF-α)/0.5 μM staurosporine/0.05 μM taxol or by UV irradiation at a dose of 120 J/m2. The broad-spectrum caspase inhibitor N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone was used at 20 μM. Cells were treated with 0.5 μM of the phosphatase inhibitor okadaic acid for 2 h. Cell death was measured by flow-cytometric analysis of hypodiploid nuclei stained with 20 μg/ml propidium iodide or annexin V–propidium iodide double stainin, according to the manufacturer's instructions (Roche Applied Science).
Phosphatase Treatment of BimEL. Jurkat T cells (1 × 106) were lysed in lysis buffer L containing 50 mM Hepes·KOH (pH 7.8), 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 2 mM DTT, and 0.5 mM PMSF. The lysate was dialyzed against 90 mM sodium citrate buffer (pH 4.8) containing 1 mM DTT, 1 mM PMSF, and the human protease inhibitor mixture (Sigma). After the addition of 1 unit of potato acid phosphatase (PAP; Sigma), the lysate was incubated at 30°C for 15 min and then analyzed.
Retroviral Infection and Cell Death Assay. Flag-tagged wild-type or truncated BimEL (tBimEL) cloned into the pBabe-puro retroviral vector were transfected into the Phoenix retrovirus packaging cells (11, 12) with Lipofectamine-Plus reagent (Invitrogen). We mixed 1 ml of the viral supernatant with an equal volume of 1 × 106 per ml Jurkat cells 2 days later, and polybrene was added to a final concentration of 4 μg/ml. The infected Jurkat cells were selected with puromycin (0.5 μg/ml) in the media 2 days after infection. Viable puromycin-resistant cells were counted every 2 or 3 days during the subsequent 8-day period.
In Vivo and in Vitro Binding Assays. For the in vivo Bcl-2-binding assay, constructs expressing the Flag-tagged wild-type or tBimEL were transfected into Jurkat T cells by electroporation. Cells were lysed in the modified lysis buffer L, containing 500 mM NaCl, 2 days after transfection. Precleared lysate was incubated with the anti-Flag agarose beads (Sigma) at 4°C for 2 h. After extensive washes in the same buffer, Flag-tagged BimEL and its associated Bcl-2 were eluted with the Flag peptide. The samples were subjected to Western blotting with anti-Bim and anti-Bcl-2 antibodies.
The in vitro tubulin-binding assay (8) was carried out by incubating 1 μg of purified tubulin (Sigma) with GST-BimL or GST-BimEL fusion proteins (1 μg) immobilized on glutathione agarose beads for 20 min at room temperature in 50 μl of binding buffer B (20 mM Tris, pH 7.5/500 mM NaCl/10% glycerol/0.2 mM EDTA/1 mM DTT/1 mM PMSF). After extensive washes with the same buffer, tubulin associated with GST-BimL or GST-BimEL was eluted with the SDS/PAGE sample buffer and detected by anti-tubulin Western blotting.
The microtubule-binding assay was performed essentially as described with some modifications (13). Jurkat cells (1 × 106) were lysed in lysis buffer L. The cleared lysate treated with or without PAP was incubated at 37°C for 20 min in the presence of 5 μg/ml taxol to reassemble microtubules. Apyrase was left out of the reaction to reduce the indirect docking of Bim onto microtubules through the motor proteins. The reaction mixture was subsequently loaded on top of a 40% sucrose cushion. Microtubules and their associated proteins were isolated by ultracentrifugation at 40,000 rpm for 20 min.
In Vitro Cleavage of BimEL. In vitro cleavage of BimEL in Jurkat cell lysate was carried out at 37°C for 4 h with recombinant caspase-3 (100 units; Calbiochem) by following the manufacturer's instructions. In vitro cleavage coupled with microtubule assembly was carried out by incubating the cleared Jurkat cell lysate (from 1 × 106 cells) with 5 μg/ml taxol at 37°C for 20 min. After microtubules were assembled, 100 units of recombinant caspase-3 (Calbiochem) were added. After a further incubation at 37°C for 4 h, the reaction mixture was subjected to ultracentrifugation.
Activated Mouse Primary T Cells. T cell suspensions from female C57BL/6 mice lymph nodes were cultured in RPMI medium supplemented with 10% FBS in plates coated with 5 μg/ml anti-CD3 and 1 μg/ml anti-CD28 for 2 h.
Results
Both Phosphorylated and Unphosphorylated BimEL Exist in Jurkat T Cells. To elucidate the mechanism of Bim-controlled apoptosis in the immune system, the regulation of Bim activity during apoptosis in T cells was investigated. In Jurkat T cell lysate, Western blotting with the rabbit polyclonal antibodies directed against the full-length BimEL (a generous gift from X. Luo and X. Wang, Howard Hughes Medical Institute and University of Texas Southwestern Medical Center, Dallas) revealed a doublet with mobility similar to that of the Flag-tagged recombinant BimEL (rBimEL) in an SDS gel (Fig. 1A, lanes 1 and 3). The antibodies did not detect the other two Bim isoforms (BimL or BimS) in the lysate, although they are reactive with the rBimL and BimS proteins (lane 2 and data not shown). Because phosphorylation often controls the activity of the Bcl-2 family members (14), we suspected that the doublet detected by immunoblotting might represent the phosphorylated and unphosphorylated forms of BimEL. In support of this idea, incubation of Jurkat cell lysate with PAP converted the upper band of the doublet into the lower one (Fig. 1 A, lanes 4 and 5). In contrast, treatment of Jurkat cells with the phosphatase inhibitor okadaic acid produced the opposite effect of turning the lower band into the upper one (lanes 6 and 7). Thus, the upper band most likely contained the pBimEL.
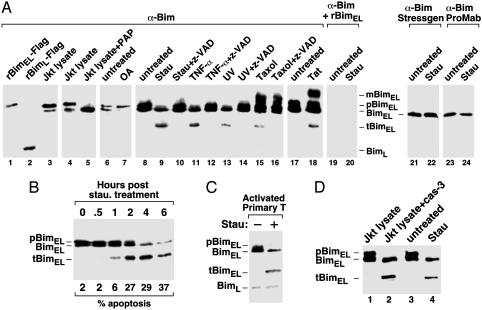
Fig. 1.
Caspase cleavage of pBimEL in T cells at an early stage of apoptosis. (A) Caspase cleavage of pBimEL in apoptotic Jurkat T cells. Anti-Bim Western blotting was performed with the rabbit polyclonal antibodies directed against the full-length BimEL to examine the purified Flag-tagged rBimEL and Flag-tagged rBimL (lanes 1 and 2), Jurkat cell lysates incubated with (lane 5) or without (lanes 3 and 4) PAP, and lysates from Jurkat cells subjected to the various treatments as indicated (lanes 7–20). In lanes 19 and 20, the antibody solution was preincubated with purified rBimEL before being used in Western blotting. In lanes 21–24, Western blotting was performed with anti-Bim antibodies obtained from StressGen Biotechnologies (Victoria, Canada) and ProMab Biotechnologies (Mountain View, CA), respectively. (B) Cleavage of pBimEL at an early stage of apoptosis. Lysates of Jurkat cells treated with staurosporine for the indicated periods of time (hr) were analyzed by anti-Bim Western blotting. The percentage of apoptotic cells was determined by flow-cytometric analysis and is indicated at the bottom. (C) Apoptosis-induced cleavage of pBimEL in activated mouse primary T cells. Anti-Bim Western blot analysis of lysates from activated mouse primary T cells treated with or without staurosporine. (D) pBimEL cleaved in vitro by recombinant caspases-3 comigrates with tBimEL from apoptotic Jurkat cells as indicated by anti-Bim Western blotting. Lanes 1 and 2, lysates from healthy Jurkat cells were incubated with or without recombinant caspase-3. Lanes 3 and 4 show lysates from Jurkat cells treated with or without staurosporine. Stau, staurosporine; OA, okadaic acid; mBimEL, modified BimEL.
Caspase Cleavage of pBimEL at an Early Stage of Apoptosis. Interestingly, treatment of Jurkat cells with several apoptosis stimuli, such as the cytotoxic drug staurosporine, UV irradiation, and the death-inducing cytokine TNF-α, resulted in the disappearance of the upper band and the appearance of a new faster-migrating band of ≈22 kDa (Fig. 1 A, lanes 9, 11, and 13). This effect was completely blocked by N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (lanes 10, 12, and 14), a broad-spectrum caspase inhibitor, suggesting that the ≈22-kDa fragment was probably a caspase-cleaved product of pBimEL (tBimEL). To confirm the specificity of the polyclonal antibodies used in immunoblotting, the antibody solution was preincubated with the purified rBimEL before its incubation with the membrane. This process completely abrogated the detection of both the doublet and the ≈22-kDa fragment (Fig. 1 A, lanes 19 and 20), indicating that these proteins indeed originated from BimEL. It is worth noting that the anti-Bim antibodies purchased from StressGen Biotechnologies and ProMab Biotechnologies detected neither the pBimEL nor the cleaved tBimEL (Fig. 1 A, lanes 21–24).
A time-course analysis revealed that tBimEL began to appear in Jurkat T cells 1 h after the treatment with staurosporine, at which time only ≈6% cells were undergoing apoptosis (Fig. 1B). The caspase cleavage of BimEL at an early stage of apoptosis suggested that tBimEL was not merely an end product generated in dead cells but, rather, may contribute to the execution and progression of apoptosis from the beginning.
Different from staurosporine, UV, and TNF-α, both the microtubule-perturbation drug taxol and the HIV-1 Tat protein, which was shown recently to induce apoptosis by disrupting microtubule dynamics (8), caused not only the caspase-dependent cleavage but also a caspase-independent modification of BimEL. This finding was illustrated by the appearance of a slow-moving BimEL species in the gel (mBimEL; Fig. 1 A, lanes 15, 16, and 18). The nature and significance of this modification remain to be investigated. We suspect that it may be responsible for the observed strong dependence on Bim for both taxol- and Tat-induced apoptosis in T cells (5, 8).
Finally, it is important to point out that the caspase cleavage of the pBimEL was not restricted to Jurkat T cell line. Induction of apoptosis in activated mouse primary T cells by staurosporine also caused the cleavage of the pBimEL (Fig. 1C).
In Vitro Cleavage of pBimEL by Recombinant Caspase-3. Because the apoptotic stimuli activating either the mitochondria pathway (staurosporine, UV, taxol, and Tat) or the death receptor pathway (TNF-α) were all capable of inducing the cleavage of pBimEL, the downstream effector caspases that are common to both pathways were probably responsible for this effect. Consistent with this idea, incubation of recombinant caspases-3 (Calbiochem) with the lysate of healthy Jurkat cells yielded a cleaved pBimEL fragment that comigrated with the tBimEL generated in vivo from apoptotic Jurkat cells (Fig. 1D). This finding further confirmed the 22-kDa fragment as a caspase-cleaved product of pBimEL.
Caspase Cleavage of BimEL After Asp-13. To determine the cleavage site in BimEL, a series of N- and C-terminal deletion mutants of BimEL with truncations after several candidate Asp residues were produced and their mobility in an SDS gel was compared with that of the proteolytic fragment of pBimEL from apoptotic cells (Fig. 2A). Deletion of the first 7 or the last 21 amino acids from BimEL produced protein fragments that migrated slower than the caspase-cleaved 22-kDa tBimEL. tBimEL, however, comigrated with the N-terminal deletion mutant BimELΔN13 lacking amino acids 2–13 (Fig. 2 A, lanes 2 and 3). Further deletion to the next Asp at position 53 or 157 produced much smaller BimEL fragments (data not shown). Further support for the existence in BimEL of a caspase-cleavage site after Asp-13 came from the observation that mutation of Asp-13 to Asn (D13N) in rBimEL abolished its cleavage by caspase-3 in vitro, whereas mutation of Asp-7 to Asn (D7N) had no effect (Fig. 2B).
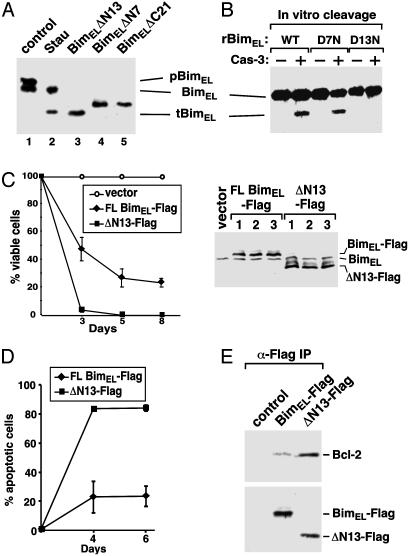
Fig. 2.
N-terminal cleavage of BimEL after Asp-13 enhances its interaction with Bcl-2 and induction of apoptosis. (A) In vivo caspase-cleaved tBimEL comigrates with rBimELΔN13 in SDS gel. BimEL truncated after the various Asp residues were expressed in transfected 293T cells. Their mobility was compared with that of tBimEL from staurosporine (Stau)-treated Jurkat cells by Western blotting. (B) Mutation of Asp-13 to Asn in BimEL prevents cleavage by caspase-3. Anti-Bim Western blot analysis of recombinant wild-type (WT) and mutant (D7N and D13N) BimEL before (–) or after (+) the incubation with recombinant caspase-3. (C) Expression of ΔN13 greatly reduces cell survival. Jurkat cells infected with retroviruses expressing full-length Flag-tagged BimEL (FL BimEL-Flag), Flag-tagged ΔN13 (ΔN13-Flag), or nothing (vector) were selected with puromycin at 2 days after infection. The percentage of surviving cells was determined over the subsequent 8 days, and the average from three independent infections is shown (Left). Flag-tagged BimEL and Flag-tagged ΔN13 produced in Phoenix retrovirus packaging cells from three independent transfections were analyzed by anti-Bim Western blotting, which also detected the endogenous BimEL (Right). (D) ΔN13 has greater apoptotic activity than wild-type BimEL. Plasmids expressing Flag-tagged BimEL or Flag-tagged ΔN13 were electroporated into Jurkat cells. Apoptosis was measured by flow-cytometric analysis of annexin V-positive and propidium iodide-negative cells. (E) ΔN13 binds more Bcl-2 than Flag-tagged BimEL or control. Flag-tagged BimEL, Flag-tagged ΔN13, and their associated Bcl-2 were immunoprecipitated (α-Flag IP) from the lysates of transfected Jurkat cells and analyzed by Western blotting as indicated.
Cleaved BimEL Has Greater Apoptotic Activity. BimEL has been identified as an apoptosis inducer. However, its potency is significantly lower than that of the other two alternatively spliced isoforms, BimL and BimS (9). We next investigated whether the proapoptotic activity of BimEL could be enhanced by the removal of the 13 amino acids at its N terminus. Phoenix retrovirus packaging cells (11) were transfected with the empty pBabe-puro vector (12) or pBabe-puro expressing full-length BimEL or ΔN13. The produced viruses were then used to infect Jurkat T cells and the surviving puromycin-resistant cells were counted over a period of 8 days. With the survival rate of cells infected with the empty vector set at 100%, sustained expression of both full-length BimEL and BimELΔN13 reduced the number of surviving cells. However, when the infection efficiency was adjusted to about the same level as reflected by the expression of the two BimEL proteins in the packaging cells (Fig. 2C Right), ΔN13 was found to be much more efficient in reducing the number of viable cells than the full-length BimEL (Fig. 2C Left). Detection of apoptotic cells by annexin V–propidium iodide double staining (Roche) revealed that ΔN13 produced from the transfected plasmids caused significantly more apoptosis than did the full-length BimEL (Fig. 2D). Thus, the proapoptotic activity of BimEL can be enhanced by the cleavage of the 13 amino acids at its N terminus.
Cleaved BimEL Binds More Efficiently to Bcl-2. Bim has been shown to induce apoptosis by interacting with the prosurvival Bcl-2 family members through its BH3 region (9). Because ΔN13 demonstrated higher apoptotic activity than the full-length BimEL, we examined whether this was because of its increased binding to Bcl-2. Constructs expressing the Flag-tagged full-length BimEL and ΔN13 were electroporated into Jurkat T cells, and Bcl-2 associated with the immunoprecipitated BimEL proteins was analyzed by Western blotting. When the expressions of BimEL and ΔN13 were normalized to a similar level, the amount of Bcl-2 associated with ΔN13 was significantly higher than that with the full-length BimEL (Fig. 2E). Thus, the N terminus of BimEL appeared to have an inhibitory effect on the BimEL–Bcl-2 interaction. The removal of the N-terminal 13 residues may expose the BH3 region for a more efficient binding to Bcl-2.
Direct Interactions of Tubulin and Microtubules with BimEL but Not BimL. We asked how, if only the pBimEL could undergo caspase cleavage during apoptosis, the phosphorylation of BimEL might control this process. The observation that the bacteria-produced rBimEL, which presumably was unphosphorylated, could be cleaved by caspase-3 in vitro (Fig. 2B) suggests that the BimEL phosphorylation was not essential for the cleavage process per se. This observation raises a possibility that the phosphorylation may release BimEL from a certain kind of sequestration, making BimEL accessible to caspase cleavage.
What sequesters the unphosphorylated BimEL and protects it from caspase cleavage? BimEL and BimL have been shown to interact with the dynein light chain 8 (LC8), which is part of the microtubular dynein motor complex (13). Apoptotic stimuli provoked the release of BimL and LC8, allowing BimL to associate with Bcl-2-like proteins (13). Although BimEL and BimL interact with LC8 (13) and Bcl-2 (9) with similar efficiency, BimL is much more apoptotic than BimEL (9). It is possible that BimEL is sequestered to microtubules more tightly than BimL by means of additional interactions, which may prevent its migration to the mitochondria.
In support of this hypothesis, purified tubulin (Sigma) was found to interact directly with immobilized GST-BimEL but not GST-BimL (Fig. 3A). An in vivo interaction of tubulin with Flag-tagged BimEL, but not Flag-tagged BimL, was also detected in anti-Flag immunoprecipitates derived from transfected 293T cells (Fig. 3B). These data indicate that BimEL bound directly to tubulin through a region (amino acids 42–97) that is missing from BimL.
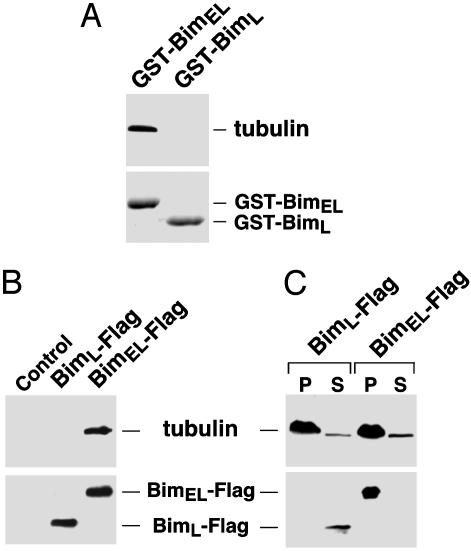
Fig. 3.
BimEL is sequestered to microtubules by means of its direct interaction with tubulin. (A) BimEL, but not BimL, binds directly to tubulin in vitro. Immobilized GST-BimEL and GST-BimL were incubated with purified tubulin. The Bim-associated tubulin was detected by anti-α-tubulin Western blotting. (B) BimEL, but not BimL, binds to tubulin in vivo. Anti-Flag immunoprecipitates from lysates of 293T cells transfected with Flag-tagged BimEL (BimEL-Flag) or BimL (BimL-Flag) were subjected to Western blotting to detect the Bim-associated tubulin. (C) BimEL, but not BimL, binds to polymerized microtubules. Cleared lysates of 293T cells transfected with Flag-tagged BimEL or Flag-tagged BimL were incubated with taxol and then subjected to ultracentrifugation. The levels of α-tubulin and Bim in the pellet (P) and the supernatant (S) were determined by Western blotting.
To determine whether BimEL could also bind to polymerized microtubules, we performed a microtubule assembly assay in 293T cell lysate containing the transfected Flag-tagged BimEL or Flag-tagged BimL. The modified assembly conditions (see Methods) reduced the binding of motor proteins to microtubules and, as a result, the indirect docking of Bim onto microtubules through the motors. On ultracentrifugation to separate the polymerized microtubules and their associated proteins from the rest of the cell lysate, BimEL, but not BimL, was found to associate with microtubules (Fig. 3C).
pBimEL Escapes Sequestration by Microtubules. Microtubule-associated proteins (MAPs) interact with the negatively charged tubulin tails through their positively charged tubulin-binding regions. Phosphorylation of MAPs disrupts these interactions (15). We next asked whether, like other MAPs, pBimEL could dissociate from microtubules, leading to its cleavage by caspases. First, microtubule-stabilizing drug taxol was added to cleared Jurkat cell lysate pretreated with or without PAP, or to cell lysate prepared from Jurkat cells treated with or without the phosphatase inhibitor okadaic acid. Ultracentrifugation was then performed to separate polymerized microtubules and their associated BimEL in the pellet from free tubulin and other soluble proteins in the supernatant. Both fractions, as well as the sample, were analyzed by Western blotting before ultracentrifugation (Fig. 4A, T). Without the PAP treatment, the majority of the unphosphorylated BimEL remained bound to microtubules in the pellet, whereas all of the pBimEL resided in the supernatant. After the PAP treatment, unphosphorylated BimEL migrated from the supernatant to the pellet to associate with the polymerized microtubules. In contrast to PAP, okadaic acid converted the unphosphorylated BimEL to the phosphorylated form, which dissociated from microtubules and stayed in the supernatant. Together, these two complementary approaches revealed a preference of microtubules to bind to the unphosphorylated BimEL.
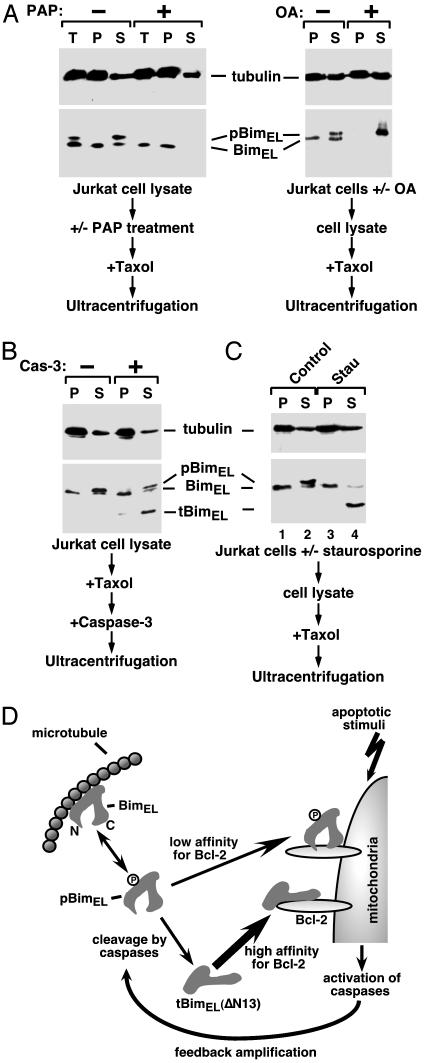
Fig. 4.
pBimEL escapes sequestration by microtubules and becomes accessible to caspase cleavage. (A) pBimEL is released from microtubules. Jurkat cell lysate preincubated with or without PAP (Left) or lysate from Jurkat cells treated with or without okadaic acid (Right) was incubated with taxol and then subjected to ultracentrifugation. α-Tubulin and Bim in the pellet (P), supernatant (S), or lysate before centrifugation (T) were detected by Western blotting. (B) The association with microtubules prevents BimEL from cleavage by caspase-3 in vitro. Jurkat cell lysate was incubated with taxol to assemble microtubules. Recombinant caspase-3 was then added to the reaction. After ultracentrifugation, α-tubulin and Bim were analyzed as in A. (C) pBimEL is released from microtubules and cleaved by caspases in apoptotic Jurkat cells. Lysate from Jurkat cells treated with or without staurosporine (Stau) was subjected to the same analysis as in A.(D) Diagram depicting the activation of a positive feedback apoptotic pathway involving the release of pBimEL from microtubules and caspase cleavage of pBimEL. The cleaved tBimEL(ΔN13) binds more Bcl-2 and has greater apoptotic activity. tBimEL may or may not contain the phosphorylation sites, which have not been mapped in the full-length BimEL. See the text for details.
pBimEL Is Much More Accessible to Caspase Cleavage. To determine whether the phosphorylation-mediated release of BimEL from microtubules would lead to an enhanced caspase cleavage of this protein, we first compared the efficiency of cleavage between the free and the microtubule-bound BimEL by recombinant caspase-3 in vitro (Fig. 4B). After the assembly of microtubules in Jurkat cell lysate, caspase-3 was incubated with the mixture. The reaction was then subjected to ultracentrifugation to separate microtubules from free tubulin. Compared with only a weak cleavage of the microtubule-bound, unphosphorylated BimEL, a much more efficient cleavage of the free pBimEL was observed (Fig. 4B). This conclusion was supported further by the result obtained in staurosporine-treated Jurkat cells, in which only the pBimEL released from microtubules was cleaved (Fig. 4C).
Discussion
Bim is the major physiological antagonist of the prosurvival proteins in B and T lymphocytes (5). It functions by maintaining hematopoietic homeostasis and precluding autoimmunity (5). By performing experiments in Jurkat and activated mouse primary T cells, in which expression of the BimEL isoform is predominant, we have discovered a mechanism by which BimEL participates in the control of apoptosis (Fig. 4D). BimEL was found to exist in both the phosphorylated and unphosphorylated forms in healthy cells, and the unphosphorylated form was sequestered to microtubules by means of a direct interaction with tubulin. This interaction depended on a region in BimEL that is missing in BimL, allowing BimEL to be more tightly sequestered to microtubules and, consequently, less available for Bcl-2 binding than BimL. This finding may explain why BimEL is generally less apoptotic than BimL (9), despite the fact that both can bind to the dynein LC8 (13) and Bcl-2 (9) with similar efficiency.
Phosphorylation of BimEL caused it to dissociate from microtubules. The freed BimEL, however, was mostly inactive because it displayed a low affinity for Bcl-2 (Fig. 4D). On the induction of apoptosis, the pBimEL released from microtubules was processed by a caspase(s) to generate an N-terminally truncated fragment (ΔN13) with an increased affinity for Bcl-2 and a significantly enhanced apoptotic activity. Because apoptotic stimuli activating either the death receptor pathway (TNF-α) or the mitochondria pathway (UV, staurosporine, Taxol, and HIV-1 Tat) are all capable of inducing the cleavage of BimEL, it is likely that a caspase(s) that is common to both pathways is responsible for this event. Although caspase-3 has been shown to cleave BimEL in vitro, it is important to point out that the caspase(s) that is in charge in vivo remains to be elucidated.
What could the caspase cleavage of BimEL contribute to the overall control of apoptosis in T cells? Several pieces of data suggest that the cleavage is not required for the initiation of apoptosis. First of all, the induction of apoptosis did not cause any enhancement in BimEL phosphorylation (as reflected by similar levels of unphosphorylated BimEL in healthy and apoptotic cells; Fig. 4C) or release of BimEL from microtubules (Fig. 4C, compare lanes 1 and 3). More importantly, the cleavage of BimEL depended on the activation of the downstream caspases. Although the cleavage of BimEL does not trigger the onset of apoptosis, it may in fact contribute to a positive feedback amplification of apoptotic signals (Fig. 4D). This idea is supported by the demonstration that the cleaved tBimEL occurred very early during the course of apoptosis and was unlikely to be a simple end product generated in dead cells (Fig. 1B). Furthermore, tBimEL was more active than the full-length protein in targeting Bcl-2 and inducing apoptosis (Fig. 2). It is worth noting that this signal-amplification mechanism initiated by the cleavage of BimEL seems to apply only to mature T cells, whereas in immature T cells undergoing apoptosis, an increased expression of BimEL and BimL has been observed (6). It is unclear what causes such a change in BimEL regulation and whether this change is biologically significant for T cell development.
It is interesting to note that although the signal-amplification effect caused by caspase cleavage of the pBimEL is different from the reported role of the BimL isoform in sensing and initiating the apoptosis pathway (13), similarities can be detected in the control of the activities of the two isoforms. In human breast carcinoma cell line MCF-7 and mouse IL-3-dependent promyelocytic cell line FDC-P1, certain apoptotic stimuli can lead to the phosphorylation of BimL by the Jun N-terminal kinase (16) and the subsequent release of BimL from the sequestration by the microtubular dynein motor complex. This process enables BimL to translocate to the mitochondria and target Bcl-2 or its homologues to initiate apoptosis (13).
The signal-amplification function of the caspase-cleaved BimEL is similar to a previously described amplification activity displayed by another BH3-only protein Bid. Induction of death receptor-initiated apoptosis triggers the activation of caspase-8, which cleaves the inactive cytosolic form of Bid into a truncated Bid (tBid). tBid then translocates to mitochondria and displays higher affinity for antiapoptotic Bcl-2 (17, 18). Whereas the Bid-mediated amplification may be more restricted to the death receptor pathway, BimEL appears to function in a more general manner to amplify death signals initiated from both the mitochondria- and the death receptor-pathways. Future experiments are needed to shed more light on the posttranslational regulation of BimEL and the biological consequences of the BimEL-mediated signal-amplification process.
Acknowledgments
We thank Drs. X. Wang and X. Luo for the anti-Bim antibodies. We also thank Drs. A. Strasser, J. Allison, A. Winoto, K. Luo, and their laboratories for valuable reagents, technical help, and expert advice. This work was supported by grants from the National Institutes of Health (AI-41757), the American Cancer Society (RSG-01-171-01-MBC to Q.Z.), and the U.S. Army Breast Cancer Predoctoral Fellowship (DAMD17-02-1-0321 to D.C.).
Abbreviations: BH, Bcl-2 homology; LC8, light chain 8; PAP, potato acid phosphatase; pBimEL/L, phosphorylated BimEL/L; tBimEL, truncated BimEL; TNF-α, tumor necrosis factor α.
References
- 1.Puthalakath, H. & Strasser, A. (2002) Cell Death Differ. 9, 505–512. [DOI] [PubMed] [Google Scholar]
- 2.Wang, X. (2001) Genes Dev. 15, 2922–2933. [PubMed] [Google Scholar]
- 3.Huang, D. C. & Strasser, A. (2000) Cell 103, 839–842. [DOI] [PubMed] [Google Scholar]
- 4.Bouillet, P., Cory, S., Zhang, L. C., Strasser, A. & Adams, J. M. (2001) Dev. Cell 1, 645–653. [DOI] [PubMed] [Google Scholar]
- 5.Bouillet, P., Metcalf, D., Huang, D. C., Tarlinton, D. M., Kay, T. W., Köntgen, F., Adams, J. M. & Strasser, A. (1999) Science 286, 1735–1738. [DOI] [PubMed] [Google Scholar]
- 6.Bouillet, P., Purton, J. F., Godfrey, D. I., Zhang, L. C., Coultas, L., Puthalakath, H., Pellegrini, M., Cory, S., Adams, J. M. & Strasser, A. (2002) Nature 415, 922–926. [DOI] [PubMed] [Google Scholar]
- 7.Hildeman, D. A., Zhu, Y., Mitchell, T. C., Bouillet, P., Strasser, A., Kappler, J. & Marrack, P. (2002) Immunity 16, 759–767. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., Wang, M., Zhou, S. & Zhou, Q. (2002) EMBO J. 21, 6801–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Connor, L., Strasser, A., O'Reilly, L. A., Hausmann, G., Adams, J. M., Cory, S. & Huang, D. C. (1998) EMBO J. 17, 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Reilly, L. A., Cullen, L., Visvader, J., Lindeman, G. J., Print, C., Bath, M. L., Huang, D. C. & Strasser, A. (2000) Am. J. Pathol. 157, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grignani, F., Kinsella, T., Mencarelli, A., Valtieri, M., Riganelli, D., Lanfrancone, L., Peschle, C., Nolan, G. P. & Pelicci, P. G. (1998) Cancer Res. 58, 14–19. [PubMed] [Google Scholar]
- 12.Morgenstern, J. P. & Land, H. (1990) Nucleic Acids Res. 18, 3587–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puthalakath, H., Huang, D. C., O'Reilly, L. A., King, S. M. & Strasser, A. (1999) Mol. Cell 3, 287–296. [DOI] [PubMed] [Google Scholar]
- 14.Strasser, A., O'Connor, L. & Dixit, V. M. (2000) Annu. Rev. Biochem. 69, 217–245. [DOI] [PubMed] [Google Scholar]
- 15.Lee, G. (1993) Curr. Opin. Cell Biol. 5, 88–94. [DOI] [PubMed] [Google Scholar]
- 16.Lei, K. & Davis, R. J. (2003) Proc. Natl. Acad. Sci. USA 100, 2432–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo, X., Budihardjo, I., Zou, H., Slaughter, C. & Wang, X. (1998) Cell 94, 481–490. [DOI] [PubMed] [Google Scholar]
- 18.Li, H., Zhu, H., Xu, C. J. & Yuan, J. (1998) Cell 94, 491–501. [DOI] [PubMed] [Google Scholar]