Abstract
Histone deacetylase (HDAC) inhibitors (HDACi) cause cancer cell growth arrest and/or apoptosis in vivo and in vitro. The HDACi suberoylanilide hydroxamic acid (SAHA) is in phase I/II clinical trials showing significant anticancer activity. Despite wide distribution of HDACs in chromatin, SAHA alters the expression of few genes in transformed cells. p21WAF1 is one of the most commonly induced. SAHA does not alter the expression of p27KIPI, an actively transcribed gene, or globin, a silent gene, in ARP-1 cells. Here we studied SAHA-induced changes in the p21WAF1 promoter of ARP-1 cells to better understand the mechanism of HDACi gene activation. Within 1 h, SAHA caused modifications in acetylation and methylation of core histones and increased DNase I sensitivity and restriction enzyme accessibility in the p21WAF1 promoter. These changes did not occur in the p27KIPI or ε-globin gene-related histones. The HDACi caused a marked decrease in HDAC1 and Myc and an increase in RNA polymerase II in proteins bound to the p21WAF1 promoter. Thus, this study identifies effects of SAHA on p21WAF1-associated proteins that explain, at least in part, the selective effect of HDACi in altering gene expression.
Keywords: suberoylanilide hydroxamic acid, multiple myeloma, Myc, RNA polymerase
Chromatin structure plays a major role in regulating the expression of the genetic information encoded in DNA (1). DNA is packaged into nucleosomes, the repeating units of chromatin, composed of ≈146 base pairs of two superhelical turns of DNA wrapped around an octamer core of pairs of histones, H2A, H2B, H3, and H4. There is increasing evidence for the importance of covalent, posttranslational modifications in gene regulation. These modifications include acetylation of lysines, methylation of lysines and arginines, and phosphorylation of serines (2–4). Accumulation of acetylated histones is associated with a neutralization of the lysine-positive charge leading to an alteration in chromatin conformation, which may provide for greater access to promoter regions of genes for transcription factor complexes (5).
Histone deacetylases (HDACs) and histone acetyltransferases are the enzymes that determine, in part, the pattern of histone acetylation (6–9). The pattern of acetylation of histones as well as the pattern of methylation and phosphorylation of histones may represent a “code” that is recognized by non-histone complexes involved in the regulation of gene expression (1–3, 10–12).
A number of chemically diverse agents have been discovered, including the hydroxamic acids suberoylanilide hydroxamic acid (SAHA) (13) and trichostatin A (TSA) (14), which inhibit HDAC activity. Crystallographic analysis has shown that SAHA and TSA interact directly with the catalytic site of HDAC like protein and inhibit its enzymatic activity (15). Inhibiting HDAC activity with SAHA and related agents alters gene expression, but, contrary to what might be anticipated by the wide distribution of HDACs in chromatin, there is not a global alteration in gene transcription. Rather, inhibition of HDAC activity by SAHA or TSA results in an increase or decrease in expression of a limited number of genes, ≈2–5% of expressed genes in various transformed cell lines (16–23). Among these genes, p21WAF1 is selectively increased in its expression in several types of transformed cells cultured with SAHA or TSA (17, 18, 21–23). The HDAC inhibitor (HDACi)-induced increase in p21WAF1 expression appears to play a major role in arresting transformed cell growth. Our previous studies found that the increase in p21WAF1 transcription is associated with accumulation of acetylation of the gene-associated histones (17). Furthermore, alteration of the chromatin structure in the transcribed region of a gene has been shown to be associated with induced expression of that gene by transcription factor complexes containing RNA polymerase II (24, 25).
There are three classes of HDAC enzymes. Class I deacetylases include HDACs 1, 2, 3, and 8, are related to yeast RPD3 deacetylase, and have molecular masses of 22–55 kDa. Class II deacetylases include HDACs 4, 5, 6, 7, 9, and 10, are related to yeast Hda1 deacetylase, and have molecular masses of 120–130 kDa (26–28). HDAC 11 contains conserved residues in the catalytic core region shared by both class I and II enzymes. The third class of HDACs are the nicotine adenine dinucleotide-dependent Sir 2 family of deacetylases, which differ from class I and II HDACs in that they are not inhibited by TSA, SAHA, or related compounds (28). There is now abundant evidence that HDACs are not redundant in function. For example, class I HDACs are found almost exclusively in the nucleus, whereas class II HDACs shuttle between the nucleus and cytoplasm on certain cellular signals (7, 28, 29). Targeted disruption of HDAC1 results in embryonic lethality despite increased expression of HDACs 2 and 3 (29). Alterations in histone acetyltransferases and HDACs occur in many cancers (30–33).
A broad spectrum of transformed cells are sensitive to SAHA-induced growth inhibition in in vitro and in vivo studies (8, 27, 28). Tumor cells are much more sensitive to SAHA than are normal cells (34). SAHA is in phase I and II clinical trials for the treatment of various cancers and has shown anticancer activity at doses that are well tolerated by patients (35, 36). These preclinical results and the clinical trials showed that SAHA targeted transformed cells in preference to normal cells.
In the present study using a human multiple myeloma cell line, ARP-1, we have examined the changes in the p21WAF1 promoter caused by SAHA. This gene is expressed at low or almost undetectable levels in ARP-1 cells and is rapidly induced by SAHA. The HDACi caused changes in the acetylation and methylation of p21WAF1 promoter-associated histones and increased the DNase I sensitivity and restriction enzyme accessibility of the gene. There was a marked decrease in HDAC1 and Myc and an increase in RNA polymerase II in the protein complex bound to the proximal region of the p21WAF1 promoter region. The alterations in the protein complexes associated with the promoter region of p21WAF1 occurred within 3 h of culture of ARP-1 cells with SAHA, as did the increase in p21WAF1 mRNA and protein. The p27KIPI gene is actively expressed and the ε-globin gene is silent in ARP-1 cells, and the expression of neither gene is altered by HDACi, nor are the patterns of acetylation or methylation of the histones H3 and H4 associated with these genes. These findings may describe the basis of the selective effect of SAHA in altering gene expression, and, in turn, on inducing growth arrest of tumor cells.
Experimental Procedures
Cell Culture. The human multiple myeloma cell line ARP-1 was generously provided by J. Hardoc (Arkansas Cancer Research Center, Little Rock). ARP-1 cells were cultured in RPMI medium 1640 as described (37). Cells were grown in suspension and subcultured every 3–4 days in complete RPMI medium 1640 and seeded at a density of 2 × 105 cells per ml for cultures with SAHA (in concentrations indicated) (13). Cell density and viability were determined as described (38).
Histone Isolation and Immunoblotting Analyses. ARP-1 cells (1 × 107) were cultured without or with different concentrations of SAHA for the indicated times. The cells were recovered by centrifugation, and core histone proteins were extracted as described (13). Histone concentration was determined by using Bio-Rad reagent according to the manufacturer's protocol (1 μg of total histone protein was used for analysis on SDS/15% PAGE gels) (39). Multiple gels were run simultaneously for Western blot analysis and gel code staining (Pierce) that was used as a histone protein-loading control (40). Histone proteins were transferred from gels to Hybond-P nylon membranes (Amersham Pharmacia Biotech) and analyzed with specific histone antibodies. The signal of the horseradish peroxidase-conjugated secondary antibody was detected by using Super-Signal West Pico detection system (Pierce).
Antibodies. The antibodies to the acetylated, methylated, and phosphorylated histones were purchased from Upstate Biotechnology (Lake Placid, NY). The following antibodies were used in this study (their catalog numbers are indicated): anti-diacetylated histone H3 (H3, K9/K14, 06-599); anti-tetraacetylated H4 (H4, K5/8/12/16, 06-866); anti-H3 acetylated K9 (06-942); anti-H3 acetylated K14 (06-911); anti-H4 acetylated K5, K8, K12, and K16 (06-759, 06-760, and 06-762, respectively); anti-H3 phosphorylated S10 (06-570); anti-H3 phosphorylated S10 plus acetylated K14 (067-081); anti-H3 methylated K4 (07-030); and anti-H3 methylated K9 (07-212). Anti-HDAC-1 was purchased from Upstate Biotechnology, and anti-HDAC2 (382153) was obtained from Calbiochem (San Diego). Anti-Sp1 (sc14027x), anti-myc (sc-51764), anti-RNA polymerase II (sc-899), barrier-to-autointegration factor 155 (10756X), anti-brm-related gene 1 (sc-10768X), anti-cAMP-responsive element binding protein (CREB)-binding protein (sc-7300X), anti-p300 (sc-584X), and anti-GCN5 (sc-20698) antibodies were purchased from Santa Cruz Biotechnology.
Nuclear Extract Preparation and Protein Analyses. For analysis of non-histone proteins, nuclear extract preparations were performed as described in ref. 40. Nuclear proteins were analyzed by immunoblotting (39) with specific primary antibodies and horseradish peroxidase-labeled secondary antibody as described above.
RNA Isolation and Northern Blotting. ARP-1 cells were cultured and recovered by centrifugation, and RNA was prepared as described (13). Northern blot hybridizations were performed by using a 32P-labeled 695-bp human p21WAF1 cDNA fragment. The membranes were stripped and reprobed with 383 bp of 32P-labeled Pstl-digested mouse p27KIP1 cDNA, 32P-labeled β-actin cDNA (ATCC 65128), and a 32P-labeled 50-mer 18S rRNA oligonucleotide, sequentially.
Chromatin Immunoprecipitation (ChIP) Analyses. ARP-1 cells (seeded at 2–2.5 × 105 cells per ml) were cultured with or without 2.0 μM SAHA for the indicated time points, up to 24 h. For ChIP assays of transcription factor complexes, cells were cultured without or with 2 μM SAHA for 3 h. Protein–protein, protein–RNA, and protein–DNA were crosslinked by addition of 0.4% formaldehyde (Fisher) to cells that were in culture medium at room temperature for 10 min. The crosslinking was stopped by adding glycine at a final concentration of 0.125 M. Chromatin was broken to the average size, 400 bp, by sonication (Fisher). ChIP and PCR amplification were performed as described, except that 27 PCR cycles were performed for histone ChIP samples and 32 PCR cycles were performed for the samples of transcriptional factor ChIPs. The products of 80- to 100-bp amplicons were electrophoresed in 8% native polyacrylamide gels, and the signals were quantitated on a PhosphorImager by using imagequant software as described (41).
Primers for p21 Gene PCR Amplification in ChIP Assays. Nine polynucleotide primer pairs were designed from far upstream of the promoter region to downstream of the transcriptional initiation site of the p21WAF1 gene.
For p27KIP1 gene ChIP assays, the primer pairs used were p27KIP1 BU (from –1392 to –1412; TACAATCCCGGGAAAGAACA) and p27KIP1 BD (from –1320 to –1339; GATCTTCCTTCCCAAGCACA).
For ε-globin gene promoter ChIP assays, the primer pair used were CACAGGTCATTGACCAATGACT (sense) and TTATTCTTTACTGCCGAAGTTCTGG (antisense) (41).
Preparation of Nuclei and DNase I Nuclease Digestion. The nuclei of ARP-1 cells (2 × 108 cells) were prepared based on a method described in ref. 41, and the nuclei of ARP-1 cells were resuspended in 8.5% sucrose washing buffer plus 3 mM CaCl2. Nuclei from 5 × 107 nuclei were digested with 0, 1, 2, and 4 μl of DNase I (105 units/μl, Invitrogen) for 10 min at room temperature. The digestions were stopped with 10× Stop solution (125 mM EDTA/1 mg/ml proteinase K/10% SDS) and NaCl (50 mM) and incubated at 55°C for 3 h to digest proteins. DNA was extracted and dissolved in 200 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). About 20 μl of DNA (20 μg) was further digested with SacI for 4 h, and DNA was run in 1% agarose, blotted with a 32P-labeled 360-bp SacI+AflII DNA fragment (in p21WAF1 promoter region). For DNase I hypersensitive assay of p27KIPI gene, the DNase I-digested DNA was further digested with EcoNI for 4 h at 37°C and blotted with a 32P-labeled 563-bp SmaI fragment of p27KIP1 gene promoter (from –844 to –1407).
Restriction Enzyme Accessibility Assay. A restriction enzyme accessibility assay was performed as described (42). Briefly, ARP-1 cells were harvested, and 1.5 × 107 nuclei of ARP-1 cells were prepared and digested with selected restriction enzymes as indicated (New England Biolabs) at 37°C for 30 min. The reactions were stopped, and DNA was extracted and dissolved in 400 μl of TE buffer and used for Southern blot analysis. About 100 μl of DNA was further digested with SacI to produce the parental band. Southern blot membrane was blotted with a 360-bp 32P-labeled SacI Taft II DNA fragment. The signal of Southern blot analysis was quantitated by using a PhosphorImager and imagequant (Molecular Dynamics).
Results
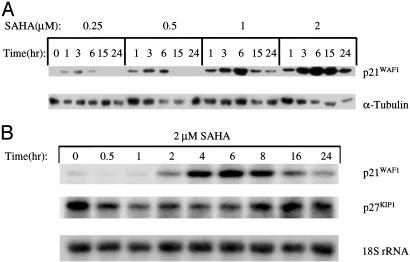
SAHA Induces p21WAF1 in ARP-1 Cells. SAHA induced a time- and dose-dependent increase in p21WAF1 protein (Fig. 1A) and mRNA levels in ARP-1 cells (Fig. 1B). The increase in p21WAF1 mRNA and protein was detected within 1 h of onset of culture. There was no detectable increase in α-tubulin protein, or in the level of p27KIP1 mRNA (Fig. 1), actin mRNA, or ε-globin mRNA (data not shown).
Fig. 1.
SAHA rapidly increases the p21WAF1 mRNA and protein levels in ARP-1 cells. (A) ARP-1 cells were cultured with the concentrations of SAHA indicated and for the times indicated. Whole-cell extracts from 107 ARP-1 cells were prepared, and 30 μg of the extract protein was loaded in SDS/15% PAGE gel and immunoblotted with antibody against p21WAF1 protein. A duplicate gel and membrane were blotted with the antibody for α-tubulin. (B) Total cellular RNA was prepared from ARP-1 cells cultured with 2 μM SAHA for the times indicated; 10 μg of RNA was electrophoresed on a 1% formaldehyde-denatured agarose gel and then transferred to a Hybond N+ nylon membrane. The membrane was hybridized with a 32P-labeled 695-bp p21WAF1 cDNA probe, then stripped and rehybridized with a 32P-labeled 383-bp PstI-digested p27KIP1 cDNA probe and a 32P-labeled 50-mer 18S rRNA oligonucleotide probe sequentially.
ARP-1 cells cultured with SAHA for 18–24 h or more undergo apoptosis. In culture with 1.0 or 2.0 μM SAHA, 70% and ≥90% of APR-1 cells had undergone apoptosis by 24 and 48 h, respectively (data not shown).
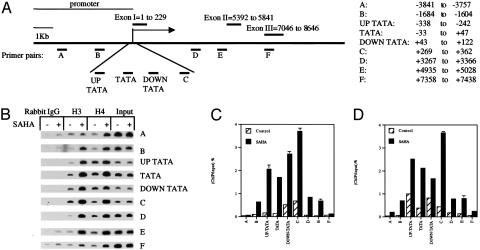
SAHA-Induced Acetylation of Histone H3 and H4 Associated with the p21WAF1 Gene. Previous studies demonstrated that SAHA, as well as other HDACi, induce the accumulation of acetylated histones in both transformed and normal cells (8, 17, 22, 43). To determine the effect of SAHA on the pattern of acetylation and methylation of histones associated with the p21WAF1 gene, we used nine primers complementary to the promoter region (–3,841 bp) to downstream of transcriptional start site (+7,438 bp) (Fig. 2A). Employing ChIP assays, we found that SAHA (2 μM) induced an increase in acetylated histones H3 and H4 in regions corresponding to –3,841 bp upstream to +7,438 bp downstream of the transcriptional initiation site the p21WAF1 gene (Fig. 2 B–D). The level of accumulation of acetylated histone H3 and H4 appeared to be highest in the region –338 bp upstream to +362 bp downstream of the transcriptional initiation site (regions that include Sp1 binding sites, TATA box, and transcriptional initiation site).
Fig. 2.
Effect of SAHA on acetylation of p21WAF1-associated histone H3 and H4. (A) A schematic graph of the p21WAF1 gene indicating the location of nine pairs of primers used for PCR amplification in ChIP assays. The position of the primer pairs designated A, B, UP-TATA, TATA, DOWN-TATA, C, D, E, and F corresponded to the base pairs indicated from upstream (–3,841 bp) to downstream (+7,438 bp) of the transcription initiation site. (B) ChIP assays were prepared by using rabbit normal IgG, anti-diacetylated H3 (K9/14-ac), and anti-tetraacetylated H4 (K5/8/12/16-ac). The samples were amplified with each of the nine pairs of primers. ARP-1 cell were cultured without (–) and with (+) 2 μM SAHA for 3 h. (C) p21WAF1 DNA corresponding to the indicated probe as a percent of the input DNA associated with histone H3. (D) p21WAF1 DNA corresponding to indicated probe as a percent of the input DNA associated with acetylated histone H4.
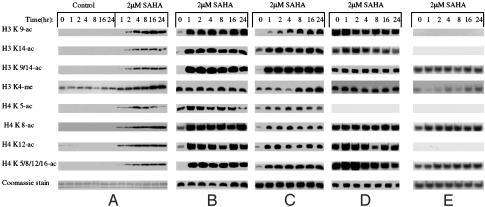
SAHA-Induced Posttranslational Modifications of Histones H3 and H4 Associated with the p21WAF1 Gene. We next examined the patterns of acetylation and methylation of specific lysines in N-terminal tails of histones H3 and H4 associated with the p21WAF1 gene of ARP-1 cells cultured with 2 μM SAHA for up to 24 h. In these studies, we first determined the effect of SAHA on the genome-wide histones. SAHA caused an increase in accumulation of acetylated lysine 9 and 14 of H3 after 2 h, reaching peak levels at 16 h (Fig. 3A). Accumulation of acetylated lysines 5, 8, and 12 of H4 were detected by 1 h and increased to peak levels by 4–8 h. Lysine 4 of H3 was methylated in cells cultured without HDACi. SAHA increased the level of dimethylation of lysine 4 of H3 over the 24-h period of culture (Fig. 3A). SAHA also increased the level of trimethylation of lysine 4 of H3. There was little or no detectable methylation of this lysine in cells without SAHA. The global histone acetylation and methylation patterns reflect the entire genome-associated histones.
Fig. 3.
SAHA-induced modifications of specific lysine residues of histone H3 and H4. (A) ARP-1 cells (2.5 × 105 cells per ml) were cultured without or with 2 μM SAHA for the indicated times. Histones were prepared and analyzed as detailed in Experimental Procedures. The primer histone lysine and serine antibodies used for Western blot analysis are indicated. (B) ChIP analyses of histone H3 and H4 modifications in p21WAF1 proximal promoter region (TATA box primer pair, –52 to +28) of ARP-1 cells at zero time and cultured with 2 μM SAHA for the times indicated. One set of ChIP analyses was performed with the antibodies indicated. (C) ChIP analyses of histone H3 and H4 modifications in p21WAF1 distal promoter region (primer pair B, –1,703 to –1,623) of ARP-1 cells used for B.(D) ChIP analyses of histone modifications in p27KIP1 promoter (from –1,320 to –1,412) prepared from ARP-1 cells used for B and C. The antibodies were used for ChIP assays as for A–C except for anti-H3 K9 mc, H4 K5 ac, and H4-K 16 ac. (E) ChIP analyses of histones modification in ε-globin promoter (see Experimental Procedures) prepared from ARP-1 cells used in B–D. No signal was detected with antibody to H3, K9ac.
We then examined the p21WAF1 promoter-associated pattern of lysine acetylation and methylation in the histones H3 and H4. Two primer pairs were used in ChIP amplification assays; one corresponded to –33 bp to +47 bp covering the TATA box and close to Sp1 binding site, referred to as the proximal promoter region (Fig. 2 A; see also Experimental Procedures). The other primer pair corresponded to –1,684 bp to –1,604 bp upstream of transcriptional initiation site, close to a p53 consensus sequence referred to as the distal promoter region (Fig. 2 A). Within 1 h of culture with 2 μM SAHA there was an increase in expression of the p21WAF1 gene (Fig. 1), associated with an increase in the level of acetylation of lysines 9 and 14 of H3 and lysines 5, 8, and 12 of H4 in both the proximal and distal promoter regions of the p21WAF1 gene. Higher levels of acetylation of lysines 9 and 14 of H3 and lysines 5, 8, and 12 of H4 occurred earlier in the proximal promoter region than in the distal promoter region or in the global histones (Fig. 3 A–C).
The p27KIPI gene is actively expressed in ARP-1 cells, and the HDACi does not alter its expression. Lysines 9 and 14 of H3 and lysines 8 and 12 of H4 were acetylated, and lysine 4 of H3 was methylated, in histones associated with the p27KIPI promoter before culture with SAHA. The HDACi did not alter this pattern of posttranslational modification of the histones (Fig. 3D).
The ε-globin gene is not expressed in ARP-1 cells, and the HDACi does not induce expression of this gene (data not shown). In ARP-1 cells cultured without SAHA, in the ε-globin-associated histones, there was no detectable acetylation of lysine 9 of H3. Lysine 14 of histone H3 and lysine 8 of histone H4 were acetylated, and lysine 4 of histone H3 was methylated. Culture with 2 μM SAHA for up to 24 h did not alter these patterns of histone acetylation and methylation (Fig. 3E).
Taken together, these findings indicate that SAHA selectively alters the pattern of posttranslational modification of lysines of H3 and H4 associated with the p21WAF1 promoter. Such selective changes in histone modifications could play an important role in the induction of p21WAF1 expression by the HDACi.
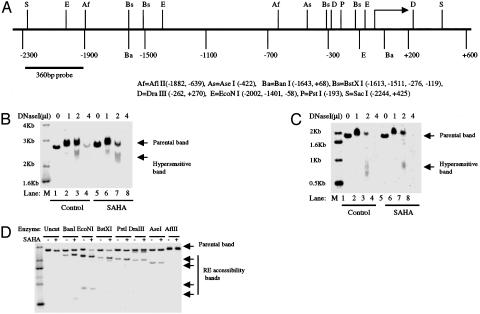
SAHA Causes Increased DNase I Sensitivity and Restriction Enzyme Accessibility of p21WAF1 Promoter Region. Acetylation of histone proteins neutralizes the positive charge on lysine residues, which presumably leads to a more “open” chromatin structure (1). We examined this question by determining the effect of SAHA on the DNase I sensitivity of the p21WAF1 gene. The DNase 1 sensitivity of the p21WAF1 gene of ARP-1 cells cultured without and with 2 μM SAHA for 3 h was assayed by digesting nuclei with increasing amounts of DNase I (Fig. 4B). SAHA caused an increased sensitivity of the p21WAF1 gene to DNase I. At 4 μl of DNase I (420 units/ml), the p21WAF1 DNA from cells cultured with SAHA was completely digested compared with partial digestion of the DNA from control cells. At 1 and 2 μl of DNase I, the DNA of the p21WAF1 gene from cells cultured with SAHA showed greater amounts of digestion than did controls.
Fig. 4.
DNase I sensitivity and restriction enzyme accessibility of the p21WAF1 gene from ARP-1 cells cultured with SAHA. (A) A schematic graph of p21WAF1 promoter region indicating the recognition sites of the restriction enzymes used for accessibility assay and the location of the probe used for Southern blot hybridization (360 bp of SacI-to-AflII fragment). Restriction enzymes are represented with a single letter or two letters, and the recognition sites are shown as numbers with reference to the transcriptional start site as +1. The subscript identifies the restriction enzymes. (B) DNase I sensitivity assay of the p21WAF1 promoter of ARP-1 cells cultured with or without 2 μM SAHA for 3 h. “Parental band” is the SacI fragment of p21WAF1 promoter (2,669 bp; –2,244 bp to 4,425 bp). “DNase hypersensitive” band is a smear (2.2–2.4 kb). Lane M, 32P-labeled 1-kb DNA ladder (Invitrogen). Lanes 1–4, nuclei of ARP-1 cells digested with increasing DNase I (Invitrogen, 105 units/μl) volume (0, 1, 2, and 4 μl of DNase I, respectively) for 10 min at room temperature. Lanes 5–8, nuclei of ARP-1 cells cultured with 2 μM SAHA for 3 h; the nuclei were digested with increasing DNase I volume (0, 1, 2, and 4 μl of DNase I, respectively) for 10 min at room temperature. (C) DNase I hypersensitivity assay of p27KIP1 promoter of ARP-1 cells with or without the treatment of 2 μM SAHA for 3 h. The analyses were preformed as indicated for B. (D) Restriction enzyme accessibility assays of chromatin of p21WAF1 promoter of ARP-1 cells cultured without (–) or with 2 μM SAHA for 3 h. Nuclei were prepared and digested with AflII, AseI, BanI, BstXI, DraIII, EcoNI, and PstI and uncut, as indicated, at 37°C for 30 min. Total DNA was extracted, and 20 μg of genomic DNA was used for a 1% agarose gel and transferred to a nylon membrane. Southern blot was performed with a 32P-labeled 360-bp fragment of p21WAF1 promoter fragment as a probe. “Parental band” shows the 2667-bp SacI fragment of p21WAF1.
The p27KIPI DNA isolated from ARP-1 cells cultured without or with SAHA for 3 h showed similar patterns of sensitivity to digestion by DNase I (Fig. 4C). These results suggest that SAHA leads to a more open chromatin structure in the p21WAF1 gene associated with induction of its transcription. SAHA did not alter the accessibility of the p27KIPI gene to DNase, a gene being actively transcribed in ARP-1 cells.
To further define the effect of SAHA on p21WAF1 chromatin structure, we examined the sensitivity of the gene to several restriction enzymes. Seven restriction enzymes were used that had recognition sites from –1,882 bp upstream to downstream of the transcriptional start site of the p21WAF1 gene (Fig. 4A). SAHA increased the sensitivity of the p21WAF1 gene to restriction enzymes for sites –643 bp upstream (EcoNI, BanI, and BstX I) to downstream (BanI) of the transcriptional start site (Fig. 4 A and D). SAHA did not alter the sensitivity at AflII, the upstream site. These data further indicate that SAHA induces a more open structure of chromatin in the region of the p21WAF1 gene.
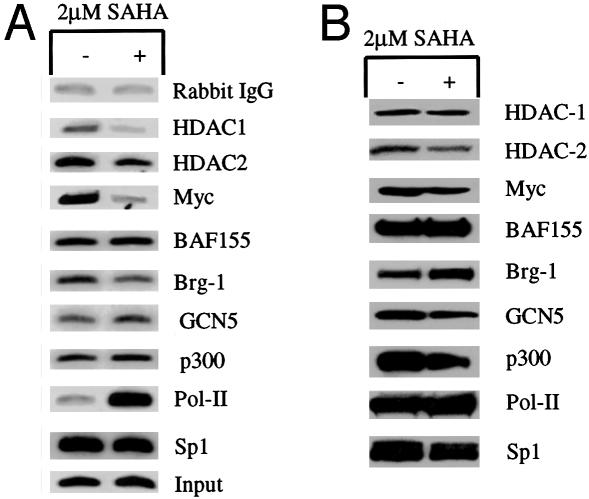
SAHA Induces Alterations in the Components of p21WAF1-Associated Proteins. We next examined whether SAHA caused changes in the components of the protein complexes bound to the p21WAF1 promoter that might be involved in the HDACi-induced transcription of the gene. Using the ChIP assay, we found that SAHA caused an ≈85% decrease in HDAC1 protein bound to the p21WAF1 promoter region (Fig. 5A). There was no detectable difference in the amount of HDAC1 protein in the nuclear lysates of cells cultured without or with SAHA (Fig. 5B). There was little decrease in HDAC2 in the promoter-bound proteins. SAHA also induced a marked decrease in c-myc protein and a >4-fold increase in RNA polymerase II associated with the p21WAF1 promoter-bound proteins. SAHA did not alter the amount of histone acetyltransferases, GCN5 or p300, or the amount of the chromatin remodeling factor, barrier-to-autointegration factor 155 (BAF155), but there was a decrease in brm-related gene 1 (Brg1), another chromatin remodeling factor associated with p21WAF1 promoter. There was no detectable difference in the levels of Myc, BAF155, Brg-1, GCN5, p300, or RNA polymerase II proteins in the nuclear lysates of cells cultured without or with SAHA (Fig. 5B).
Fig. 5.
Induced alterations in components of the p21WAF1 promoter-associated proteins. (A) Antibodies anti-HDAC1, anti-HDAC2, anti-Myc, anti-barrier-to-autointegration factor 155, anti-brm-related gene 1, anti-GCN5, anti-RNA polymerase II, anti-p300, and anti-Sp1 were used for ChIP assays. PCR amplification was done with p21WAF1 promoter TATA box region primers labeled with 32P (see Fig. 2). PCR products were analyzed as described in Experimental Procedures. ARP-1 cells were cultured without (–) and with (+)2μM SAHA for 3 h. Chromatin (0.05%)-extracted DNA (Input) was used for PCR amplification as chromatin loading. (B) ARP-1 cells were grown without (–) or with (+)2 μM SAHA for 3 h, and nuclear extracts were prepared. The level of each component was analyzed in Western blots by using the indicated antibodies.
Discussion
The HDACi SAHA rapidly induces the expression of p21WAF1 but does not alter the expression of p27KIPI, α-tubulin, or actin genes or induce the expression of the ε-globin gene in ARP-1 cells, a human multiple myeloma cell line. In this study, we found that SAHA rapidly induced changes in the p21WAF1 gene and associated proteins (including acetylation of lysines 9 and 14 of histone H3 and lysines 5, 8, and 12 of histone H4), increased the sensitivity of the gene to DNase I, and increased accessibility to restriction enzymes. Lysine 4 of H3 associated with the p21WAF1 gene was methylated in cells before and during culture with SAHA.
The p27KIPI gene is actively transcribed in ARP-1 cells and is not altered in its expression by SAHA. The pattern of p27KIPI promoter-associated acetylation and methylation of histone H3 and H4 was not altered by the HDACi. Histone H3 was methylated at lysine 4 and acetylated at lysine 9 before culture with SAHA. The ε-globin gene is silent in ARP-1 cells and is not induced by the HDACi. SAHA did not induce a change in the pattern of acetylation or methylation of histones H3 or H4 associated with the ε-globin gene promoter. Furthermore, the changes detected in acetylation in the p21WAF1-associated histones were not reflected in the genome-wide pattern of histone modifications. The acetylation of histone lysine residues and consequent neutralization of the basic lysine charge was associated with structural changes in chromatin of the p21WAF1 gene, as evidenced by the increase in DNase I sensitivity and restriction enzyme accessibility of the promoter region of the gene. SAHA did not alter the DNase I sensitivity of the p27KIPI gene. These findings indicate that SAHA has a selective effect on the p21WAF1 gene and associated proteins.
SAHA induced the acetylation of H3 lysine 9, whereas H3 lysine 4 methylation was not altered in the p21WAF1 promoter-associated histones. The present findings are of particular interest in that methylation of H3 lysine 9 has been reported to be largely associated with repression of gene expression, whereas methylation of H3 lysine 4 is often associated with active gene expression (44). It has been hypothesized that histone modifications acting alone, sequentially, or in combination moderate the code recognized by non-histone protein complexes that regulate gene expression (2–4, 10–12, 44). HDACs do not bind directly to DNA but are recruited to DNA by multiprotein complexes (7). In analyzing the components of the protein complex bound to the proximal promoter region of the p21WAF1 gene, we found that SAHA caused disassociation of HDAC1 from the promoter within3hof culture. There was no detectable change in HDAC1 protein in the nuclear lysates. This loss of HDAC1 may play a role in SAHA-induced increase in acetylation of the histones.
SAHA caused other changes in the composition of the protein bound to the proximal region of the p21WAF1 promoter that are consistent with induced expression of the gene. There was marked decrease in c-myc protein. It has been shown that c-myc can be recruited directly to the p21WAF1 promoter by Miz-1 and blocks the induction of the gene by various activators (45). There was also the rapid recruitment of RNA polymerase II, increasing 3-fold within 1 h and 4-fold by 3 h of culture with inducer. The HDACi-induced decrease in c-myc and recruitment of RNA polymerase II are likely major factors leading to the rapid increase in expression of the p21WAF1 gene. Although HDACs are broadly distributed in the chromatin of tumor and normal cells, the HDACi SAHA has shown marked selectivity against cancer cells compared with normal cells. SAHA induces growth arrest, differentiation, or apoptosis of tumor cells in culture at concentrations that have no toxicity for normal cells (34). SAHA inhibits cancer cell growth in tumor-bearing animals with little or no toxicity (28, 38, 43, 46) and has significant anticancer activity in clinical trials at doses well tolerated by patients (35, 36). One can speculate that the selective effect of SAHA on inducing a gene such as p21WAF1 whose expression is suppressed in many tumor cells explains, in part, the favorable therapeutic index of SAHA.
Acknowledgments
We are grateful to Drs. Andrew Koff and Xinhua Zhu (Memorial Sloan–Kettering Cancer Center) for providing human p21WAF1 and mouse p27KIP1 cDNA plasmids, Dr. Toshiyuki Sakai (Kyoto Prefectural University of Medicine, Kyoto) for providing the p27KIP1 promoter plasmid, and Dr. Wafik S. El-Deiry (University of Pennsylvania, Philadelphia) and Dr. Bert Vogelstein (The John Hopkins University, Baltimore) for providing the p21WAF1 promoter plasmid. This work was supported, in part, by grants from the National Cancer Institute (CA-0974823), the Kleberg Foundation, the DeWitt Wallace Fund for the Memorial Sloan–Kettering Cancer Center, and the Susan and Jack Rudin Foundation, and by the David H. Koch Prostate Cancer Research Award.
Abbreviations: ChIP, chromatin immunoprecipitation; HDAC, histone deacetylase; HDACi, HDAC inhibitor; SAHA, suberoylanilide hydroxamic acid; TSA, trichostatin A.
References
- 1.Felsenfeld, G. & Groudine, M. (2003) Nature 421, 448–453. [DOI] [PubMed] [Google Scholar]
- 2.Spotswood, H. T. & Turner, B. M. (2002) J. Clin. Invest. 110, 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 4.Zhang, Y. & Reinberg, D. (2001) Genes Dev. 15, 2343–2360. [DOI] [PubMed] [Google Scholar]
- 5.Allfrey, V. G. (1966) Proc. Can. Cancer Conf. 6, 313–335. [PubMed] [Google Scholar]
- 6.Johnson, C. A. & Turner, B. M. (1999) Semin. Cell. Dev. Biol. 10, 179–188. [DOI] [PubMed] [Google Scholar]
- 7.Khochbin, S., Verdel, A., Lemercier, C. & Seigneurin-Berny, D. (2001) Curr. Opin. Genet. Dev. 11, 162–166. [DOI] [PubMed] [Google Scholar]
- 8.Marks, P. A., Rifkind, R. A., Richon, V. M., Breslow, R., Miller, T. & Kelly, W. K. (2001) Nat. Rev. Cancer 1, 194–202. [DOI] [PubMed] [Google Scholar]
- 9.Roth, S. Y., Denu, J. M. & Allis, C. D. (2001) Annu. Rev. Biochem. 70, 81–120. [DOI] [PubMed] [Google Scholar]
- 10.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 11.Turner, B. M. (2000) Bioessays 22, 836–845. [DOI] [PubMed] [Google Scholar]
- 12.Agalioti, T., Chen, G. & Thanos, D. (2000) Cell 111, 381–392. [DOI] [PubMed] [Google Scholar]
- 13.Richon, V. M., Webb, Y., Merger, R., Sheppard, T., Jursic, B., Ngo, L., Civoli, F., Breslow, R., Rifkind, R. A. & Marks, P. A. (1996) Proc. Natl. Acad. Sci. USA 93, 5705–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida, M., Kijima, M., Akita, M. & Beppu, T. (1990) J. Biol. Chem. 265, 17174–17179. [PubMed] [Google Scholar]
- 15.Finnin, M. S., Donigian, J. R., Cohen, A., Richon, V. M., Rifkind, R. A., Marks, P. A., Breslow, R. & Pavletich, N. P. (1999) Nature 401, 188–193. [DOI] [PubMed] [Google Scholar]
- 16.Van Lint, C., Emiliani, S. & Verdin, E. (1996) Gene Expr. 5, 245–253. [PMC free article] [PubMed] [Google Scholar]
- 17.Richon, V. M., Sandhoff, T. W., Rifkind, R. A. & Marks, P. A. (2000) Proc. Natl. Acad. Sci. USA 97, 10014–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vrana, J. A., Decker, R. H., Johnson, C. R., Wang, Z., Jarvis, W. D., Richon, V. M., Ehinger, M., Fisher, P. B. & Grant, S. (1999) Oncogene 18, 7016–7025. [DOI] [PubMed] [Google Scholar]
- 19.Xiao, H., Hasegawa, T. & Isobe, K. (1999) J. Cell. Biochem. 73, 291–302. [PubMed] [Google Scholar]
- 20.Butler, L. M., Zhou, X., Xu, W.-S., Scher, H. I., Rifkind, R. A., Marks, P. A. & Richon, V. M. (2002) Proc. Natl. Acad. Sci. USA 99, 11700–11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, L., Sowa, Y., Sakai, T. & Pardee, A. B. (2000) Oncogene 19, 5712–5719. [DOI] [PubMed] [Google Scholar]
- 22.Sambucetti, L. C., Fischer, D. D., Zabludoff, S., Kwon, P. O., Chamberlin, H., Trogani, N., Xu, H. & Cohen, D. (1999) J. Biol. Chem. 274, 34940–34947. [DOI] [PubMed] [Google Scholar]
- 23.Sowa, Y., Orita, T., Minamikawa, S., Nakano, K., Mizuno, T., Nomura, H. & Sakai, T. (1997) Biochem. Biophys. Res. Commun. 241, 142–150. [DOI] [PubMed] [Google Scholar]
- 24.Orphanides, G. & Reinberg, D. (2000) Nature 407, 471–475. [DOI] [PubMed] [Google Scholar]
- 25.Belotserkovskaya, R., Oh, S., Bondarenko, V. A., Orphanides, G., Studitsky, V. M. & Reinberg, D. (2003) Science 301, 1090–1093. [DOI] [PubMed] [Google Scholar]
- 26.Grozinger, C. M. & Schreiber, S. L. (2002) Chem. Biol. 9, 3–16. [DOI] [PubMed] [Google Scholar]
- 27.de Ruijter, A. J., van Gennip, A. H., Caron, H. N., Kemp, S. & van Kuilenburg, A. B. (2003) Biochem. J. 370, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marks, P. A., Miller, T. & Richon, V. M. (2003) Curr. Opin. Pharmacol. 3, 344–351. [DOI] [PubMed] [Google Scholar]
- 29.Lagger, G., O'Carroll, D., Rembold, M., Khier, H., Tischler, J., Weitzer, G., Schuettengruber, B., Hauser, C., Brunmeir, R., Jenuwein, T. & Seiser, C. (2002) EMBO J. 21, 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cress, W. D. & Seto, E. (2000) J. Cell Physiol. 184, 1–16. [DOI] [PubMed] [Google Scholar]
- 31.Timmermann, S., Lehrmann, H., Polesskaya, A. & Harel-Bellan, A. (2001) Cell Mol. Life Sci. 58, 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, C., Fu, M, Mani, S., Wadler, S., Senderowicz, A. M. & Pestell, R. G. (2001) Front. Biosci. 6, D610–D629. [DOI] [PubMed] [Google Scholar]
- 33.Jones, P. A. & Baylin, S. B. (2002) Nat. Rev. Genet. 3, 415–428. [DOI] [PubMed] [Google Scholar]
- 34.Qiu, L., Kelso, M. J., Hansen, C., West, M. L., Fairlie, D. P. & Parsons, P. G. (1999) Br. J. Cancer 80, 1252–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly, W. K., O'Connor, O. A. & Marks, P. A. (2002) Exp. Opin. Invest. Drugs 11, 1695–1713. [DOI] [PubMed] [Google Scholar]
- 36.Kelly, W. K., Richon, V. M., O'Connor, O., Curley, T., MacGregor-Curtelli, B., Tong, W., Klang, M., Schwartz, L., Richardson, S., Rosa, E., et al. (2003) Clin. Cancer Res. 9, 3578–3588. [PubMed] [Google Scholar]
- 37.Siegel, D. S., Zhang, X., Feinman, R., Teitz, T., Zelenetz, A., Richon, V. M., Rifkind, R. A., Marks, P. A. & Michaeli, J. (1998) Proc. Natl. Acad. Sci. USA 95, 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler, L. M., Agus, D. B., Scher, H. I., Higgins, B., Rose, A., Cordon-Cardo, C., Thaler, H. T., Rifkind, R. A., Marks, P. A. & Richon, V. M. (2000) Cancer Res. 60, 5165–5170. [PubMed] [Google Scholar]
- 39.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed., pp. 18.60–18.75.
- 40.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1998) Current Protocols in Molecular Biology (Wiley, New York), pp. 12.1.1–12.1.6.
- 41.Gui, C. Y. & Dean, A. (2001) Mol. Cell. Biol. 21, 1155–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gui, C. Y. & Dean, A. (2003) Proc. Natl. Acad. Sci. USA 100, 7009–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munster, P. N., Troso-Sandoval, T., Rosen, N., Rifkind, R. A., Marks, P. A. & Richon, V. M. (2001) Cancer Res. 61, 8492–8497. [PubMed] [Google Scholar]
- 44.Fischle, W., Wang, Y. & Allis, C. D. (2003) Nature 425, 475–479. [DOI] [PubMed] [Google Scholar]
- 45.Seoane, J., Le, H. V. & Massague, J. (2002) Nature 419, 729–734. [DOI] [PubMed] [Google Scholar]
- 46.Johnstone, R. W. (2002) Nat. Rev. Drug Discov. 1, 287–299. [DOI] [PubMed] [Google Scholar]