Abstract
Ameloblastin (AMBN) was originally described as a tooth-specific extracellular matrix protein, but current data have shown that AMBN is present in many different tissues of mesenchymal origin. The identification of regulatory elements in the promoter region of the Ambn gene would assist in identifying potential mesenchymal-specific transcriptional factors. In this study we subcloned a 3,788-bp region upstream (and a 54-bp region downstream) of the mouse Ambn transcriptional start site into a LacZ reporter construct and called this construct 3788-Ambn-lacZ. In silico analysis of the 3,788-bp Ambn promoter region identified 50 potential cis-regulatory elements, 29 of which are known to be functional in cell populations of mesenchymal origin. The reporter construct was activated in transfected bone marrow cells, and the promoter activity was induced in cell cultures following addition of recombinant AMBN, interferon-γ, serotonin, or dexamethasone. We discuss the relative significance of the potential cis-acting gene-regulatory elements of Ambn in relation to bone morphogenesis. Knowledge of Ambn gene-regulatory elements will be of importance when developing strategies for bone repair and replacement in a clinical surgical setting.
Keywords: ameloblastin, bone, mesenchymal cells, promoter elements
Ameloblastin (AMBN) was first detected in secretory-stage ameloblasts (1, 2) and later in the Hertwig’s epithelial root sheath (HERS) cells and in differentiating odontoblasts (3, 4). AMBN expression has more recently been detected in trauma-induced reparative dentin (5), in bone tissue during early stages of bone formation (6), in cultured mesenchymal hard tissue cells, and in precursor cells from the blood and bone marrow (7). AMBN expression is temporally and spatially restricted during tooth development (8) and diminishes with tooth eruption (9). AMBN expression tends to phase out during the differentiation of bone cells and after completion of bone remodelling (6, 7).
The biological function of AMBN remains obscure, and it is now documented that AMBN expression is not restricted to enamel formation (5, 6, 10). In odontogenesis, Ambn is believed to be implicated in enamel biomineralization (11–13) and in interactions between the ameloblasts and the extracellular matrix (14–16). It has been suggested that AMBN could act as a signal molecule in epithelial–mesenchymal interactions (4, 17), and AMBN is presumed to have ‘growth factor’ activity during periodontal ligament formation and regeneration (18). The expression level of enamel proteins is several hundred times lower in odontoblasts than in secretory ameloblasts (16, 19), and the level of AMBN expression during bone formation is also much lower than during amelogenesis (6). This may argue for a possible role of AMBN as a signalling molecule involved in orchestrating cellular events, such as cell proliferation, differentiation, and mineralization, rather than as a structural protein of the extracellular matrix. This assumption is further supported by recent results showing that recombinant AMBN can induce proliferation in stem cells and osteoblasts, osteoclast differentiation, and the expression of genes involved in inflammation and repair in osteoblasts (20).
Weak Ambn promoter activity has been demonstrated in mesenchymal cells (gingival and pulp fibroblasts) transfected with a previously characterized promoter (1,616 bp upstream region of the transcriptional start site) (21). The authors found that both positive and negative cis-acting regions are involved in Ambn transcriptional regulation, and demonstrated that bone-related Runt transcription factor 2 (Runx2) interacts with osteoblast-specific element 2 (OSE2) in the region upstream of the Ambn promoter in ameloblast-like cells (22). However, they concluded that the Ambn promoter is inactive in mesenchymal cells.
Based on the observations that AMBN is expressed in mesenchymal tissues, we wanted to study the region further upstream of the reported Ambn promoter (21), seeking to identify potential bone-related transcriptional regulatory elements. Here we describe the cloning and activation of the 3,788-bp Ambn promoter in bone marrow-derived stromal cells, and describe potential transcriptional factor-binding sites involved in bone development, maintenance, and repair.
Material and methods
Generation of Ambn reporter gene construct
A mouse Ambn promoter/LacZ reporter construct was prepared, consisting of 3,788 bp of Ambn DNA upstream of the transcriptional start site. PCR was used to generate two overlapping DNA fragments (Fragment 1 and Fragment 2) containing the 5′-flanking region upstream of the Ambn promoter from mouse (BALB/c) genomic DNA (catalogue # 6650-1; Clontech Laboratories, Mountain View, CA, USA (Fig. 1). The primers used were as follows:
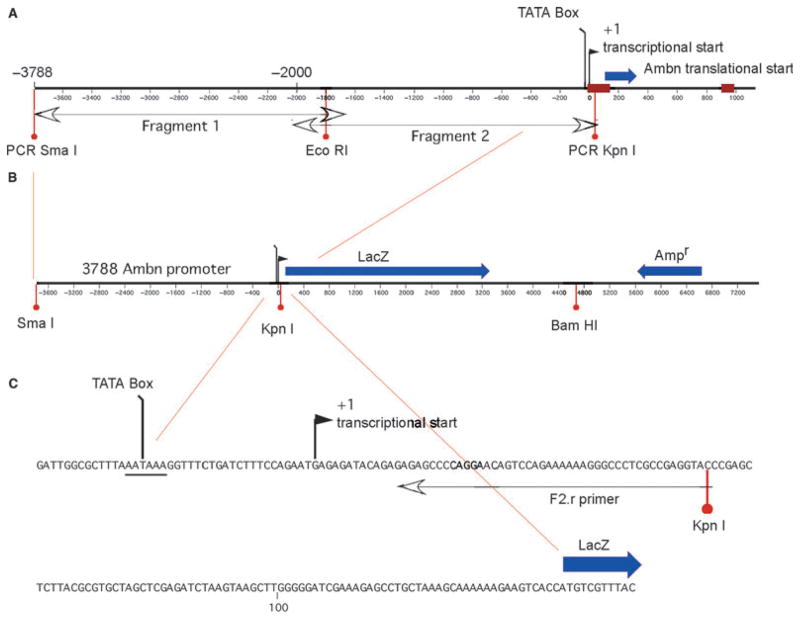
Fig. 1.
The mouse ameloblastin promoter construct (3788-Ambn-lacZ), showing the two overlapping DNA fragments 3,788 bp upstream of the transcriptional start site (A) driving the LacZ reporter gene (B). The sequence from the TATA box through the transcriptional start site and to the LacZ gene is highlighted (C).
Fragment 1, forward primer, 5′-ACCCGGGTGGGG CTCAAGGAACTCTACAGAAAG;
Fragment 1, reverse primer, 5′-GGAGTTGAAAGT AAGGGAGAATGTGAACC;
Fragment 2, forward primer, 5′-ACCCGGGCTAAGATTGAAAACTGGACAGCCACTACAG; and
Fragment 2, reverseprimer, 5′-GGGTACCTCGGCGAGGGCCCTTTTTTCTGGACTGTTCCTGGGGCTCTC.
Selected endonuclease restriction sites (CCCGGG-SmaI, GGTACC-KpnI, and GGGCCC-ApaI) were included in the primer (underlined regions) to permit uncomplicated sub-cloning steps. PCR reactions were performed using 2 ng μl−1 of mouse genomic DNA, 45 μl of Platinum PCR SuperMix High Fidelity, (Invitrogen, Carlsbad, CA, USA), 10 μM each primer, and 35 cycles each of 30 s of denaturation at 94°C, 1 min of annealing at 58°C, 2 min 30 s of elongation at 72°C and a final elongation step at 72°C for 7 min. The PCR reaction products were confirmed by separation on a 0.8% agarose gel. The resulting fragments of 2,135 (Fragment 1) and 2,099 bp (Fragment 2) were subcloned into a plasmid vector (pCR 2.1-TOPO; Invitrogen) and their entire nucleotide sequence was determined to confirm their identity. The two PCR-derived Ambn DNA products were ligated to one another across a shared restriction site and subcloned into the pβgal-Basic (Clontech) vector at the SmaI–KpnI positions of the multicloning site (Fig. 1). The pβgal-Basic vector contains the reporter gene LacZ and lacks promoter and enhancer sequences. The resulting DNA insert was resequenced in both directions to reconfirm its identity. The plasmid was amplified and purified using caesium chloride–ethidium bromide gradient centrifugation in a high-speed centrifuge at 190,000 × g. The band of DNA was recovered, washed, and its mass determined before being used for cell transfection (23).
Transcription factor analysis
Using in silico analysis, the 3,788-bp Ambn promoter sequence was screened for potential cis-regulatory binding sites by Web-based prediction of regulatory elements, using TRANSFAC (https://portal.biobase-international.com/cgi-bin/portal/login.cgi), latest version May 2009 (24) and the complementing database TRANSCompel (25). The TRANSFAC database on eukaryotic transcriptional regulation comprises data on transcription factors, their target genes, and their regulatory binding sites. By using MRS at EMBnet Norway and UniProtKB (http://www.uniprot.org/uniprot/), conceptual transcription factor proteinbinding sites were identified. Relevant potential transcription factors and their cis-acting DNA-regulatory elements that are related to their function during bone formation were identified using published literature (references listed in Table S1, A1–8).
Cell cultures
The M2-10B4 is a stromal cell line (ATCC #CRL-1972; American Type Culture Collection, Manassas, VA, USA) derived from the bone marrow of a (C57BL/6J × C3H/HeJ) F1 mouse (26). The cells were cultured in RPM1 1640 (PAA Laboratories, Pasching Austria) containing 10% fetal calf serum (FCS) (PAA Laboratories), 100 U ml−1 of penicillin, and 100 μg ml−1 of streptomycin (Sigma, St Louis, MO, USA). The cells were cultured in six-well plates and maintained in a humidified 95% air/5% CO2 atmosphere at 37°C.
Transfection with Nucleofector II system
For each well in a six-well plate, 2 × 106 cells were prepared for transfection according to the manufacturer’s instructions for adherent cell lines, and were resuspended in Nucleofector solution T (Amaxa, Lonza, Köln, Germany). As a control the transcription activity of the transfected construct was knocked down by cotransfection with a pool of three target-specific Ambn small interfering RNAs (siRNAs) (sc-44945; Santa Cruz Biotechnology, Santa Cruz, CA, USA) mapping to 4q13.3 against the Ambn mRNA sequence (NCBI gene ID 258) that is also partially contained in the 3788-Ambn-lacZ construct, upstream of the LacZ gene. DNA and cells were mixed in the Amaxa cuvette and placed directly in the nucleofector device, according to the manufacturer’s instructions. The cell suspensions were removed immediately from the cuvette by adding prewarmed medium, added to a six-well plate containing prewarmed medium, and incubated at 37°C, in a humidified atmosphere of 95% air/5% CO2. The transcription activity of the transfected construct was modulated by transfection of cells without DNA.
After 24 h, the cells were stimulated with four different reagents to study potential regulation of Ambn promoter activity: recombinant rat AMBN (rAMBN) (4,10) (10 μg ml−1); serotonin creatine sulphate complex (5-HT) (Sigma) (10 μM); dexamethasone (Dex) (Sigma) (10−8 M); and recombinant interferon-γ (rIFN-γ) (Sigma) (50 ng ml−1). Unstimulated cells were cultured in cell culture medium (RPMI 1640; PAA Laboratories) with or without BSA (Sigma) (100 μg ml−1).
mRNA isolation
Cells were disrupted in lysis/binding buffer [100 mM Tris–HCl, pH 8.0, 500 mM LiCl, 10 mM EDTA, pH 8.0, 0.5 mM dithiothreitol (DTT), and 1% SDS]. mRNA was isolated using magnetic beads – (oligo(dT)25 – as described by the manufacturer (Dynal, Oslo, Norway). Beads containing mRNA were resuspended in 10 mM Tris–HCl, pH 8.0, and stored at −70°C until use. One microlitre of the mRNA-containing solution was applied directly to obtain a first-strand cDNA using the iScript cDNA Synthesis kit that contains both oligo(dT) and random hexamer primers (Bio-Rad, Hercules, CA, USA).
Real-time RT-PCR
Real time RT-PCR reactions were performed and monitored using iCycler iQ (Bio-Rad). The 2X iQ SYBR Green Supermix was based on iTaq DNA polymerase (Bio-Rad). cDNA samples of M2-10B4 cells were analyzed for the mouse Ambn gene using the forward primer 5′-GCGTTTCCAAGAGCCCTGATAAC-3′ and the reverse primer 5′-AAGAAGCAGTGTCACATTTCCTGG-3′, resulting in a 366-bp product, and transfected M2-10B4 cells were analyzed for the LacZ gene using 5′-GCATTTTCCGTGACGTCTCGT-3′ as the forward primer and 5′-AACTCGCCGCACATCTGAACT-3′ as the reverse primer, resulting in a product of 130 bp. The amplification program consisted of a preincubation step for denaturation of the template cDNA (3 min, 95°C), followed by 50 and 40 cycles for Ambn and LacZ, respectively, consisting of a denaturation step (15 s, 95°C), an annealing step (30 s, 54°C and 60°C, Ambn and LacZ, respectively), and an extension step (30 s, 72°C). After each cycle, fluorescence was measured at 72°C. A negative control without cDNA template was run in each assay. To allow relative quantification after PCR, standard curves were constructed from the reactions for each target and for two housekeeping genes – glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and β-actin – by plotting cycle threshold (Ct) values, that is, the cycle number at which the fluorescence signal exceeds background vs. log cDNA dilution. The Ct readings for each of the unknown samples were used to calculate the amount of either the target or the housekeeping gene relative to the standard. The amount of mRNA for the target gene was normalized relative to the expression of the two housekeeping genes. The primers used for the housekeeping genes are as follows: Gapdh forward primer, 5′-ACCCAGAAGACTGTGGATGG-3′ and reverse primer 5′-CACATTGGGGGTAGGAACAC-3′; β-actin forward primer, 5′-GCTTCTTTGCAGCTCCTTCGT-3′ and reverse primer 5′-ATATCGTCATCCATGGCGAAC-3′, resulting in products of 171 and 64 bp, respectively. PCR products were subjected to a melting curve analysis on the iCycler and subsequently to 2% agarose/Tris-acetate-EDTA (TAE) gel electrophoresis to confirm amplification specificity, melting temperature (Tm), and amplicon size.
Statistics
Statistical comparisons between the treatments were performed using the non-parametric Mann–Whitney rank sum test and analysis of variance (ANOVA) on ranks; however, when the data were considered to be normally distributed after testing for normality and equal variance, Student’s t-tests were used. Differences were considered statistically significant at P < 0.05.
Results
Characterization of the 3,788-bp Ambn promoter
A combination of activators and repressors has been previously suggested to influence Ambn promoter activity (Table S1, A9). We identified 50 different potential positive and negative regulatory elements in the 3,788 Ambn promoter region using TRANSFAC, TRANSCompel, and published sequence motifs (25). The potential regulatory elements identified are active in almost all tissue types and serve to modulate the gene-expression levels in relation to embryonic developmental stage (Table S1, A10–26), and osteogenesis (Table S1, A16, A18, B1–12 and D1–12).
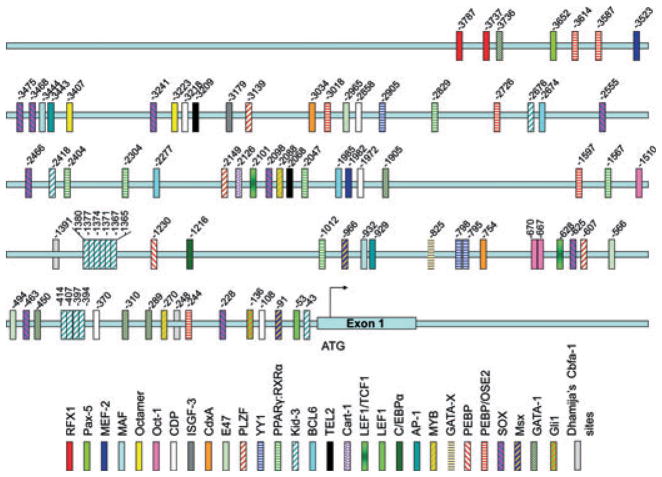
At least 29 of the regulatory elements identified are described as being related to osteogenesis (Fig. 2), and include genes operating during cell cycle control and differentiation of bone-related cells (Table S1, C1–11). Table S2 lists the identified nucleotide sequences, the binding-site name (gene name) and potential transcription factors, position, and strand where the sites were located, and the score for the sequence match to the databases of TRANSFAC and TRANSCompel.
Fig. 2.
Twenty nine regulatory elements related to bone formation were identified and localized in the 3788-Ambn-lacZ construct. Abbreviations: RFX1, MHC class II regulatory factor RFX1; Pax-5, paired box protein; MEF-2, myocyte-specific enhancer factor 2; MAF, V-maf musculoaponeuretic fibrosarcoma oncogene homolog (avian); Octamer, NONO, non POU-domain-containing octamer binding protein; Oct-1, Pou2f1 POU-domain class 2; CDP, CCAT displacement protein (= Cux1); ISGF-3, signal transducer and activator of transcription 1-alpha/beta (transcription factor ISGF-3 components p91/p84) = STAT-1; CdxA, Cdx1/CDX1 (a caudal-type homeobox intestine-specific transcription factor); E47, E12/E47 basic helix–loop–helix transcription factor (the E2A gene product); PLZF, promylocytic leukemia zinc finger; YY1, Yin and Yang 1 (synonym NMP1); PPARγ:RXRα, peroxisome proliferator-activated receptor gamma; retinoic acid receptor alpha; Kid-3, kidney, ischemia, and developmentally regulated protein 3 (= Zinc finger protein 354c and AJ18); BCL6, B-cell lymphoma 6 protein homolog; TEL2, telomere length regulation protein TEL2 homolog; Cart-1, cartilage homeoprotein 1 (= ALX 1); LEF1/TCF1, lymphoid enhancer-binding factor 1/T-cell-specific transcription factor 1; LEF1, lymphoid enhancer-binding factor 1; C/EBPα, CCAAT enhancer binding protein alpha/gamma homolog; AP-1, transcription factor AP-1 (also known as activator protein 1 and proto-oncogene c-jun, gene name: Jun); MYB, V-maf musculoaponeurotic fibrosarcoma oncogene homolog; GATA-X, GATA binding factor, erythroid transcription factor; PEBP, polyoma enhancer binding protein 2; PEBP/OSE2, Cbfa1/Runx2 binding motif; SOX, transcription factor Sox (SRY-related HMG box); Msx, homeobox protein MSX (Msh homeobox-like protein) (Hox-7) (Hox-7.1); GATA-1, GATA binding factor 1, erythroid transcription factor; Gli1, glioma-associated oncogene homolog.
In addition, we manually identified six potential OSE2 sites where Runx2 is known to bind (Table S1, A2) (Listed as ‘manual’ in Table S2). Four of these elements are located upstream of the 1,616-bp Ambn promoter region previously described. Runx2 is reported to bind the sequences CACCAA, AACCAC, CTCCAA, ACCCAC, AACCTC, and CACCAT (Table S1, A1–2), and the positions of these OSE2 elements present in the 3,788-bp Ambn promoter region are noted.
Nine DNA-binding sites for SRY-related HMG boxes (Sox) proteins with the consensus DNA-binding sequence (A/T)AACAA(T/A) (Table S1, A3), described as the core binding element for all Sox proteins (Table S1, A3 and E1), were identified. However, various enhancers and cofactors have to be present and correctly assembled to activate gene expression of Sox proteins (Table S1, A3 and E2). Two Msx consensus DNA-binding motifs were detected with the core motif ATAATTG(G/C) (Table S1, A4), and four sites for GATA-binding factor 1 (GATA-1), with the consensus deoxynucleotide sequence AGATAA, were also identified (Table S1, A5). One member of the family of transcription factors glioma-associated oncogene homolog (Gli-1), with the DNA-binding site GAGCCTGCA, was identified (Table S1, A6–7).
Kid-3 motifs, also known as AJ18 (Table S1, A18, D7, D11), were repeated 13 times; two of the repeats were located upstream of the 1,616-bp Ambn promoter region. Kid-3/AJ18 is related to early bone development (Table S1, A18, C9).
Furthermore, peroxisome proliferator-activated receptor gamma; retinoic acid receptor alpha (PPARγ:RXRα) regulatory elements were identified and repeated six times; four repeats were located upstream of the 1,616-bp Ambn promoter region. PPARs are mostly known as being involved in lipid metabolism and fat storage (Table S1, F1–2); however, PPARγ has recently been reported to be involved in bone metabolism (Table S1, B10, C8, F3).
The rest of the identified regulatory elements are repeated two or three times, except for osteocalcin gene (OG-2) and CCAT displacement protein (CDP), which are repeated five times. All of the regulatory elements were more or less evenly distributed throughout the 3,788-bp region, which suggests that the upstream region of the 1,616-bp Ambn promoter region is not specifically related to bone metabolism.
Transcriptional activation of the mouse 3,788-bp Ambn promoter
The potential Ambn promoter activity was tested in a mesenchymal cell line (M2-10B4), and was evaluated after stimulation with rAMBN, IFN-γ, DEX, or 5-HT. These reagents were previously demonstrated to have an effect on mesenchymal stem cells.
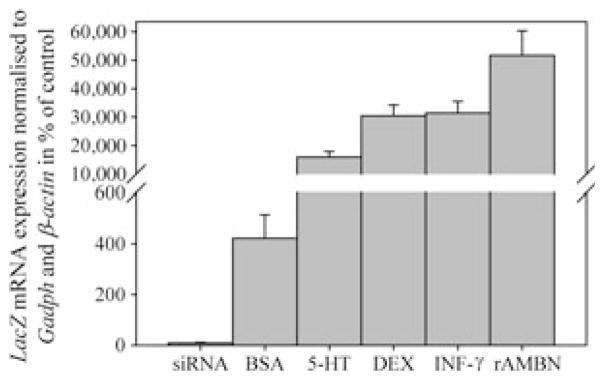
Cells transfected with the 3,788-bp Ambn–LacZ construct expressed LacZ mRNA (Fig. 3), verified by gel electrophoresis (Fig. 4). The strongest expression of LacZ mRNA was found in transfected cells stimulated with rAMBN (51,700 ± 8,600; (P = 0.029) compared with the unstimulated control (Fig. 3). Transfected cells incubated with 5-HT, Dex, and IFN-γ also had enhanced expression of LacZ mRNA (15,900 ± 2,000, 30,400 ± 3,800, and 31,500 ± 4,000, respectively) (P = 0.029 for all) compared with the unstimulated control (100%) (Fig. 3). The transfected cells stimulated with BSA showed a modestly enhanced expression of LacZ mRNA(400 ± 92) (P = 0.029) (Fig. 3). The cotransfection with ameloblastin siRNA reduced, by ninefold (P = 0.029), the LacZ mRNA expression level (Fig. 3). The size of the products corresponded with the predicted PCR products (LacZ, 130 bp; and GAPDH, 171 bp), and cells without the Ambn–lacZ construct did not express LacZ mRNA, as confirmed by gel electrophoresis (Fig. 4). These results suggest that the 3,788-bp Ambn promoter construct is active in mesenchymal cells, and is up-regulated by rAMBN, IFN-γ, DEX, and 5-HT.
Fig. 3.
Effect of ameloblastin small inhibitory RNA (siRNA), BSA (100 μg ml−1), serotonin (5-HT), dexamethasone (DEX), recombinant interferon-γ (rIFN-γ) and recombinant ameloblastin (rAMBN) on the expression of LacZ mRNA in transfected mouse bone marrow stromal cells (M2-10B4). The results are calculated relative to the expression of glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA and β-actin and are presented as a percentage of the expression of LacZ in unstimulated transfected cells.
Fig. 4.
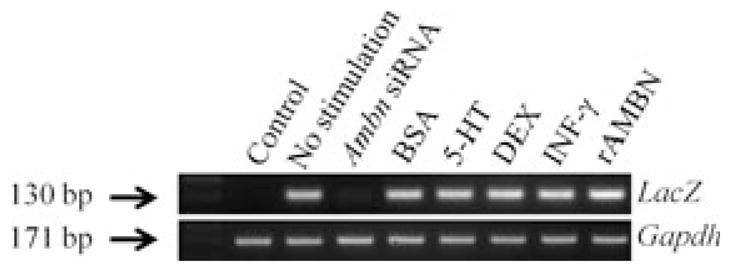
Gel electrophoresis confirming the LacZ and glyceraldehyde- 3-phosphate dehydrogenase (Gapdh) PCR products of 130 and 171 bp, respectively, in the stromal bone marrow stem cells (M2-10B4). 5-HT, serotonin; Ambn siRNA, ameloblastin small inhibitory RNA; DEX, dexamethasone; rAMBN, recombinant ameloblastin; rIFN-γ, recombinant interferon-γ.
Discussion
Several studies have reported the expression of AMBN in mesenchymal cells and tissues. For example, AMBN has been reported in pulpal cells during early odontogenesis, in differentiating odontoblasts (4), and during trauma-induced reparative dentin formation (5). AMBN expression has similarly been reported during intramembranous and endochondral ossification, within the extracellular matrix of the cartilage, in the perichondrium (6), and in various cultured mesenchymal hard tissue cells, as well as in precursor cells from the blood and bone marrow (7).
It is based on these data that we wanted to confirm AMBN expression in mesenchymal cells and characterize the promoter region, choosing to do so using DNA further upstream than the previously characterized 1,616-bp Ambn promoter (21).
Using in silico analysis we identified more than 50 different potential regulatory elements (both positive and negative) in the 3,788-bp Ambn promoter region. The identified regulatory elements are known to be active in almost all types of tissues and are related to embryonic development, mesenchymal differentiation, haematopoietic differentiation, and cell cycle control (Table S1). Of these 50 elements, 29 are involved in bone formation. The regulatory elements related to bone metabolism are evenly distributed throughout the 3,788-bp Ambn promoter region, and are not specifically located upstream of the 1,616-bp promoter region, as we hypothesized.
We identified a Kid-3 regulatory element, for transcription of Kid-3/AJ18 protein, which was repeated 13 times. The Kid-3/AJ18 is a zinc finger protein expressed temporally and spatially in developing mouse tissues, and is reported to have the highest expression in brain, kidney, and bone of mouse embryos (27). Kid-3/AJ18 has been shown to be strongly expressed in chondrocytes and osteoblasts, and also in teeth of embryonic and 4-wk-old mice, in precursor and mature ameloblasts, odontoblasts, and cementoblasts, respectively, in addition to alveolar bone (27). The regulatory Kid-3/AJ18 element has previously been shown to have partial sequence identity with the OSE2-binding site and it is found to interact with Runx2 (28, 29).
Osteoblast-specific element 2 was previously identified in the Ambn promoter (21). In our analysis we located six OSE2 sequences, two of which were identified upstream of the start site (at positions −244 and −1597), which is probably the same as reported previously (21). Runx2 is involved in bone and tooth development and is essential for maturation of osteoblasts (30) in both intramembranous and endochondral ossification (31). Runx2 determines the linage of osteoblastic cells from multipotent mesenchymal cells, enhances osteoblast differentiation at an early stage, and inhibits osteoblast differentiation at a late stage (32). However, different roles of Runx2 are indicated in bone and tooth development (33).
Regulatory elements related to bone metabolism also include the binding site for the Gli family of transcription factors reported to play critical regulatory roles in mediating Sonic hedgehog (Shh) signalling for embryonic development and the bone morphogenetic protein 2 (BMP2) response to hedgehog signalling (Table S1, D1–2). Binding to zinc-finger transcription factor GATA-1 is identified in osteoblast differentiation (Table S1, B8, C2), and the high-mobility-group protein lymphoid enhancer-binding factor 1 (LEF1) and the T-cell-specific transcription factor 1 (TCF1) are reported to form heterodimers with β-catenin for expression of Wnt-responsive genes (Table S1, B5). Activation of canonical Wnt signalling pathway components is known to stimulate osteoblast differentiation from mesenchymal stem cells, enhances proliferation and expansion of lineage-committed preosteoblasts, and stimulates mature osteoblasts to secrete the bone resorption inhibitor, osteoprotegerin (Table S1, D4).
Several of the observed regulatory elements are related to an early regulation of bone and tooth development. Examples of transcription factors identified to regulate early osteogenesis are Kid-3, Runx2, TCF/LEF, Sox, myocyte enhancer factor 2 (Mef2), and Gli1, as well as the promyelocytic leukaemia zinc finger protein 145 (PLZF), all of which are listed in Table S1, D5–6, A15, B3, B6, D2, D7–12. A regulatory element known to bind the transcription factor osterix (Osx), which has a function downstream of Runx2, is not detected, and may indicate that the promoter region contains binding sites for early regulators of osteoblast differentiation.
Furthermore, we found that the 3,788-bp Ambn promoter region was activated in transfected bone marrow cells, and that expression was further enhanced in cells stimulated with rAMBN, 5-HT, DEX, and IFN-Y. Our group has previously shown that rAMBN (20) and 5-HT (34) enhance proliferation and differentiation of mesenchymal stem cells. The rIFN-Y-enhanced promoter activity in the M2-10B4 cell line may be linked to our previous findings, where rAMBN strongly induced the IFN-Y pathway in osteoblasts (20) and DEX is a known factor in osteogenic differentiation (35), and strengthens the suggestion that AMBN may act as a signalling molecule in developing hard tissues (5, 10, 14, 18, 20, 36).
Our results indicate that the Ambn promoter has multiple cis-regulatory elements upstream of the previously characterized Ambn promoter region (21), many of which could be related directly to bone formation. Thirteen Kid-3/AJ18 and six OSE-2 regulatory elements distributed in the 3,788-bp promoter region suggest that the Ambn promoter has the potential to regulate both teeth and bone metabolism. Further work is necessary to confirm if the identified sequences are functional in a DNA–protein interaction study.
Supplementary Material
Acknowledgments
We want to thank Hong Jun Wang at the Centre for Craniofacial Molecular Biology at the University of Southern California, Los Angeles, USA, for his kind help, George Magklares at http://www.no.embnet.org for his help with TRANSFAC information, and MedProbe, Oslo, Norway for use of the Nucleofector II equipment.
Footnotes
Conflicts of interest – The authors declare no conflicts of interest.
Additional Supporting information may be found in the online version of this article:
Table S1. References related to regulatory elements and/or transcription factors.
Table S2. Localization of potential cis regulatory sequences according to transcription start site.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Lee SK, Krebsbach PH, Matsuki Y, Nanci A, Yamada KM, Yamada Y. Ameloblastin expression in rat incisors and human tooth germs. Int J Dev Biol. 1996;40:1141–1150. [PubMed] [Google Scholar]
- 2.Krebsbach PH, Lee SK, Matsuki Y, Kozak CA, Yamada KM, Yamada Y. Full-length sequence, localization, and chromosomal mapping of ameloblastin. A novel tooth-specific gene. J Biol Chem. 1996;271:4431–4435. doi: 10.1074/jbc.271.8.4431. [DOI] [PubMed] [Google Scholar]
- 3.Fong CD, Slaby I, Hammarstrom L. Amelin: an enamel-related protein, transcribed in the cells of epithelial root sheath. J Bone Miner Res. 1996;11:892–898. doi: 10.1002/jbmr.5650110704. [DOI] [PubMed] [Google Scholar]
- 4.Fong CD, Cerny R, Hammarstrom L, Slaby I. Sequential expression of an amelin gene in mesenchymal and epithelial cells during odontogenesis in rats. Eur J Oral Sci. 1998;106 (Suppl 1):324–330. doi: 10.1111/j.1600-0722.1998.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 5.Spahr A, Lyngstadaas SP, Slaby I, Haller B, Boeckh C, Tsoulfidou F, Hammarstrom L. Expression of amelin and trauma-induced dentin formation. Clin Oral Investig. 2002;6:51–57. doi: 10.1007/s00784-001-0139-y. [DOI] [PubMed] [Google Scholar]
- 6.Spahr A, Lyngstadaas SP, Slaby I, Pezeshki G. Ameloblastin expression during craniofacial bone formation in rats. Eur J Oral Sci. 2006;114:504–511. doi: 10.1111/j.1600-0722.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 7.Tamburstuen MV, Reseland JE, Spahr A, Brookes SJ, Kvalheim G, Slaby I, Snead ML, Lyngstadaas SP. Ameloblastin expression and putative autoregulation in mesenchymal cells suggest a role in early bone formation and repair. Bone. 2011;48:406–413. doi: 10.1016/j.bone.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SK, Kim SM, Lee YJ, Yamada KM, Yamada Y, Chi JG. The structure of the rat ameloblastin gene and its expression in amelogenesis. Mol Cells. 2003;15:216–225. [PubMed] [Google Scholar]
- 9.Torres-Quintana MA, Gaete M, Hernandez M, Farias M, Lobos N. Ameloblastin and amelogenin expression in posnatal developing mouse molars. J Oral Sci. 2005;47:27–34. doi: 10.2334/josnusd.47.27. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura Y, Slaby I, Spahr A, Pezeshki G, Matsumoto K, Lyngstadaas SP. Ameloblastin fusion protein enhances pulpal healing and dentin formation in porcine teeth. Calcif Tissue Int. 2006;78:278–284. doi: 10.1007/s00223-005-0144-2. [DOI] [PubMed] [Google Scholar]
- 11.Uchida T, Murakami C, Dohi N, Wakida K, Satoda T, Takahashi O. Synthesis, secretion, degradation, and fate of ameloblastin during the matrix formation stage of the rat incisor as shown by immunocytochemistry and immunochemistry using region-specific antibodies. J Histochem Cytochem. 1997;45:1329–1340. doi: 10.1177/002215549704501002. [DOI] [PubMed] [Google Scholar]
- 12.Nanci A, Zalzal S, Lavoie P, Kunikata M, Chen W, Krebsbach PH, Yamada Y, Hammarstrom L, Simmer JP, Fincham AG, Snead ML, Smith CE. Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J Histochem Cytochem. 1998;46:911–934. doi: 10.1177/002215549804600806. [DOI] [PubMed] [Google Scholar]
- 13.Brookes SJ, Kirkham J, Shore RC, Wood SR, Slaby I, Robinson C. Amelin extracellular processing and aggregation during rat incisor amelogenesis. Arch Oral Biol. 2001;46:201–208. doi: 10.1016/s0003-9969(00)00121-7. [DOI] [PubMed] [Google Scholar]
- 14.Cerny R, Slaby I, Hammarstrom L, Wurtz T. A novel gene expressed in rat ameloblasts codes for proteins with cell binding domains. J Bone Miner Res. 1996;11:883–891. doi: 10.1002/jbmr.5650110703. [DOI] [PubMed] [Google Scholar]
- 15.Paine ML, Wang HJ, Luo W, Krebsbach PH, Snead ML. A transgenic animal model resembling amelogenesis imperfecta related to ameloblastin overexpression. J Biol Chem. 2003;278:19447–19452. doi: 10.1074/jbc.M300445200. [DOI] [PubMed] [Google Scholar]
- 16.Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Begue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur J Oral Sci. 1998;106:963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- 18.Zeichner-David M, Chen LS, Hsu Z, Reyna J, Caton J, Bringas P. Amelogenin and ameloblastin show growth-factor like activity in periodontal ligament cells. Eur J Oral Sci. 2006;114 (Suppl 1):244–253. doi: 10.1111/j.1600-0722.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 19.Nagano T, Oida S, Ando H, Gomi K, Arai T, Fukae M. Relative levels of mRNA encoding enamel proteins in enamel organ epithelia and odontoblasts. J Dent Res. 2003;82:982–986. doi: 10.1177/154405910308201209. [DOI] [PubMed] [Google Scholar]
- 20.Tamburstuen MV, Reppe S, Spahr A, Sabetrasekh R, Kvalheim G, Slaby I, Syversen U, Lyngstadaas SP, Reseland JE. Ameloblastin promotes bone growth by enhancing proliferation of progenitor cells and by stimulating immunoregulators. Eur J Oral Sci. 2010;118:451–459. doi: 10.1111/j.1600-0722.2010.00760.x. [DOI] [PubMed] [Google Scholar]
- 21.Dhamija S, Liu Y, Yamada Y, Snead ML, Krebsbach PH. Cloning and characterization of the murine ameloblastin promoter. J Biol Chem. 1999;274:20738–20743. doi: 10.1074/jbc.274.29.20738. [DOI] [PubMed] [Google Scholar]
- 22.Dhamija S, Krebsbach PH. Role of Cbfa1 in ameloblastin gene transcription. J Biol Chem. 2001;276:35159–35164. doi: 10.1074/jbc.M010719200. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russel DW, editors. Molecular cloning, a laboratory manual. Cold Spring Harbour Laboratory Press; NY, USA: 2001. p. 16. [Google Scholar]
- 24.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemoine FM, Humphries RK, Abraham SD, Krystal G, Eaves CJ. Partial characterization of a novel stromal cell-derived pre-B-cell growth factor active on normal and immortalized pre-B cells. Exp Hematol. 1988;16:718–726. [PubMed] [Google Scholar]
- 27.Jheon A, Chen J, Teo W, Ganss B, Sodek J, Cheifetz S. Temporal and spatial expression of a novel zinc finger transcription factor, AJ18, in developing murine skeletal tissues. J Histochem Cytochem. 2002;50:973–982. doi: 10.1177/002215540205000711. [DOI] [PubMed] [Google Scholar]
- 28.Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, Bae SC. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20:8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jheon AH, Ganss B, Cheifetz S, Sodek J. Characterization of a novel KRAB/C2H2 zinc finger transcription factor involved in bone development. J Biol Chem. 2001;276:18282–18289. doi: 10.1074/jbc.M010885200. [DOI] [PubMed] [Google Scholar]
- 30.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 31.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 32.Komori T. Requisite roles of Runx2 and Cbfb in skeletal development. J Bone Miner Metab. 2003;21:193–197. doi: 10.1007/s00774-002-0408-0. [DOI] [PubMed] [Google Scholar]
- 33.James MJ, Jarvinen E, Wang XP, Thesleff I. Different roles of Runx2 during early neural crest-derived bone and tooth development. J Bone Miner Res. 2006;21:1034–1044. doi: 10.1359/jbmr.060413. [DOI] [PubMed] [Google Scholar]
- 34.Gustafsson BI, Thommesen L, Stunes AK, Tommeras K, Westbroek I, Waldum HL, Slordahl K, Tamburstuen MV, Reseland JE, Syversen U. Serotonin and fluoxetine modulate bone cell function in vitro. J Cell Biochem. 2006;98:139–151. doi: 10.1002/jcb.20734. [DOI] [PubMed] [Google Scholar]
- 35.Mori K, Shioi A, Jono S, Nishizawa Y, Morii H. Dexamethasone enhances In vitro vascular calcification by promoting osteoblastic differentiation of vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 1999;19:2112–2118. doi: 10.1161/01.atv.19.9.2112. [DOI] [PubMed] [Google Scholar]
- 36.Veis A, Tompkins K, Alvares K, Wei K, Wang L, Wang XS, Wang L, Wang XS, Brownell AG, Jengh SM, Healy KE. Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. J Biol Chem. 2000;275:41263–41272. doi: 10.1074/jbc.M002308200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.