Abstract
Mobile DNAs have had a central role in shaping our genome. More than half of our DNA is comprised of interspersed repeats resulting from replicative copy and paste events of retrotransposons. Although most are fixed, incapable of templating new copies, there are important exceptions to retrotransposon quiescence. De novo insertions cause genetic diseases and cancers, though reliably detecting these occurrences has been difficult. New technologies aimed at uncovering polymorphic insertions reveal that mobile DNAs provide a substantial and dynamic source of structural variation. Key questions going forward include the how and how much new transposition events affect human health and disease.
Human Retroelements
High copy number repeats reflecting mobile DNA integrations comprise a large fraction of genomes in a wide variety of organisms. The proportions of mobile DNAs in genomes is highly variable among species, and each eukaryote has a specific complement of recently active transposable elements (TEs). Transposons are thus key genetic features distinguishing related species.
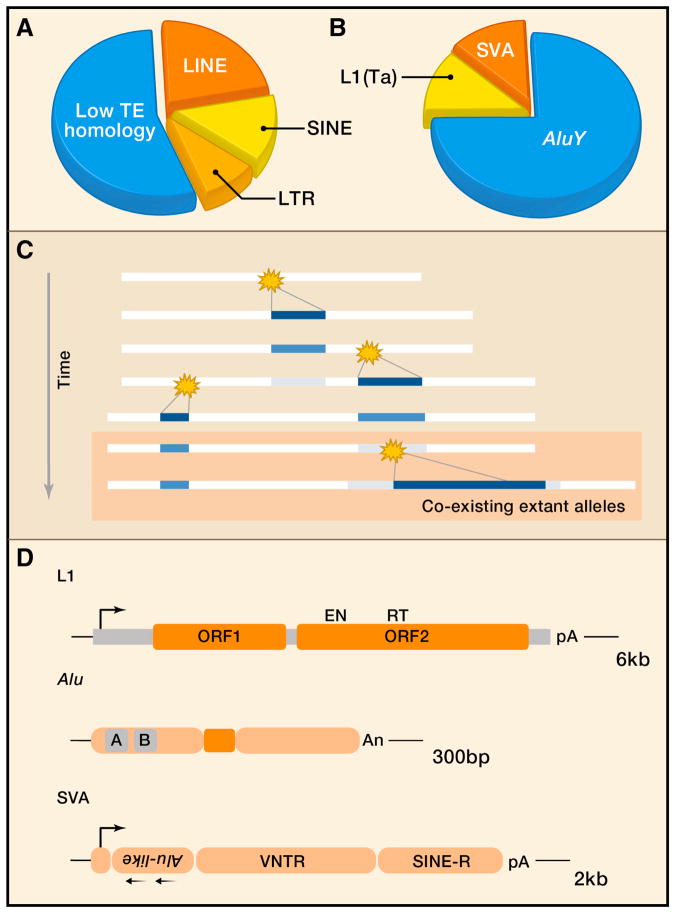
Humans are no exception. Like most other mammals, the landscape of our genome reflects a long history of activity of retrotransposons known as Class I transposable elements. These elements replicate by a copy and paste mechanism, producing mRNA-like intermediates which are reverse transcribed by an element-encoded enzyme. In contrast, Class II DNA transposons employ a cut and paste mechanism, directly moving DNA segments from one location to another. Although DNA transposons are not active in humans, a co-opted DNA cut and paste system is involved in recombination events that generate lymphocyte antigen binding diversity (Agrawal et al., 1998). The retrotransposons that have recently made significant contributions to the human genome include long and short interspersed repeats (termed LINEs and SINEs, respectively) and long terminal repeat elements (LTR elements). In the current genome assembly, about 45% of our total DNA is recognizable as having homology to consensus sequences of retroelements (Figure 1A) (Jurka et al., 2005; Lander et al., 2001; Smit et al., 1996–2010). The true contribution of retroelements to the human genome is likely to be considerably larger. A new computational approach reliant on de novo recognition of high abundance oligonucleotides recognizes many smaller fragments of elements accrued over hundreds of millions of years of vertebrate evolution and estimates that repeats comprise nearly two thirds of our total genome (de Koning et al., 2011).
Figure 1.
Human retrotransposons. A. Composition of the human genome with respect to high copy number repeats. Data are from the RepeatMasker analysis of the hg19 human genome assembly (Genome Reference Consortium GRCh37). The illustration shows fractions of the genome derived from the major orders (Wicker et al., 2007) of retrotransposons. The remaining 55% of the genome bears low homology to TEs, although substantial portions may be derived from mobile DNA. B. Transposon types illustrated as fractions of total ongoing activity. AluY are the most prolific source of new insertions at one de novo germ line insertion per 20 births; L1 and SVA are thought to be comparable in current activity, responsible for one insertion per 100–200 births. C. Schematic showing an accumulation of interspersed repeat insertions over time. New integrations are stochastic events in individuals (star), such that co-existence with the antedating ‘empty’ allele occurs in the population initially. In the schematic, two alleles are present currently, reflecting presence and absence of the most recent insertion [a retrotransposon insertion polymorphism (RIP)]. As persisting insertions age, they become ‘fixed’ or invariant in the population and decrease in sequence likeness to similar elements (black to gray). D. Structure of the most active retroelements in human genomes. L1 LINEs have a CpG rich 5′UTR with an internal RNA polII promoter (5′ rightward arrow), two open reading frames encoding ORF1p and ORF2p (pink segments), and a 3′ UTR with a polyadenylation (pA) sequence. The ORF2 reading frame encodes endonuclease (EN) and reverse transcriptase (RT) domains. Alu elements are derived from 7SL ribosomal RNAs; they have two internal ‘monomer’ sequences with a centrally located A-rich sequence and an RNApolIII promoter (A and B, gray). The sequence ends in multiple adenosines (An). SVAs are composites of other repeats, from left to right, a CCCTCTn repeat, two tandem Alu-like sequences in antisense (leftward arrows), a VNTR (variable number tandem repeat) region, and a SINE-R region with HERV homology. Sequence suggests RNApolII driven transcription (5′ rightward arrow), and there is a 3′ AAUAAA sequence (pA).
A relatively recent or ‘young’ transposon insertion sequence bears high homology to currently active template elements; older insertions accrue changes resulting in divergence of their sequences from the family consensus (Figure 1C). Although rates vary, in humans, sequence divergence of interspersed repeats of about 12–18% has occurred over the last 100 million years (Lander et al., 2001). L1 LINEs and Alu SINEs date to about 150 and 80 million years, respectively, and were preceded by expansions of L2 LINEs and MIR SINEs. In contrast, currently active retrotransposons include a subset of L1 with about 0.8% divergence from the consensus and elements it mobilizes. When a L1 LINE, SINE, or SVA retrotransposon insertion occurs in or is passed to the germ line, the locus can be inherited with it present or absent; these are colloquially referred to as the “filled” versus “empty” alleles. The “empty” allele antedates the insertion event; it is the ancestral allele. If autosomal, an insertion may be homozygous or heterozygous in an individual. Such polymorphic insertions are categorized as a subtype of “indel” structural variants, though no deletion event is relevant for these non-LTR retrotransposons. We consider these as biallelic polymorphisms herein, disregarding subsequent nucleotide changes within the inserted sequence for simplicity.
Most of the repetitive landscape of our genome reflects integration events that became homozygous in ancestral species. Species-specific insertions are responsible for a minor though notable portion of the difference between our genome and that of the common chimpanzee, Pan troglodytes(Hedges et al., 2004, 2005 #792, Mills, 2006 #815). In total, transposable element insertions are responsible for about 16 Mb of sequence difference in a pairwise comparison of the genomes; that amounts to 0.5% of either genome. For context, approximately 35 million nucleotide substitutions (1.2% of either genome) distinguish the species. Since divergence, humans and chimps have accrued similar numbers of species-specific L1 and SVA inserts whereas humans accumulated nearly three times as many Alu elements. The human genome contains approximately 2000 species-specific LINEs [L1], 8000 species-specific inserts of dependent elements [7000 Alu and 1000 SVA], and 73 LTRs [mostly HERV-K solo LTRs]. For each type, a limited repertoire of recently active transposon subfamilies is responsible for the expansion in humans. Subfamilies are defined by internal transposon sequence, as described further below. For example, AluYa5 and AluYb8 subfamilies predominate among human specific SINEs; L1HS are chiefly responsible for LINEs unique to humans.
There are three highly active human retrotransposon familes today (Figure 1B,1D). All require a combination of host factors and proteins encoded by L1 LINEs to continuously reenter our genomes (see Ostertag and Kazazian, 2001a for a review of human L1), and thus L1 transposons are termed the autonomous elements. A full-length L1 is 6 kilobases long. Following transcription by RNA polymerase II, translation of the two open reading frames occurs, producing ORF1p and ORF2p. Association of the L1 transcript with these proteins occurs in ribonucleic acid particles (RNPs) (Hohjoh and Singer, 1996), and cis preference is seen for these interactions (Kulpa and Moran, 2006; Wei et al., 2001). ORF1p is required for L1 transposition and functions as a chaperone protein or single-strand RNA binding protein (reviewed in (Martin, 2010)). ORF2p has two recognized enzymatic domains, an endonuclease (Feng et al., 1996) and reverse transcriptase (Mathias et al., 1991). L1 retrotransposition is diagrammed in Figure 2. The ORF2p-encoded endonuclease mediates a DNA nick in the host genome, and the resulting 3′-OH is extended by the reverse transcriptase to make a nuclear DNA copy of the L1 RNA template; this is termed target primed reverse transcription (TPRT). The precise steps for resolution of this structure are not known, although the breakpoint of the second DNA strand is staggered such that a small sequence of the target site is duplicated flanking the insertion. The second strand appears capable of annealing internally in the L1 RNA and initiating a second RT reaction, a process termed twin priming. This can result in inversions and truncations of 5′ L1 sequence, and may be responsible for the preponderance of L1 insertions that have lost 5′ sequence(Ostertag and Kazazian, 2001b). In vitro assays using a recombinant transposon have been key to identifying critical sequences required for transposition (reviewed in (Rangwala and Kazazian, 2009)). These place a marker cassette in antisense orientation with respect to the transposon; it is interrupted by an intron spliced in sense, such that cells with an integration event acquire a functional marker. Similar systems can be used for Alu insertions (Dewannieux et al., 2003) or introduced in transgenic mice to provide a system for studying host control (An et al., 2006; Heidmann and Heidmann, 1991; Ostertag et al., 2002).
Figure 2.
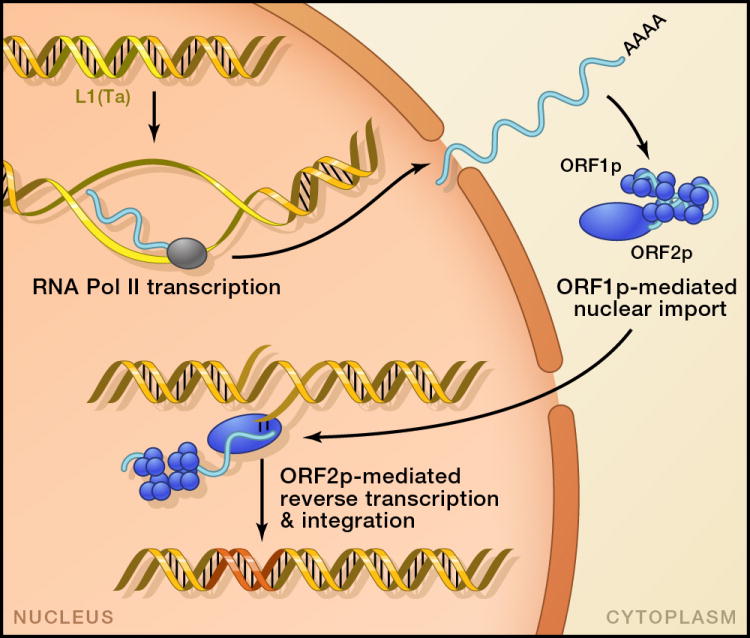
L1 LINE propagation. L1 retrotranspositition requires RNA and two proteins encoded by L1; they assemble into ribonucleic acid particles during translation. Reverse transcription of the RNA is coupled to insertion as it initiates from the 3′-OH of the broken strand (target primed reverse transcription, TPRT). Resolution of the structure results in target site duplication (TSD).
In primates, Alu elements co-opt L1 machinery (Dewannieux et al., 2003), and their de novo insertion rate has rivaled or exceeded that of L1 ((Cordaux et al., 2006; Shen et al., 1991); see (Batzer and Deininger, 2002) for a review of Alu). Alu SINEs are named for an internal AluI restriction enzyme recognition site (Houck et al., 1979). Like SINEs in other species, Alu elements are derived from preexisting cellular RNAs, originating from the 7SL ribosome complex (Ullu and Weiner, 1984). Alu sequences are transcribed by RNA polymerase III, which recognizes an internal 5′ promoter as well as external sequences and reads through beyond the element 3′ polyA end (Ullu and Weiner, 1985). Alu elements are about 300bp in length and have two distinct internal ‘monomer’ sequences with a centrally located A-rich sequence [A5TACA6](Figure 1D). They interact with L1 encoded ORF2p during insertion so as to leave a target site duplication, and L1 ORF1p is dispensable for Alu mobilization (Dewannieux et al., 2003).
A third family of active retrotransposons in humans is termed SVA, for their multipartite structure reflecting SINE-R, VNTR and Alu components (see (Hancks and Kazazian, 2010) for a review of SVA). SVA are mobilized by L1 proteins (Hancks et al., 2011; Raiz et al., 2011). Compared to L1 and Alu, SVAs are structurally more heterogeneous; they range in size from 700bp to 4kb with a canonical 2kb element (Figure 1D). SVAs also exhibit more RNA complexity, with numerous splice isoforms capable of retrotransposition and a strong propensity to relocate neighboring 5′ or 3′ sequence during transposition, incorporating this in the SVA RNA by upstream transcript initiation or polyA read-through. Compared to other active repeats, they constitute a small amount of our total DNA; there are about 3700 intervals in our genome with homology to SVA (Jurka, 2000; Smit et al., 1996–2010).
Human LTR retroelements consist of three classes of endogenous proviruses, class I (gamma retroviruses), class II (beta retroviruses), and class III (spuma retroviruses) (Mager and Medstrand, 2003). Full length human endogenous retrovirus (HERV) elements structurally resemble exogenous retroviruses, with recognizable gag, pol, and, in some cases, env genes. Fragments are also frequently found in the genome as single or ‘solo’ LTRs. These result from insertion followed by a recombination between the long terminal repeats which deletes the intervening proviral sequences. The mechanism of LTR transposition is completely independent of L1, and ERVs may be transmitted horizontally. HERV elements are named with single letter amino acid abbreviations denoting the host tRNA co-opted as a primer for reverse transcription (Cohen and Larsson, 1988). Human endogenous retroviruses are transpositionally inert, with the possible exception of HERV-K (HML2), a family of class II elements. HERV-K (HML2) proviruses capable of templating infectious particles and new insertions have not been found, although they can be reconstituted by recombining together sequences from existing elements (Dewannieux et al., 2006; Lee and Bieniasz, 2007). One of the best pieces of evidence for recent HERV-K activity is that polymorphic elements persist in human populations (i.e., there are loci where an individual may carry an “empty” allele; a ‘solo’ LTR allele; and/or a full-length proviral insertion allele) (Moyes et al., 2007).
Hiding in the Haystack: Identifying Retroelements in the Genome
For most geneticists, genomic repeats are nuisances – sequences of extraordinarily high copy number which overwhelm hybridization based assays, introduce artifacts in PCR amplifications, and generate unmappable reads. Historically, our appreciation for the importance of polymorphic repeats has lagged behind other areas of genomics. Today, this view is changing rapidly as investigators target polymorphisms for sequencing or tailor sequence analysis pipelines for transposon discovery.
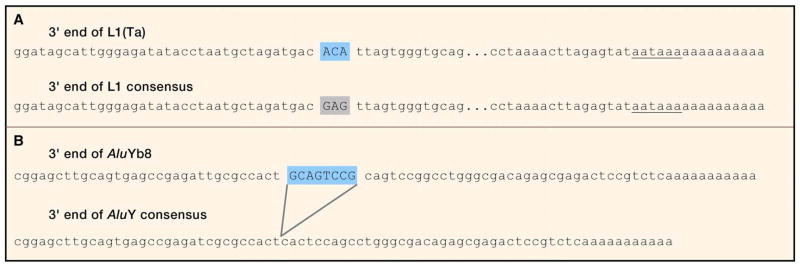
Approaches for targeted recovery of insertion sites of ‘young’ repeat subfamilies, which are most likely to be polymorphic, have been PCR-based methods also known as ‘hemispecific’ or ‘one-sided’ amplifications. They amplify known (repeat) sequence and neighboring, unique DNA sequence. The reactions gain specificity by exploiting internal repeat sequences characteristic of recently expanding subfamilies. For human L1, this means two or three contiguous nucleotides in the 3′UTR of the element characteristic of the pre-Ta and Ta subsets, respectively (Ta, for transcribed subset a) (Skowronski et al., 1988) (Figure 3A). This allows selective amplification of L1 insertion sites which date from about 2 million years ago and are partly polymorphic in human populations (Myers et al., 2002) while preventing amplification of older “fixed present” L1 insertions, which outnumber intended targets nearly a thousand fold. Examples of these approaches include subtractive suppression PCR (Badge et al., 2003; Buzdin et al., 2003), random decamer PCR (Sheen et al., 2000), and ligation-mediated PCR (Pornthanakasem and Mutirangura, 2004). These ‘first generation’ methods reduced the complexity of amplicons so that L1 insertion variants could be identified by electrophoresis by suppressing recovery of L1(Ta)s shared between samples and/or imposing limits on adjacent sequences. These successfully identified small numbers of polymorphic insertions.
Figure 3.
A. Families of L1 LINEs have expanded over evolutionary time in primate genomes in a singular succession of retrotransposition “waves”; that is, elements have accumulated in a specific order, with L1PA5 insertions preceding L1PA4 preceding L1PA3, L1PA2 and L1PA1 (Boissinot and Furano, 2001). For the last two million years, continued L1 activity is largely owed to the L1(Ta) subset (L1PA1). A 3 nucleotide sequence in the 3′UTR of L1(Ta) can be used to physically distinguish its members from sequences of older L1 LINEs in the human genome; this can lend specificity to L1(Ta) mapping PCRs. B. Similarly, insertion of a 8 base pair sequence near the 3′ end of AluYb8/9 subfamilies are characteristic sequences which can be used to specifically recover insertion sites of these elements.
Advances in genomic technologies have led to far more comprehensive methods for discovering retrotransposon insertion polymorphisms (RIPs). For L1 insertion mapping, ligation mediated PCR known as vectorette PCR, can be used to comprehensively recover L1(Ta) subset insertions in the human genome. The amplicons begin near one transposon end and extend to adjacent, mappable DNA sequence. The resulting complex mixture of PCR products can be resolved by either hybridization to genomic tiling microarrays (transposon insertion profiling by microarray, TIP-chip (Huang et al., 2010; Wheelan et al., 2006)), or next generation sequencing (TIP-seq). Different PCR methods have also been used in tandem with next generation sequencing methods for L1 mapping (Ewing and Kazazian, 2010; Iskow et al., 2010). Comprehensive L1(Ta) profiling of an individual by TIP-chip encompassed 323 insertions included in the reference genome and uncovered 191 novel candidate insertions (Huang et al., 2010). Many previously unreported L1 insertion positions could then be verified by site-specific PCR and the precise insertion site shown by Sanger sequencing. Whole genome L1(Ta) insertion profiles of 15 individuals using TIP-chip indicated a predilection for insertions in AT-rich regions and no evidence of insertions strongly targeting or avoiding particular gene loci or chromosomes. Although SINEs are more prevalent in genomes and more heterogeneous in terms of active families, mapping Alu insertions is tractable to similar approaches. These have included TIP-chip applications for AluYa and AluYb (Huang et al, 2010), linker-mediated PCRs with amplicon sequencing for AluYa (Iskow et al., 2010), and AluYb8/9 insertion recovery by a ligation mediated PCR termed mobile element scan (ME-scan) (Witherspoon et al., 2012). ME-scan includes a hybridization based purification of desired AluYb8/9-containing fragments followed by their amplification. AluYb insertion sites lend themselves especially well to family-specific amplification as they have an eight base pair insertion very close to the 3′ element end (Figure 3B). Finally, an array hybridization-based enrichment, termed retrotransposon capture sequencing (RC-seq), has been recently reported and applied to somatic insertion discovery, as we consider further below (Baillie et al., 2011).
Genomic scale approaches for polymorphism discovery are highly effective and will have special utility for clinical hypothesis testing in the future. However, they have been quickly outpaced by in silico computational methods for general cataloging of common repeat polymorphisms. By in silico methods, we mean that computational pipelines rather than sequencing methods are tailored for repeat discovery. These studies include, for example, two published comparisons of genome assemblies, a pairwise analysis of the Human Genome Project hg17 assembly verses the Celera build as reported by Konkel et al. (Konkel et al., 2007) and, more recently, an hg18 comparison to Venter HuRef by Xing et al. (Xing et al., 2009). Xing et al. considered all major transposon families in their analysis, and identified 584 HuRef specific Alu, 52 L1, and 14 SVA insertions, in general agreement with the numbers of polymorphic L1 described in the study by Konkel and colleagues. These tallies underscore how much any individual varies from the reference genome. These numbers are dwarfed by the RIPs being found in sequencing projects designed to capture human genetic diversity. The original study of 36 International HapMap project samples provided an early indication that such approaches would be fruitful, returning hundreds of new mobile element insertions by recognizing repeat homologies in sequences proximal to putative indel sites (Bennett et al., 2004). Today, the 1000 Genomes project has provided the single richest source of new RIPs (Stewart et al., 2011). Stewart et al. described polymorphic repeats in about 180 individual genomes using two types of datasets, paired-end short read sequences (Illumina) and longer sequences which were bisected for analysis (Roche/454). Novel RIPs were called as clusters of sequence fragments in which one end showed mobile element homology while the other end was uniquely mappable to the reference assembly a sufficient distance away from known reference elements. In a separate complementary analysis, reference elements were deemed polymorphic if evidence of an ‘empty allele’ was found. This identified 6229 polymorphic Alus (4499 non-reference), 998 polymorphic L1s (792 non-reference), and 153 variant SVAs (79 non-reference). The L1 number is in fairly good agreement with an independently developed algorithm from an analysis of 307 individual genomes (Ewing and Kazazian, 2011). Some of the same individuals were independently analyzed for Alu indels by Hormozdiari et al. as part of an eight person study (Hormozdiari et al., 2010). Although limited in total numbers of people, this encompassed a broad sampling of ethnicities, and sequencing coverage for each person was relatively high, potentially explaining identification of 4342 novel Alus with very high experimental validation rates.
‘Second generation’ wet bench and in silico methods for transposon discovery have gone beyond providing the promise of a more complete catalog of common polymorphisms. They have helped us realize the level of mobile DNA activity in modern humans. Several groups have used numbers of new insertions discovered to estimate the rate of occurrence of non-parental insertions which are thereafter heritable polymorphisms. Xing et al. based their estimate on the number of non-reference mobile DNA insertions found in Venter’s genome and used a SNP based clock measuring time to the most recent common ancestor between him and the reference build. This led them to the expectation that one in 21 people would have a new Alu, one in 212 would have a new L1, and one in 916 would have a new SVA (Xing et al., 2009). Using a similar calculation based on TIP- hip, we estimated a rate of one de novo L1 insertion in 108 individuals (Huang et al., 2010). A distinct method of predicting L1 polymorphism rates estimates 1 de novo L1 in 140 people (Ewing and Kazazian, 2010). These transposition rate estimates combined with population sizes can be used to project overall numbers of human insertion alleles. For L1, this may be as many as 12,000 insertions with allelic frequencies greater than 0.05. For Alu, a de novo insertion rate of 1 in 20 would result in 65,000 segregating insertions in populations of the same frequency. Thus, we can expect ‘common variant’ RIP lists will continue to grow in the near future.
A catalog of repeat polymorphisms, the dbRIP, is maintained by Ping Liang and colleagues at Brock University (Wang et al., 2006). Lengthier lists of common polymorphic transposon insertions are a fundamental beginning, though it is important to establish a contextual view of these genetic variants. These will include integrating RIP sites with information about other sources of genetic variation (SNPs and structural variants) and epigenetic states. Both should be promoted by the development of high throughput RIP genotyping methods capable of distinguishing homozygous and heterozygous states. Several approaches for direct assays are in development, though concerted efforts to find tagging SNPs of ‘empty’ and ‘filled’ alleles may prove an effective alternative. We also speculate intrarepeat sequence polymorphisms – though overlooked by most genetic variation studies today – will also gain consideration.
Active transposons and their control
RIPs do not enter genomes to rest in peace. L1 and Alu continue to expand their numbers in human genomes today, though the pace of Alu accumulation may be slower than historical rates (Britten, 1994; Shen et al., 1991). Insertions can serve as templates for multiple ‘daughter’ elements but can easily be blocked from establishing themselves as active in populations by either genetic drift or negative selection resulting in allele loss or inactivation via mutation. As stressed above, a narrowly defined set of repeat subfamilies are responsible for sustaining all ongoing retrotransposition in humans. Several recent studies indicate that among the transposon groups defined by sequence variation, a relatively small number of L1 and Alu elements drive the process.
Inactive L1 elements are frequently recognizable because they are 5′ truncated or have mutated open reading frames preventing production of functional ORF1p and ORF2p. Intact elements are actively limited in genomes, and establishing a highly active element in a population may be rare. Full-length L1(Ta)s are subject to negative selection, and active ones may be ephemeral in populations (Boissinot et al., 2006; Boissinot et al., 2001). Among potentially active L1(Ta) elements in the reference genome, in vitro retrotransposition assays indicate that a relatively small group disproportionally dominates aggregate transposition potential (Brouha et al., 2003). Building on the observation of active or ‘hot L1s’ in the reference genome, Beck et al. (Beck et al., 2010) recently assayed activity levels specifically in novel (non-reference) L1 insertions in human populations. These relatively uncommon element copies (i.e., low frequency alleles) were recovered using a fosmid paired- end DNA sequencing strategy to identify large indels. Impressively, the majority of 68 full-length L1s found in fosmid libraries from six individuals proved ‘hot’ in culture. While expected that highly active elements would be responsible for ongoing insertions of L1, Alu, and SVA, the observation changed how we view the repository of L1 activity. It provides evidence that each diploid genome – though harboring on the order of a hundred competent L1s – has a complement of ‘hot’ elements. Each of the latter is relatively recently derived (averaging about 1 million years in age) and occurs with low allele frequency. The model implies that because the number of highly active elements is so small, substantial individual variation in retrotransposition activity may exist.
The level of similarity a given L1 element shares with a ‘hot L1’ consensus sequence can serve as a predictor of L1 activity (Brouha et al., 2003). Exactly what the sequence variants are with the most impact and their mechanisms of action may provide insight into transposition mechanisms or host control. It is possible to engineer super active versions of human and mouse L1 by recoding the open reading frames (An et al., 2006; Han and Boeke, 2004). The synthetic murine recoded L1 (L1 ORFeus) exhibits 200-fold increased activity in in vitro retrotransposition assays compared to its native counterpart. In designing L1 ORFeus, we altered about one quarter of the nucleic acid sequence, changing codons to match the favored trinucleotide usage of highly expressed mammalian genes. This did not change ORF1p or ORF2p protein sequence, and was meant to mitigate DNA sequence features such as high overall adenosine content which may compromise transcriptional read through of L1 (Han et al., 2004).
Distinct intrinsic and surrounding sequence features also influence the relative activity of Alu repeats. Network phylogenetic approaches show that an oligopoly of ‘master’ sequences is responsible for the bulk of Alu element expansion, with each active subfamily of Alu (Ya5a2, Ya8, Yb9, Yc1) having a singular ‘master’ template sequence central to its network of daughter insertions. Though nearly 80% of the daughter insertions appear to have directly derived from the ‘master’ Alu, a few sequences of each subfamily are further removed and reflect activity of ‘secondary source’ elements (Cordaux et al., 2004). Studies of the AluYb subfamily add to this picture. Although these elements are highly active in humans today, their origins antedate rapid phases of their expansion by as much as 20 million years. This observation is the foundation for the concept of ‘stealth drivers’, elements that are quiescent enough to gain allelic frequency but also capable of templating relatively more active progeny (Han et al., 2005). Cis acting factors unique to the integration site seem especially critical when considering the potential of any individual Alu sequence for activity, including a requirement for 3′ RNApolIII terminator sequences that are not encoded within the elements (Aleman et al., 2000).
Once inserted, transposons encounter host pressures at the level of DNA sequence – selection favoring the empty allele over the insertion allele or mutational inactivation of the transposon – as well as pathways to repress mobile DNA insertions through mechanisms such as RNA editing and epigenetic silencing (Bogerd et al., 2006; Esnault et al., 2005; Muckenfuss et al., 2006; Stenglein and Harris, 2006).
Epigenetic silencing relies on a variety of chromatin modifications. Among the best studied host control mechanisms is a specialized small RNA inhibition pathway that ultimately controls de novo DNA methylation, repressesing mobile DNA expression in the male germ line. Central are the Piwi-interacting RNAs (piRNAs), short 26–31 nucleotides long RNAs with similarity to transposon control loci first characterized in Drosophila (Aravin et al., 2006; Aravin et al., 2007). Once piRNAs are generated from long precursor RNAs, they are amplified and affect transposon silencing in association with Piwi proteins. The amplification involves piRNA-guided nuclease (slicer) activities of PIWIL1 [piwi-like homolog 1, also known as MIWI (mouse piwi) or HIWI (human piwi)] and PIWIL2 [MILI (miwi-like)] (De Fazio et al., 2011; Kuramochi-Miyagawa et al., 2004; Reuter et al., 2011). Interestingly, the catalytic endonuclease activity of PIWIL4 [also known as MIWI2 (mouse) or HIWI2 (human)] is dispensable for amplification and piRNA-induced L1 LINE silencing, though other MIWI2 functions are critical in transposon silencing. MIWI2 RNPs are are enriched for secondary piRNAs antisense to transposable elements under its control (Aravin et al., 2008), and nuclear localization of MIWI2 complexes is thought to directly recruit a DNA methyltransferase regulator, DNMT3L, effectively silencing L1 (Aravin et al., 2008; Bourc’his and Bestor, 2004). Mice null for Miwi2 are male infertile, show defects in spermatogenesis with increased retrotransposon RNAs and decreased methylation of genomic retrotransposons, like animals lacking Miwi, Mili, or DNMT3L activity (De Fazio et al., 2011; Kuramochi-Miyagawa et al., 2004; Reuter et al., 2011) (Aravin et al., 2008; Bourc’his and Bestor, 2004). Other Piwi-interacting proteins are also essential for germ line transposon silencing, including components of nuage in spermatogonia and spermatocytes, and the chromatoid body in spermatids.
Cooperative roles of other methyltransferases, DNMT1, DNMT3A, and DNMT3B are important in establishing transposon methylation in embryonic stem (ES) cells, subsequent to de novo methylation (Liang et al., 2002). In mouse ES cells, the enzymatic activity is essential; targeted inactivation of DNMT1 methyltransferase activity by point mutation leads to loss of intracisternal A-particle (IAP) retrotransposon methylation and high IAP transcription (Damelin and Bestor, 2007). Recently, in vitro retrotransposition assays in human embryonic carcinoma cell (EC) lines show that these pluripotent cells actively induce heterochromatin formation in the vicinity of new transposon insertions in a histone deacetylation-dependent manner. This feature is lost upon cellular differentiation (Garcia-Perez et al., 2010). Though not a physiologic cell type, the finding suggests that like early development in mouse, there may be key windows for establishing retrotransposon repression in human development. Further strengthening the connection between methylation and retroelements silencing, Dnmt1 knockout mice are not viable, and compound Dnmt1 heterozygotes, with one hypomorphic and one null allele, develop thymic lymphomas (Gaudet et al., 2003). These tumors exhibit genome-wide hypomethylation and recurrent insertions of endogenous intracisternal A particles (IAPs) at the Notch1 locus. Out of 16 lymphomas analyzed, seven showed IAP insertions to activate the Notch1 oncogene. (Howard et al., 2008). Thus, loss of Dnmt1 provides susceptibility to endogenous retrotransposon mutagenesis affecting differentiated cells.
Another chromatin regulator important for transposon silencing in mice is LSH (lymphoid specific helicase; also known as HELLS), a member of the the SNF2/helicase family of chromatin remodelers,. LSH co- precipitates with DNMT3A and DNMT3B (Zhu et al., 2006), and with LINE, SINE, and IAP genomic loci (Huang et al., 2004) in vitro. Lsh null mice die in the perinatal period (Geiman et al., 2001) and exhibit hypomethylation of LINEs, SINEs, and IAPs (Dennis et al., 2001) and derepressive histone H3 and H4 acetylation marks in association with IAPs (Huang et al., 2004). Hypomorphs are viable, but suffer growth defects and premature aging (Sun et al., 2004).
In addition to the piRNA-piwi based mechanism for controlling germline transposon silencing, recent studies have implicated DICER produced miRNAs as important to transposon silencing in somatic cells. In a mouse model of age-related macular degeneration, Cre mediated mutation of Dicer specifically in retinal pigmented epithelium is accompanied by Alu-containing RNA accumulation and a dependent cytotoxicity (Kaneko et al., 2011). The study illustrates how repeat-containing RNAs and small RNAs can be manipulated in experimental systems, and how this can provide important perspectives on consequences of repeat expression. Note that expression of Alu RNA, rather than retrotransposition per se, appears to be disease causing. As we consider shortly, mobilome de-repressions in a host of human neoplasias have also described, though their specific role in oncogenesis is not yet as defined.
Clearly, massive derepression of transposable elements can have severe consequences, from crises in spermatogenesis to complete loss of viability. While most agree with the conventional view that ongoing retrotransposition poses a hazard for individual genomes (Dawkins, 1976), this perspective is being challenged. Evidence of somatic retrotransposition in neurons has led to the provocative proposal that such insertions might endow specific populations of cells with beneficial unpreformed genetic diversity, allowing for selection of phenotypes on a cellular scale (Muotri et al., 2005). Leveraging a recently developed sequencing method that enriches for repeats through an array capture, Faulkner and colleagues have found genomic evidence to substantiate this model (Baillie et al., 2011). The fact that these are extremely low-abundance relative to germ line insertions – often identified as single junctional reads – suggests that these somatic insertions are either cell-specific or present in tiny clonal lineages within normal neuronal tissue. Remarkably, genes containing putative somatic intronic L1 insertions were twice as likely to be differentially overexpressed in the brain as would be expected from random chance (Baillie et al., 2011). This contrasts with random positioning of new insertions, which was suggested by earlier low throughput sequencing of L1 integrations from in vitro retrotransposition assays. Whether tissues besides brain exhibit this microchimerism is an evolving area, though some studies have suggested the central nervous system is a uniquely privileged environment for transposition.
How the mobilome matters
We have introduced the concepts that highly active transposons show fitness costs associated with their intrinsic activities and that active suppression of expressed repeats in the germ line and in somatic cells is vital in mice. In considering how the human mobilome matters, we begin with the most straightforward examples of their roles in clinical genetics as insertional mutagens. In rare instances, a new insertion of L1, Alu, or SVA hits a single locus in a cell that survives to compromise the function of that gene in an individual.
The first report of this described two independently occurring L1 LINE insertions in two unrelated males with hemophilia A (Kazazian et al., 1988). Both X-chromosome insertions occurred in exon 14 of the factor VIII gene, each preventing synthesis of functional coagulation factor. Since that time, the literature has accrued a series of similar case reports, 13 L1, 33 Alu, and 4 SVA insertions causing a variety of conditions (reviewed in Belancio et al., 2008; Chen et al., 2005; Ostertag and Kazazian, 2001a). Many of these cases resulted from directed studies of a specific locus with a well-established role in disease. Most known pathogenic insertions have been exonic such that critical coding sequence is interrupted by the transposon itself. Exonic disruptions associated with deletions or locus rearrangements from transposon integration are less common. A total of 11 insertion events involving exclusively intronic sequence are listed in Table 1. Although this tally is short and skewed by ascertainment methods such as exon spanning gDNA PCRs, it is interesting to note the propensity of pathogenic insertions near splice acceptor sites at 3′ intron/5′ exon boundaries and the antisense strand bias of such Alu repeats. Overall, these reports indicate that transposon insertions provide recurrent but relatively uncommonly sources of genetic mutation in humans. As new sequencing technologies avail themselves in clinical genetics, it will be interesting to see how our understanding of disease causing insertions changes. In this light, we mention a recent report of an Alu insertion discovered through exome sequencing in retinitis pigmentosa (Tucker et al., 2011). This is the first example of a pathogenic transposon insertion ascertained by next generation sequencing, which is liable to exclude informative reads as unalignable. The authors describe this problem and underscore the need to make exome sequencing analyses inclusive of transposon insertions.
Table 1.
Pathologic intronic insertions in humans.
| retroelement | length | orientation | position in gene | gene name | abbreviation | Phenotype | mRNA studies | reference |
|---|---|---|---|---|---|---|---|---|
| Alu | ||||||||
| AluYc1 | 316bp | antisense | −52bp from the 3′ end of intron 4 | glycerol kinase | GK | benign isolated glycerol kinase deficiency | not reported | (Zhang et al., 2000) |
| AluYb9 | ~330bp | antisense | −19bp from the 3′ end of intron 18 | coagulation factor VIII | F8 | hemophilia A | exon 19 skipping | (Ganguly et al., 2003) |
| AluYa5 | 331bp | antisense | −50bp from the 3′ end of intron 7 | tumor necrosis factor receptor superfamily, member 6 | FAS | autoimmune lymphoproliferative syndrome | exon 8 skipping | (Tighe et al., 2002) |
| AluYa5 | 368bp | antisense | −19bp from the 3′ end of intron 8 | fibroblast growth factor receptor 2 | FGFR2 | Apert syndrome | ectopic exon 7/8 splicing in lieu of 7/9 | (Oldridge et al., 1999) |
| AluYa5 | 320bp | antisense | −44bp from the 3′ end of intron 5 | neurofibromatosis type 1 | NF1 | neurofibromatosis type 1 | exon 6 skipping | (Wallace et al., 1991) |
| L1 | ||||||||
| L1(Ta) | 836bp | sense and rearranged | intron 5 | cytochrome b-245, beta polypeptide | CYBB | chronic granulomatous disease (CGD) | variable L1 exonization; exon 5 and exon 6 skipping | (Meischl et al., 2000) |
| L1(Ta) | 6kb | antisense | reported as 3′ end of intron 2 | hemoglobin, beta | HBB | beta-thalasemia | not reported | (Kimberland et al., 1999) |
| L1(Ta) | 1.2kb | sense | −24bp from the 3′ end of intron 7 | fukutin | FKTN | Fukuyama-type congenital muscular dystrophy | variable exon 7, 8, and 9 skipping | (Kondo-Iida et al., 1999) |
| L1(Ta) | 6kb | sense | intron 1 | retinitis pigmentosa 2 | RP2 | X-linked retinitis pigmentosa | no transcript detected by RT-PCR | (Schwahn et al., 1998) |
| L1(Ta) | 2.8kb | antisense and rearranged | −8bp from the 3′ end of intron 3 | ribosomal S6 kinase 2 gene | RPS6KA3 | Coffin-Lowry syndrome | exon 4 skipping | (Martinez- Garay et al., 2003) |
| SVA | ||||||||
| SVA | 2.6kb | sense | intron 1 | low density lipoprotein receptor adaptor protein 1 | LDLRAP1 | autosomal recessive hypercholesterolemia | no expression by Northern blot | (Wilund et al., 2002) |
One pathogenic 3′UTR insertion ignites hope for altering the natural history of genetic diseases caused by retroelements insertions. This SVA insertion in the 3′UTR of the Fukutin gene causes Fukuyama-type congenital muscular dystrophy (FCMD). FCMD is one of the most common autosomal recessive genetic diseases in Japan, with an incidence of about 1 in 10,000; it is diagnosed in early infancy and characterized clinically by hypotonia, muscle weakness, and central nervous system defects including delayed mental development. It is a type of α-dystroglycanopathy, with loss of O-glycolsylation on this cell membrane protein which mediates interactions with extracellular matrix. Discovered in 1998, the causative 3kb sense oriented SVA insertion is an component of the FCMD founder haplotype seen in 87% of FCMD cases and carried by about 1/88 Japanese individuals (Kobayashi et al., 1998). The insertion induces a splice donor site within the last exon of Fukutin and exonizes part of the SVA; there is a replacement of the end of the normal open reading frame (coding 38 C-terminal AA) with coding sequence for 129AA. Excitingly, in cell culture and in a murine model of the disease, these effects can be mitigated and normal splicing restored by the addition of a combination of 25-mer 2′-O- methyl phosphoramidite (2′OMePS) antisense oligonucleotides targeted to the aberrant splice acceptor, donor, and exonic splicing enhancer sites (Taniguchi-Ikeda et al., 2011).
Effects on gene function of fixed insertions or commonly occurring RIPs is an active area of ongoing research. Our working model is that mammalian repeats can act as ‘soft’ genetic variants damping levels of gene expression or altering transcript structure to partial degrees and in context-dependent manners. In the case of L1, premature polyadenylation and compromised RNA polymerase elongation are two well described mechanisms for this (Chen et al., 2006; Han and Boeke, 2005; Han et al., 2004; Perepelitsa-Belancio and Deininger, 2003). For example, sense-orientation insertions of the ORF2-encoding portions of L1 into a reporter gene intron lowered both RNA and protein production sharply, reflecting attenuated transcript elongation through the adenosine-rich strand, an effect that is not sequence specific but can be recapitulated with other L1 portions when present in sufficient total length (Han and Boeke, 2005). Insertion in the antisense orientation produced a decrease in full-length RNA that was less pronounced, and this could be ascribed primarily to prematurely polyadenylated forms. The non-random orientation of intronic L1s in our genome with respect to their encompassing genes suggests that these findings are relevant for understanding the impact of these elements in the human genome. L1s within transcript units show a biased antisense orientation, consistent with a more deleterious role of sense insertions and thus stronger negative selection. This effect is appreciable in young, low allele frequency L1(Ta) but increasingly pronounced if older, fixed L1(Ta) or older pre-Ta L1 elements are considered (Huang et al., 2010). This said, there is a dearth of specific RIPs that affect genes in known and consequential ways. An Alu indel at the angiotensin-I converting enzyme (ACE) locus is the best studied candidate given its association with serum enzyme levels (Rigat et al., 1990), though how this intronic insertion functions, or whether it is more than a proxy for some other variant, is yet unclear.
Transcriptome studies will likely demonstrate the functional impact of repeats on gene transcripts. Examples have been shown in traditional methods (Rangwala et al., 2009; Speek, 2001), and may be hugely bolstered by new sequencing technologies. Faulkner et al. recently performed Cap Analysis Gene Expression (CAGE) experiments sequencing 20-nt and 21-nt tags from the 5′ most ends of transcripts in a variety of mouse and human cells. The authors imposed a hierarchical alignment strategy in attempts to unequivocally assign genomic positions to each tag; ‘multimapping’ tags where there is ambiguity in the site comprised a minority of aligned reads. Transcript start sites (TSSs) were recognized once mapped with at least two congruous tags. A total of 275,185 TSSs in human cells, representing 31.4% of all TSSs, showed homology to repeats. The majority, about 214,000, corresponded specifically to transposable elements. Their data also suggest a high degree of spatiotemporal specificity and correlation between transposon initiated transcription and expression of proximal genes. This suggests coregulation of repeats and neighboring loci supersedes transcriptional interference.
Neoplastic conditions will rightly receive special attention as functional impacts of the human mobilome are approached (see review by Belancio et al., 2010). L1 hypomethylation in tumors is well-described in many tumor types, including breast (Alves et al., 1996), colon (Dante et al., 1992; Estecio et al., 2007; Suter et al., 2004), and prostate cancers (Cho et al., 2007; Santourlidis et al., 1999; Schulz et al., 2002) to cite a few. In the case of the latter, hypomethylation of L1 associated with preoperative prostate specific antigen levels, histopathologic measures of aggression (Gleason grade), and anatomic spread of the tumor (clinical stage); it also associated independently with cytogenetic abnormalities. Reagents for assaying protein expression of L1 ORF1p and ORF2p are not developed to the point of being useful clinical markers, though ORF1p expression has been reported in breast cancer cases. Here, a nuclear staining pattern is reported to correlate positively with incidence of both local recurrence and distal metastases and with worse overall survival (Harris et al., 2010). Repeat derepression in tumors can be envisioned to have several sequelae. One may be complete retrotransposition cycles, generating somatic L1, Alu, and SVA insertions specific to the neoplastic lineage. It is known that somatic insertions relevant to cellular transformation can occur. In a case of colon cancer, a somatic L1(Ta) insertion has been found interrupting an exon of the adenomatous polyposis coli (APC) tumor suppressor locus (Miki et al., 1992). Recently, the Devine laboratory has described somatic insertions in cases of lung cancer revealed through next generation sequencing of L1 and Alu insertion sites, though retroelement-induced driver mutations have not been described (Iskow et al., 2010). Interestingly, their work suggests somatic insertions may be tumor tissue type specific and, even within a broad neoplastic category, occur selectively in tumors with an epigenetic signature of differentially methylated gene loci. How well existing clinical diagnostic criteria have captured information about L1 activity will be interesting to understand as these studies go forward. In considering other mechanisms, L1-encoded ORF2p expression can promote DNA breaks and even perhaps specific chromosomal translocations (Lin et al., 2009) independently from completed mobile element insertion. Finally, effects on neighboring gene loci may be important aspects to understand. Specific L1 antisense initiated transcripts can act as oncogenes (Roman-Gomez et al., 2005; Weber et al., 2010). More broadly, Estecio and colleagues recently recognized that gene promoters with high transposon content are also relatively resistant to methylation in cancer cell lines and maintain higher expression levels in that context as compared to loci which are hypermethylated (Estecio et al., 2010). These data suggest that transposable element-directed epigenetic studies capable to describe the status of individual elements (fixed and polymorphic) will be valuable to integrate with our current picture of the cancer cell genome.
Concluding remarks
High copy number, self propagating repeats are landscape determining components of our DNA. Many are fixed, static and slowly eroding features, but there are also focal ‘hot spots’ – elements that demonstrate episodic activity and sites where new insertions can be found. Today, new technologies are being developed to reveal insertions, confirming that interspersed repeat polymorphisms are important sources of genetic variation in human populations and suggesting that each of us have somatic compartments genetically variegated by insertion events. Many models of how repeats may influence human phenotypes are emerging, and we look forward to delineating and altering functions of specific insertions in human disease.
Figure 4.
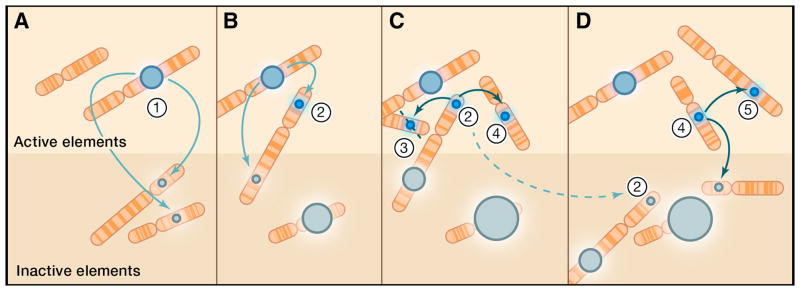
A schematic of retrotransposon dynamics. (A.) An L1 insertion (1) of moderate activity (blue) and allelic frequency (circle size) is shown (1). It templates new insertions, many of which are 5′ truncated or mutated at the time of insertion (gray denotes inactivity). The new insertions segregate mostly as neutral alleles; some accumulate to a higher allele frequency over time (larger gray circles in subsequent panels). (B.) A new insertion is generated that is highly competent or ‘hot’ for retrotransposition (2, blue) because of features intrinsic to its sequence or by its location in an expression-permissive target site. (C.) This ‘hot’ L1 templates others; there may be a tendency to template other full-length, ‘hot’ progeny insertions. ‘Hot’ elements are continually lost through negative selection (3, interrupted diagonal), or (D.) mutation (2, interrupted arrow from C). However, some remain to potentiate other retrotransposition events (4). It is envisioned that these ‘hot’ elements are relatively transient in genomes due to purifying selection against them, do not achieve high allele frequency, but are responsible for the bulk of transposition in modern humans.
Acknowledgments
This work was supported by grants P01 CA16519 and RC1 HG005359 (to JDB) and K08 CA134746 and a Career Award for Medical Scientists from the Burroughs Wellcome Foundation (to KHB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kathleen H. Burns, Email: kburns@jhmi.edu.
Jef D. Boeke, Email: jboeke@jhmi.edu.
References
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Aleman C, Roy-Engel AM, Shaikh TH, Deininger PL. Cis-acting influences on Alu RNA levels. Nucleic Acids Res. 2000;28:4755–4761. doi: 10.1093/nar/28.23.4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves G, Tatro A, Fanning T. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene. 1996;176:39–44. doi: 10.1016/0378-1119(96)00205-3. [DOI] [PubMed] [Google Scholar]
- An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, Triplett C, Boeke JD. Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci U S A. 2006;103:18662–18667. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Badge RM, Alisch RS, Moran JV. ATLAS: a system to selectively identify human-specific L1 insertions. Am J Hum Genet. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ‘bout retroelements in cancer. Semin Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci U S A. 2006;103:9590–9594. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. 2001;18:2186–2194. doi: 10.1093/oxfordjournals.molbev.a003765. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Britten RJ. Evidence that most human Alu sequences were inserted in a process that ceased about 30 million years ago. Proc Natl Acad Sci U S A. 1994;91:6148–6150. doi: 10.1073/pnas.91.13.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdin A, Ustyugova S, Gogvadze E, Lebedev Y, Hunsmann G, Sverdlov E. Genome-wide targeted search for human specific and polymorphic L1 integrations. Hum Genet. 2003;112:527–533. doi: 10.1007/s00439-002-0904-2. [DOI] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum Mol Genet. 2006;15:2146–2156. doi: 10.1093/hmg/ddl138. [DOI] [PubMed] [Google Scholar]
- Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease7 dependent retrotranspositional events causing human genetic disease. Hum Genet. 2005;117:411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211:269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- Cohen M, Larsson E. Human endogenous retroviruses. Bioessays. 1988;9:191–196. doi: 10.1002/bies.950090603. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Hedges DJ, Batzer MA. Retrotransposition of Alu elements: how many sources? Trends Genet. 2004;20:464–467. doi: 10.1016/j.tig.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Damelin M, Bestor TH. Biological functions of DNA methyltransferase 1 require its methyltransferase activity. Mol Cell Biol. 2007;27:3891–3899. doi: 10.1128/MCB.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dante R, Dante-Paire J, Rigal D, Roizes G. Methylation patterns of long interspersed repeated DNA and alphoid repetitive DNA from human cell lines and tumors. Anticancer Res. 1992;12:559–563. [PubMed] [Google Scholar]
- Dawkins R. The Selfish Gene. Oxford University Press; 1976. [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Estecio MR, Gallegos J, Vallot C, Castoro RJ, Chung W, Maegawa S, Oki Y, Kondo Y, Jelinek J, Shen L, et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res. 2010;20:1369–1382. doi: 10.1101/gr.107318.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, et al. LINE-1 Hypomethylation in Cancer Is Highly Variable and Inversely Correlated with Microsatellite Instability. PLoS ONE. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011;21:985–990. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human47 specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci U S A. 2010;107:11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Dunbar T, Chen P, Godmilow L, Ganguly T. Exon skipping caused by an intronic insertion of a young Alu Yb9 element leads to severe hemophilia A. Human Genetics. 2003;113:348–352. doi: 10.1007/s00439-003-0986-5. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, Hammer GD, Collins KL, O’Shea KS, Menendez P, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–773. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Geiman TM, Tessarollo L, Anver MR, Kopp JB, Ward JM, Muegge K. Lsh, a SNF2 family member, is required for normal murine development. Biochim Biophys Acta. 2001;1526:211–220. doi: 10.1016/s0304-4165(01)00129-5. [DOI] [PubMed] [Google Scholar]
- Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- Han K, Xing J, Wang H, Hedges DJ, Garber RK, Cordaux R, Batzer MA. Under the genomic radar: the stealth model of Alu amplification. Genome Res. 2005;15:655–664. doi: 10.1101/gr.3492605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH., Jr SVA retrotransposons: Evolution and genetic instability. Semin Cancer Biol. 2010 doi: 10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CR, Normart R, Yang Q, Stevenson E, Haffty BG, Ganesan S, Cordon-Cardo C, Levine AJ, Tang LH. Association of Nuclear Localization of a Long Interspersed Nuclear Element-1 Protein in Breast Tumors with Poor Prognostic Outcomes. Genes Cancer. 2010;1:115–124. doi: 10.1177/1947601909360812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, Callinan PA, Cordaux R, Xing J, Barnes E, Batzer MA. Differential alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann O, Heidmann T. Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell. 1991;64:159–170. doi: 10.1016/0092-8674(91)90217-m. [DOI] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Alkan C, Ventura M, Hajirasouliha I, Malig M, Hach F, Yorukoglu D, Dao P, Bakhshi M, Sahinalp SC, et al. Alu repeat discovery and characterization within human genomes. Genome Res. 2010;21:840–849. doi: 10.1101/gr.115956.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck CM, Rinehart FP, Schmid CW. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979;132:289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- Huang CR, Schneider AM, Lu Y, Niranjan T, Shen P, Robinson MA, Steranka JP, Valle D, Civin CI, Wang T, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Fan T, Yan Q, Zhu H, Fox S, Issaq HJ, Best L, Gangi L, Munroe D, Muegge K. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 2004;32:5019–5028. doi: 10.1093/nar/gkh821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum Mol Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-Iida E, Nomura Y, Segawa M, Yoshioka M, Saito K, Osawa M, et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- Kondo-Iida E, Kobayashi K, Watanabe M, Sasaki J, Kumagai T, Koide H, Saito K, Osawa M, Nakamura Y, Toda T. Novel mutations and genotype-phenotype relationships in 107 families with Fukuyama-type congenital muscular dystrophy (FCMD) Hum Mol Genet. 1999;8:2303–2309. doi: 10.1093/hmg/8.12.2303. [DOI] [PubMed] [Google Scholar]
- Konkel MK, Wang J, Liang P, Batzer MA. Identification and characterization of novel polymorphic LINE-1 insertions through comparison of two human genome sequence assemblies. Gene. 2007;390:28–38. doi: 10.1016/j.gene.2006.07.040. [DOI] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Medstrand P. Retroviral repeat sequences. In: Cooper D, editor. Nature encyclopedia of the human genome. Hampshire: United Kingdom Macmillan Publishers; 2003. pp. 57–63. [Google Scholar]
- Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010;7:706–711. doi: 10.4161/rna.7.6.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garay I, Ballesta MJ, Oltra S, Orellana C, Palomeque A, Molto MD, Prieto F, Martinez F. Intronic L1 insertion and F268S, novel mutations in RPS6KA3 (RSK2) causing Coffin-Lowry syndrome. Clin Genet. 2003;64:491–496. doi: 10.1046/j.1399-0004.2003.00166.x. [DOI] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Meischl C, Boer M, Ahlin A, Roos D. A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur J Hum Genet. 2000;8:697–703. doi: 10.1038/sj.ejhg.5200523. [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- Moyes D, Griffiths DJ, Venables PJ. Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet. 2007;23:326–333. doi: 10.1016/j.tig.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Myers JS, Vincent BJ, Udall H, Watkins WS, Morrish TA, Kilroy GE, Swergold GD, Henke J, Henke L, Moran JV, et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am J Hum Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldridge M, Zackai EH, McDonald-McGinn DM, Iseki S, Morriss-Kay GM, Twigg SR, Johnson D, Wall SA, Jiang W, Theda C, et al. De novo alu-element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. Am J Hum Genet. 1999;64:446–461. doi: 10.1086/302245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, Kazazian HH., Jr A mouse model of human L1 retrotransposition. Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001a;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001b;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- Pornthanakasem W, Mutirangura A. LINE-1 insertion dimorphisms identification by PCR. Biotechniques. 2004;37:750–752. doi: 10.2144/04375BM07. [DOI] [PubMed] [Google Scholar]
- Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, Lower J, Stratling WH, Lower R, Schumann GG. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala SH, Kazazian HH., Jr The L1 retrotransposition assay: a retrospective and toolkit. Methods. 2009;49:219–226. doi: 10.1016/j.ymeth.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala SH, Zhang L, Kazazian HH., Jr Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 2009;10:R100. doi: 10.1186/gb-2009-10-9-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, Colomer D, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate. 1999;39:166–174. doi: 10.1002/(sici)1097-0045(19990515)39:3<166::aid-pros4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Schulz WA, Elo JP, Florl AR, Pennanen S, Santourlidis S, Engers R, Buchardt M, Seifert HH, Visakorpi T. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer. 2002;35:58–65. doi: 10.1002/gcc.10092. [DOI] [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, et al. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet. 1998;19:327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- Sheen FM, Sherry ST, Risch GM, Robichaux M, Nasidze I, Stoneking M, Batzer MA, Swergold GD. Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MR, Batzer MA, Deininger PL. Evolution of the master Alu gene(s) J Mol Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF, Hubley R, Green P. RepeatMasker. 1996–2010. Current Version: open-3.3.0 (RMLib: 20110920) [Google Scholar]
- Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Stewart C, Kural D, Stromberg MP, Walker JA, Konkel MK, Stutz AM, Urban AE, Grubert F, Lam HY, Lee WP, et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, Miao D, Huso DL, Arceci RJ. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 2004;18:1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis. 2004;19:95–101. doi: 10.1007/s00384-003-0539-3. [DOI] [PubMed] [Google Scholar]
- Taniguchi-Ikeda M, Kobayashi K, Kanagawa M, Yu CC, Mori K, Oda T, Kuga A, Kurahashi H, Akman HO, DiMauro S, et al. Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature. 2011;478:127–131. doi: 10.1038/nature10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe PJ, Stevens SE, Dempsey S, Le Deist F, Rieux-Laucat F, Edgar JD. Inactivation of the Fas gene by Alu insertion: retrotransposition in an intron causing splicing variation and autoimmune lymphoproliferative syndrome. Genes Immun. 2002;3(Suppl 1):S66–70. doi: 10.1038/sj.gene.6363864. [DOI] [PubMed] [Google Scholar]
- Tucker BA, Scheetz TE, Mullins RF, Deluca AP, Hoffmann JM, Johnston RM, Jacobson SG, Sheffield VC, Stone EM. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia3 related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci U S A. 2011;108:E569–576. doi: 10.1073/pnas.1108918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Weiner AM. Human genes and pseudogenes for the 7SL RNA component of signal recognition particle. EMBO J. 1984;3:3303–3310. doi: 10.1002/j.1460-2075.1984.tb02294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Weiner AM. Upstream sequences modulate the internal promoter of the human 7SL RNA gene. Nature. 1985;318:371–374. doi: 10.1038/318371a0. [DOI] [PubMed] [Google Scholar]
- Wallace MR, Andersen LB, Saulino AM, Gregory PE, Glover TW, Collins FS. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991;353:864–866. doi: 10.1038/353864a0. [DOI] [PubMed] [Google Scholar]
- Wang J, Song L, Grover D, Azrak S, Batzer MA, Liang P. dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans. Hum Mutat. 2006;27:323–329. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Kimhi S, Howard G, Eden A, Lyko F. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene. 2010;29:5775–5784. doi: 10.1038/onc.2010.227. [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan SJ, Scheifele LZ, Martinez-Murillo F, Irizarry RA, Boeke JD. Transposon insertion site profiling chip (TIP-chip) Proc Natl Acad Sci U S A. 2006;103:17632–17637. doi: 10.1073/pnas.0605450103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wilund KR, Yi M, Campagna F, Arca M, Zuliani G, Fellin R, Ho YK, Garcia JV, Hobbs HH, Cohen JC. Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum Mol Genet. 2002;11:3019–3030. doi: 10.1093/hmg/11.24.3019. [DOI] [PubMed] [Google Scholar]
- Witherspoon DJ, Xing J, Zhang Y, Watkins WS, Batzer MA, Jorde LB. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics. 2010;11:410. doi: 10.1186/1471-2164-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y, Ohta H, Abe T, Kurimoto K, Chuma S, Saitou M. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol. 2011;192:781–795. doi: 10.1083/jcb.201009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dipple KM, Vilain E, Huang BL, Finlayson G, Therrell BL, Worley K, Deininger P, McCabe ER. AluY insertion (IVS4-52ins316alu) in the glycerol kinase gene from an individual with benign glycerol kinase deficiency. Hum Mutat. 2000;15:316–323. doi: 10.1002/(SICI)1098-1004(200004)15:4<316::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi40 interacting RNA (piRNA) pathway. Proc Natl Acad Sci U S A. 2010;107:11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, Li E, Muegge K. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]