Abstract
We first review what is known about patterns of codon usage bias in Drosophila and make the following points: (i) Drosophila genes are as biased or more biased than those in microorganisms. (ii) The level of bias of genes and even the particular pattern of codon bias can remain phylogenetically invariant for very long periods of evolution. (iii) However, some genes, even very tightly linked genes, can change very greatly in codon bias across species. (iv) Generally G and especially C are favored at synonymous sites in biased genes. (v) With the exception of aspartic acid, all amino acids contribute significantly and about equally to the codon usage bias of a gene. (vi) While most individual amino acids that can use G or C at synonymous sites display a preference for C, there are exceptions: valine and leucine, which prefer G. (vii) Finally, smaller genes tend to be more biased than longer genes. We then examine possible causes of these patterns and discount mutation bias on three bases: there is little evidence of regional mutation bias in Drosophila, mutation bias is likely toward A+T (the opposite of codon usage bias), and not all amino acids display the preference for the same nucleotide in the wobble position. Two lines of evidence support a selection hypothesis based on tRNA pools: highly biased genes tend to be highly and/or rapidly expressed, and the preferred codons in highly biased genes optimally bind the most abundant isoaccepting tRNAs. Finally, we examine the effect of bias on DNA evolution and confirm that genes with high codon usage bias have lower rates of synonymous substitution between species than do genes with low codon usage bias. Surprisingly, we find that genes with higher codon usage bias display higher levels of intraspecific synonymous polymorphism. This may be due to opposing effects of recombination.
As far as is known, synonymous mutations are truly neutral with respect to natural selection.
The above quotation from King and Jukes (1) was one of the major, and more reasonable, tenets of the neutral theory of molecular evolution. With few exceptions (e.g., ref. 2), even those researchers who tended toward the selectionist view of molecular evolution were willing to concede synonymous substitutions to the neutralists. After all, such mutations do not affect the structure of the primary gene product and therefore should not be able to affect the phenotype, the level at which natural selection acts. One of the more surprising observations provided by the accumulating DNA sequence data has been the evidence that selection can and does affect synonymous substitutions.
One of the strongest pieces of evidence of the nonneutrality of synonymous substitutions is codon usage bias, the unequal usage of codons encoding the same amino acid. If synonymous substitutions are neutral and if mutations are truly random (i.e., equal probability of change to all nucleotides), then all codons coding for the same amino acid should be equally represented in a large sample of genes. Therefore, unequal usage of synonymous codons must be due to either mutation bias or selection. Drosophila has served as a model multicellular eukaryote in the study of codon usage bias (e.g., ref. 3). As we will document below, there is little evidence that mutation bias is the cause of codon usage bias in Drosophila, and thus we are left with selection as the likely candidate to explain codon usage in these flies. Here we will first review the pattern of codon usage bias in Drosophila, then present data relevant to the cause of the bias, and end by discussing the effect of codon bias on intra- and interspecific DNA variation.
Levels and Patterns of Codon Usage in Drosophila
Levels of Bias.
Several measures of the degree of codon bias for a given gene have been developed. Here we use one termed the effective number of codons, ENC (4). This is analogous to the effective number of alleles and is related to the “homozygosity” for codons—i.e., the probability that two randomly chosen synonymous codons are identical. ENC ranges from 20 if only one codon is used for each amino acid to 61 if all synonymous codons are used equally. ENC can also be calculated for individual amino acids, what we call ENC-X (X = particular amino acid). As originally formulated (4), the contributions of individual amino acids to ENC are dependent upon the number of synonymous codons—i.e., twofold degenerate amino acids can have a maximum ENC-X of 2, fourfold degenerate amino acids have a maximum ENC-X of 4, etc. To allow each amino acid to equally contribute to ENC, we scaled ENC-X to range from 0 (no bias) to 1 (maximum bias) for all amino acids, what we call sENC-X (unpublished work).
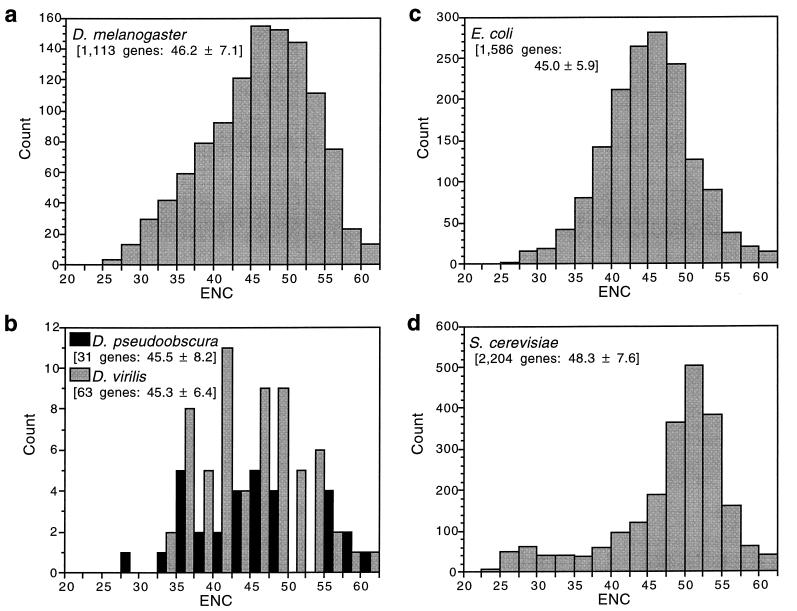
Fig. 1 shows the distribution of ENC for genes available for several species of both Drosophila and microorganisms. Bacteria and yeast have long been model organisms for the study of codon usage bias (5, 6), and it is clear from Fig. 1 that Drosophila genes are as biased as those of microorganisms. All three species of Drosophila and Escherichia coli have about equal mean ENC, which is somewhat less than for Saccharomyces cerevisiae. However, D. melanogaster has a somewhat greater proportion of very highly biased genes than does E. coli. If we consider extreme bias as an ENC of 35 or less, 8% of D. melanogaster genes and 5% of E. coli genes are in this category. Another way of seeing the same phenomenon is to note that, while having the same mean, D. melanogaster genes display a greater variance (SD) in codon usage bias than do E. coli genes (Fig. 1).
Figure 1.
Distribution of genes with various degrees of codon usage bias measured by “effective number of codons”, ENC; lower ENC is greater bias. The number of genes for each species (Drosophila melanogaster, Drosophila pseudoobscura, Drosophila virilis, Escherichia coli, and Saccharomyces cerevisiae) and the mean ENC ± SD are shown in brackets.
Phylogenetic Persistence of Bias.
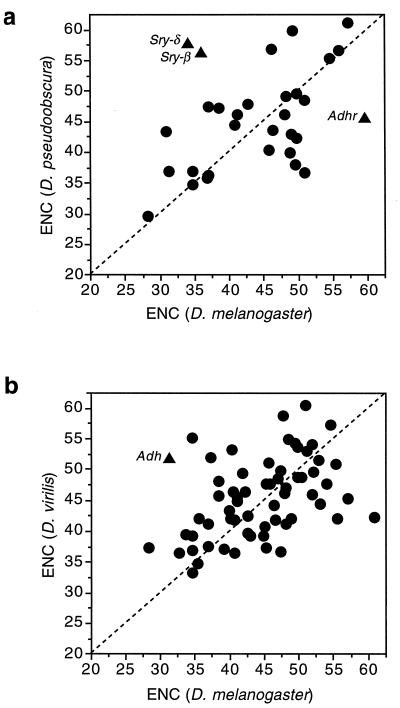
Generally, genes remain at a certain level of codon usage bias across species. Fig. 2 shows the correlations between species of Drosophila. It is important to realize that the level of divergence between the species compared is very high; Ks, the synonymous substitutions per site, is greater than 1 for most genes. This indicates enough evolutionary time has elapsed to radically change codon usage in the absence of constraints. Not only does the level of bias remain conserved, but often the actual pattern as well. One example is Alcohol dehydrogenase (Adh), which has been sequenced in more than 50 species of Drosophila. Table 1 shows the pattern of codon usage for three amino acids. The subgenera Sophophora and Drosophila diverged from each other about 50 million years ago (7), so the avoidance of particular codons in Adh, namely AUA (isoleucine), GGG (glycine), and UUA (leucine), has persisted for a very long time. It is not the case that Drosophila simply cannot use these codons; many genes do use them, an example being the very closely linked Adh-related (Adhr) gene shown in the lower part of Table 1.
Figure 2.
Correlation of codon usage bias across species. The correlation coefficient for the upper graph is 0.42 (n = 31, P = 0.01) and that for the lower graph is 0.42 (n = 63, P < 0.001).
Table 1.
Codon usage for Adh and Adhr
| Subgenus group | No. of species | Mean ENC | No. of times codon used
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isoleucine
|
Glycine
|
Leucine
|
|||||||||||||
| AUU | AUC | AUA | GGU | GGC | GGA | GGG | UUA | UUG | CUU | CUC | CUA | CUG | |||
| Adh | |||||||||||||||
| Sophophora | |||||||||||||||
| melanogaster | 9 | 31.8 ± 3.2 | 72 | 136 | 0 | 51 | 81 | 36 | 1 | 0 | 30 | 3 | 33 | 0 | 224 |
| obscura | 7 | 41.5 ± 5.8 | 64 | 93 | 3 | 37 | 87 | 9 | 0 | 1 | 22 | 6 | 16 | 1 | 125 |
| willistoni | 6 | 45.9 ± 0.7 | 79 | 53 | 0 | 67 | 33 | 8 | 0 | 2 | 99 | 7 | 13 | 3 | 32 |
| Idiomyia | |||||||||||||||
| “Hawaiians” | 10 | 43.8 ± 1.7 | 113 | 115 | 1 | 66 | 112 | 6 | 0 | 0 | 49 | 43 | 26 | 23 | 103 |
| Drosophila | |||||||||||||||
| repleta | 9 | 44.6 ± 2.7 | 77 | 144 | 4 | 35 | 89 | 29 | 2 | 2 | 23 | 16 | 36 | 6 | 124 |
| virilis | 8 | 53.5 ± 1.7 | 92 | 100 | 3 | 35 | 64 | 41 | 4 | 0 | 18 | 35 | 8 | 15 | 112 |
| Adhr | |||||||||||||||
| Sophophora | |||||||||||||||
| melanogaster | 6 | 57.1 ± 2.0 | 42 | 33 | 23 | 28 | 12 | 58 | 11 | 13 | 32 | 6 | 7 | 18 | 42 |
| obscura | 7 | 49.5 ± 3.9 | 48 | 49 | 21 | 45 | 46 | 34 | 14 | 7 | 20 | 6 | 15 | 16 | 84 |
Numbers in main body are numbers of times each codon is used in that group of species. ENC is effective number of codons, defined in the text, and is presented ±SD.
While most genes display evolutionary conservatism for codon bias, other genes do not. Fig. 2 notes a few examples of exceptions which are of some interest. First, Adh in D. virilis is quite unbiased, having an ENC of about 53, while in D. melanogaster and most other species it is quite biased. (Note that even though low in codon usage bias over all the gene, D. virilis Adh still avoids the three codons noted in Table 1, so the avoidance of these codons is not simply due to overall bias.) Adhr also varies in bias between species, being nearly totally unbiased in D. melanogaster but displaying quite high bias in D. pseudoobscura (Fig. 2). Contrariwise, Adh is more biased in D. melanogaster (ENC = 31.4) than in D. pseudoobscura (ENC = 36.7). These two genes are only a few hundred base pairs apart.
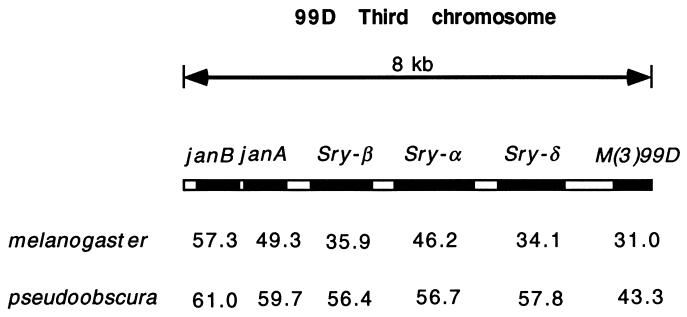
The Serendipity genes, indicated by points Sry-β and Sry-δ in Fig. 2, are also of some interest. These genes are part of a gene cluster that contains six transcriptional units in an 8-kb stretch of DNA. In Fig. 3 we compare the codon usage bias of these genes between D. melanogaster and D. pseudoobscura. Some genes in this cluster have remained relatively highly biased (e.g., the ribosomal protein gene M(3)99D) and others remain quite unbiased (e.g., janA, janB, and Sry-α). Interspersed are the two Sry genes that shift in level of codon bias between these species. There is evidence that Sry genes are expressed differently in these two species (8), which may be related to their change in level of codon usage bias.
Figure 3.
Codon usage bias (ENC) for six tightly linked genes in two species. Note how Sry-β and Sry-δ change considerably between species, while the other genes change much less.
Pattern of Codon Usage Bias.
While ENC and related measures indicate the overall bias, it is also instructive to look more closely at the pattern of codon bias. Generally, Drosophila genes with high codon usage bias have G and especially C at silent positions (9, 10). Table 2 shows the base composition at two- and fourfold degenerate synonymous sites for the approximately 10% highest and 10% lowest biased genes in D. melanogaster.
Table 2.
Base compositions among different D. melanogaster gene groups
| Gene groups | No. of genes | Base composition, %
|
|||||
|---|---|---|---|---|---|---|---|
| T4 | C4 | A4 | G4 | C2 | A2 | ||
| ≈10% highest bias | 122 | 16.2 | 51.1 | 7.7 | 25.0 | 81.0 | 8.0 |
| ≈10% lowest bias | 127 | 21.5 | 29.6 | 23.9 | 25.0 | 49.6 | 41.7 |
| Fourth chromosome | 7 | 33.3 | 18.8 | 32.3 | 15.7 | 39.9 | 63.0 |
The average base composition at fourfold degenerate sites (T4, C4, A4, and G4) and at the twofold degenerate sites (C2 and A2) are shown. C2 % and A2 % were calculated separately from the T/C and A/G twofold degenerate sites, respectively.
Do all amino acids contribute to the codon usage bias of a gene and, if so, do they all show the same pattern (i.e., an increase in C ending codons)? Comparing the individual amino acid measure, ENC-X, to overall bias of the gene, we found all amino acids contribute significantly (P < 0.0001) to the overall bias of a gene, although Asp is a clear outlier with relatively little contribution to overall codon usage bias (unpublished work). We then examined if the pattern of bias for each amino acid is similar; Table 3 shows the correlation of codon usage bias (ENC-X) of each fourfold and sixfold degenerate amino acid and the base composition at synonymous sites. As expected, for most amino acids the highest positive correlation is an increase in C as bias increases. However, there are exceptions to the preference for C. Val and Leu increase in use of G as bias increases. This pattern is very similar in D. pseudoobscura and D. virilis, a general preference for C except for Val and Leu, which prefer G in the codon third position. For Ile (threefold degenerate) and most twofold degenerate T/C amino acids the highest significant positive correlation is for an increase in C as bias increases. The exception is Asp, which shows no significant correlation in its ENC-X and base composition at the wobble position, in agreement with the previous point. For all A/G twofold degenerate amino acids, G increases as bias increases (unpublished work).
Table 3.
Codon usage bias of fourfold and sixfold degenerate amino acids
| Amino acid | Codons | Correlation coefficients of
codons
|
|||
|---|---|---|---|---|---|
| NNT | NNC | NNA | NNG | ||
| Fourfold degenerate | |||||
| Val | GTN | −0.48* | −0.04 | −0.56* | 0.71* |
| Pro | CCN | −0.36* | 0.58* | −0.31* | −0.14* |
| Thr | ACN | −0.33* | 0.76* | −0.56* | −0.27* |
| Ala | GCN | −0.17* | 0.79* | −0.60* | −0.40* |
| Gly | GGN | −0.33* | 0.63* | −0.22* | −0.45* |
| Sixfold degenerate | |||||
| Leu | CTN | −0.52* | 0.06 | −0.41* | 0.86* |
| TTR | −0.51* | −0.42* | |||
| Ser | TCN | −0.28* | 0.45* | −0.43* | 0.12* |
| AGY | −0.41* | 0.16* | |||
| Arg | CGN | 0.16* | 0.74* | −0.51* | −0.33* |
| AGR | −0.36* | −0.41* | |||
The numbers shown are the correlation coefficients between the degree of codon usage bias of the individual amino acid, ENC-X, and the frequency of the base in the third position. The largest positive correlation for each amino acid is highlighted in boldface type. ∗, Significance at P < 0.001. R = A or G; Y = T or C.
Gene Length.
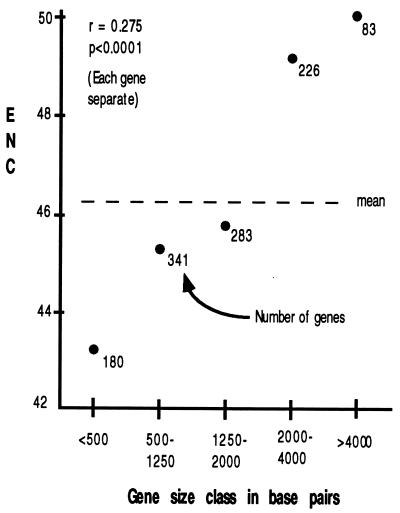
As we discuss below, some explanations of codon usage bias may be affected by the length of a gene. Does the length of a gene in D. melanogaster correlate with the degree of codon bias? To answer this, we need to be certain to use a measure of bias that itself is not biased by sample size (i.e., the number of codons in a gene). Wright (4) performed simulation studies on ENC and found little or no detectable bias with sample size; we have confirmed this finding (E.N.M., unpublished data). Fig. 4 summarizes the relationship between gene length and codon usage bias: smaller genes tend to have higher bias than do longer genes.
Figure 4.
Correlation between gene length (exons only) and codon usage bias. The correlation coefficient was calculated by using each gene separately, while the graph was constructed by lumping genes into size classes for visual clarity.
Recombination.
There is also an effect of the level of recombination on the level of codon usage bias of Drosophila genes: genes in regions of low recombination tend to have low bias (11). This is attributed to the fact that selection can act more effectively at single loci or nucleotide positions when recombination is high, the so-called Hill–Robertson (12) effect.
Causes
Mutation Bias.
There is evidence that mutation bias may affect codon usage in warm-blooded vertebrates that have mosaic genomes consisting of long stretches of A+T-rich DNA interspersed with long stretches of G+C-rich DNA. This isochore structure, as it is termed (13), is thought to be due to regional differences in mutation bias (14, 15). The observation is that genes in A+T-rich isochores tend to have A+T predominantly at silent sites, while genes in G+C-rich isochores have G+C more often at silent sites (16, 17). This is shown by a correlation between base content of introns and the exons of the same gene.
Could mutation bias account for codon usage bias in Drosophila? For three reasons this seems to be very unlikely. First, Drosophila have no isochores (18). As noted above, very tightly linked genes can be very different in codon usage bias, notably Adh and Adhr, which are only a few hundred base pairs apart, and the very dense Sry cluster. There is no correlation between base content of introns and exons of a gene (9) or only weak correlation with twofold degenerate amino acids (19); in the latter study the correlations were greatest for weakly biased genes, the opposite of what would be expected if mutation bias was a cause of codon usage bias.
A second point is that mutation bias in Drosophila is probably toward A+T, whereas codon usage bias increases with use of G and C. As we noted above, genes in regions of low recombination have less codon usage bias than genes in regions of high recombination, a phenomenon thought to be due to the ineffectiveness of selection at individual nucleotide sites. The extreme example of low recombination is the dot fourth chromosome of D. melanogaster, which exhibits no recombination. Thus, in the absence of effective selection, genes on the fourth chromosome should display base composition at synonymous sites reflective of mutation bias. The bottom line in Table 2 shows the base composition at synonymous sites for the seven fourth chromosome genes available for D. melanogaster. A and T are the most common bases at both two- and fourfold degenerate sites. Consistent with A+T mutation bias is the further observation that introns have higher A+T content than do exons in Drosophila (9, 19).
A third argument against mutation bias as a major cause of codon usage bias is that not all amino acids display the same pattern of bias. For example, if mutation bias toward C is why highly biased genes have C most frequently at synonymous sites (Table 2), then all amino acids should show this bias. But as shown in Table 3, two amino acids, Val and Leu, increase in use of G as bias increases.
Taken together, these three observations make it unlikely that mutation bias is playing a large role in maintaining codon usage in Drosophila. In the absence of such bias, we are left with some form of selection as an explanation.
Selection for Codon Usage.
The most plausible and well-documented selection-based explanation for codon usage bias is selection for efficient translation related to the relative abundance of isoaccepting tRNAs (20, 21). The evidence for this comes primarily from microorganisms, namely bacteria and yeast. Preferred codons are those that can base pair optimally with the most abundant tRNA. Generally this involves Watson–Crick pairing or, when bases are modified in the tRNA, some modifications in optimal binding occur. Codon usage bias in microorganisms is well explained by what have been called “Ikemura’s rules” (21) describing optimal binding. There are two observations which support selection for efficient translation in microorganisms: highly expressed genes have greater codon bias (presumably because selection is more intense for efficient translation of such genes), and the relative abundance of isoaccepting tRNAs do match very well the codon usage. Is there evidence that a similar mode of selection could be operating in Drosophila?
Highly expressed genes in Drosophila do tend to be highly biased in codon usage. Among the approximately 10% highest biased genes can be found larval serum proteins, larval and adult cuticle proteins, yolk proteins, chorions, actins, alcohol dehydrogenase, superoxide dismutase, lysozymes, amylases, and α- and β-tubulins; 23 of the 26 known ribosomal proteins are also in this group. Genes which have two copies that differ in level of expression have more codon usage bias in the more highly expressed copy (3).
Also similar to the situation in microorganisms, what is known of relative levels of isoaccepting tRNAs matches quite well the codon bias in Drosophila (Table 4, using data from refs. 22 and 23). Eleven of the 18 amino acids with redundant codons are shown here; for the other 7 amino acids, the anticodons of the most abundant isoaccepting tRNAs are not known. However, for 5 (Pro, Thr, Ala, Leu, and Ile) nuclear copies of tRNA with optimal binding are known, although their relative abundance has not been studied. No tRNA sequences are known for the two remaining amino acids, Cys and Gln. Therefore, in every case where relative abundance of isoaccepting tRNAs is known, there is a very good match with the most used codons.
Table 4.
Most abundant isoaccepting tRNA and preferred codons
| Amino acid | Preferred codon in highly biased genes | Most abundant tRNA anticodon
|
||
|---|---|---|---|---|
| Adult | Third instar | First instar | ||
| Val | GUG | CAC | CAC | CAC |
| Gly | GGC | GCC | GCC | GCC |
| Ser | UCC | IGA | IGA | IGA |
| Tyr | UAC | GUA | GUA | GUA |
| His | CAC | GUG | GUG | GUG |
| Asn | AAC | ? | ? | GUU |
| Asp | GAC | QUC | GUC | GUC |
| Lys | AAG | CUU | CUU | CUU |
| Glu | GAG | SUC | SUC | ? |
| Phe | UUC | GmAA | GmAA | GmAA |
| Arg | CGC | A*CG | A*CG | A*CG |
I = inosine, which binds optimally to C and U; Q = queuosine, which binds C and T about equally; S = 2-thiouridine, which binds optimally to A and G; and Gm = 2′-O-methylguanosine, which binds optimally with C in A/T-A/T-Y codons. Optimal binding rules follow Ikemura (21). Information on tRNAs is from refs. 22 and 23.
Determined from nuclear copy; most often A is modified to I in first anticodon position.
The tRNA pool/translational efficiency hypothesis to explain codon usage bias in microorganisms is thought to be due to the fact that all genes in these single-celled organisms share the same tRNA pool, and thus converge on a single optimal codon usage pattern. In multicellular eukaryotes, it is conceivable that different tissue types have different tRNA pools, perhaps adjusted to match codon usage of genes most expressed in each tissue. The evidence from Drosophila does not indicate tissue specificity in either level or pattern of codon usage bias. For example the avoidance of the three codons noted in Table 1—i.e., AUA, GGG, and UUA—is shared by genes expressed primarily in the fat body (Adh), in ovarian egg chambers (chorions), and in the midgut (Amylase), as well as occurring in genes expressed in all cell types—e.g., myosin and Glyceraldehyde-3-phosphate dehydrogenase (unpublished data).
If codon usage in Drosophila is explained by selection for efficient translation, is this consistent with the length effect documented above? Mutations to nonoptimal codons will have a greater relative effect in smaller genes compared with larger genes. Assume a nonoptimal codon requires twice as long to incorporate an amino acid as does the optimal codon. In a short gene, with say 100 codons, a mutation to a nonoptimal codon from an optimal one will increase translation time by 1%, whereas a similar mutation in a gene with 1,000 codons would increase translation time by only 0.1%. Alternatively, the length effect could be explained by the fact that highly expressed genes tend to be short. With the present data it is impossible to eliminate either explanation.
Efficiency of translation has two interrelated effects. First is speed of translation, presumably especially important for highly and/or rapidly expressed genes. A second aspect affected by codon usage is accuracy of translation—i.e., the relative rate of misincorporation of amino acids. It is known that, at least in microorganisms, nonoptimal codons misincorporate more frequently than do optimal codons (24, 25). The issue of selection for speed vs. accuracy is very difficult to disentangle, and both may well be occurring. Most misincorporation is thought to occur during the waiting time for the “search” for the ternary complex (aminoacyl-tRNA–elongation factor Tu–GTP) matching the codon being translated; the longer the wait, the higher the probability of misincorporation. Therefore genes translated fast are also translated more accurately. Akashi (26) has argued that selection for accuracy may account for at least some of the codon bias in Drosophila. He reasoned that selection would be greatest for misincorporation of amino acids at crucial functional sites in a protein. He identified such amino acids by evolutionary conservation and found that conserved amino acids had a tendency to have greater codon usage bias than do amino acids that vary across species. This may account for some codon usage bias, but it cannot explain patterns such as in Adh, where certain codons are avoided throughout a gene (Table 1).
Finally, in this section we note that the tRNA abundance/efficiency of translation hypothesis can account for the maintenance of codon usage bias in Drosophila, but this still begs the question of the origin of the bias. Did genes evolve to match tRNA pools? Or did tRNA pools evolve to match codon usage of genes? This is something of a “chicken or egg first” question which cannot be answered. It is conceivable that other factors could have initiated the codon usage bias which subsequently led to selection for adjustment of the relative levels of isoaccepting tRNAs; this could set up a feedback cycle. It seems unlikely in Drosophila that mutation bias could have been the initiating factor because, as we argued above, mutation bias is toward A+T, while codon bias is toward G+C. However, factors such as transcriptional efficiency and mRNA stability and processing could also potentially cause codon usage bias and be initiators of the selection for adjustment of tRNA pools.
Effects of Codon Usage Bias on DNA Evolution
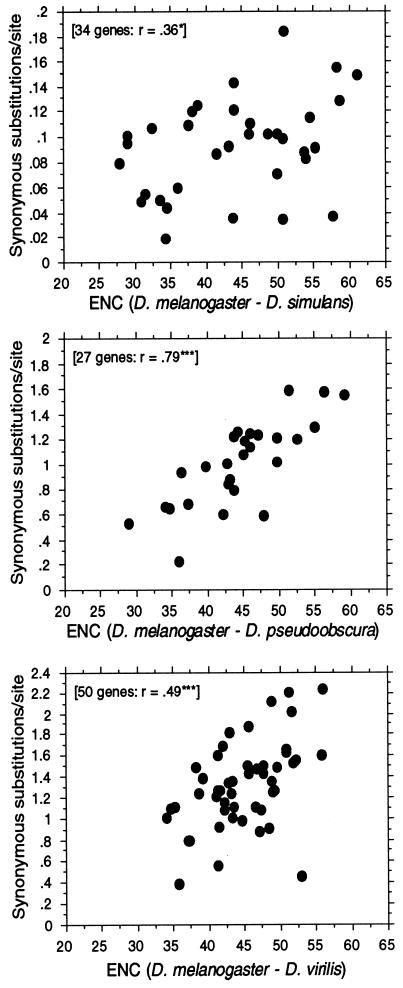
If selection constrains codon usage to differing degrees in different genes, then we expect to see some differences in evolutionary rates of change at synonymous sites. Genes with little or no codon usage bias should exhibit a higher Ks than do genes with high codon usage bias. This has been shown to be the case in both bacteria (27) and Drosophila (28). Here we update this observation by including many more genes than previously available as well as make three species comparisons at different levels of divergence (Fig. 5). Furthermore, we have developed a method (37) for estimating synonymous substitutions that corrects for base composition, and thus the patterns in Fig. 5 are not simply due to artifacts caused by base composition differences among genes. The expected correlations are best observed for the more distant pairs of D. melanogaster–D. pseudoobscura and D. melanogaster–D. virilis. We suspect the rather large scatter and weak correlation for the D. melanogaster–D. simulans pair is due simply to noise in the data because the divergence time is quite short for this pair, the Ks being on average about an order of magnitude lower than for the other two pairs. Also, it is possible sharing of polymorphic alleles in closely related species may also be obscuring the picture.
Figure 5.
Correlation between synonymous DNA divergence between species of Drosophila and codon usage bias of a gene. Numbers of synonymous substitutions per synonymous site were calculated by a method of Moriyama and Powell (37). The mean ENC of the gene from the two species was used. ∗, P = 0.05; ∗∗∗, P = 0.001.
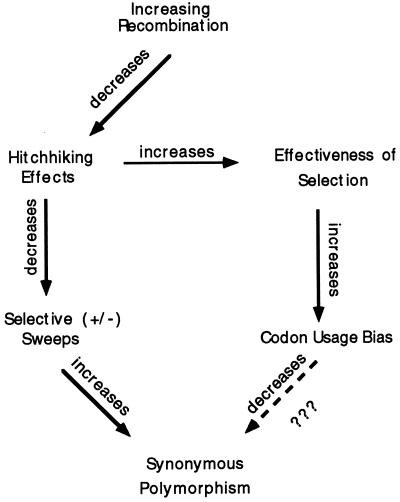
Does a similar phenomenon occur for intraspecific polymorphisms—i.e., do more highly biased genes have less synonymous polymorphism within a species? We observed the opposite for 21 genes in D. melanogaster for which data on intraspecific variation were available (29): there is a statistically significant positive correlation between codon usage bias and level of synonymous polymorphism in a gene. How can this be explained? We speculate this may be due to the effect of variation in recombination. Fig. 6 outlines the argument. Genes vary in their levels of recombination dependent upon position in the genome. Increased recombination can have two effects, one of which is to increase codon usage bias (19) and the other is to increase synonymous polymorphisms (30). Both are due to a decrease in hitchhiking effects of linked genes. As mentioned previously, selection at single nucleotide positions is more effective in regions of high recombination, thus allowing for an increase in selection for optimal codons. The effect of recombination on levels of synonymous polymorphism is thought to be due to selective “sweeps” at linked positions; such sweeps take along with them linked sites which then become less variable. Such selection may be positive when a new linked favorable mutation arises and goes to fixation (31), or may be due to negative “background selection” against deleterious mutations (32); it is not clear which of these processes best fits the data, but their effects are similar. From the observation in D. melanogaster that highly biased genes tend to have higher synonymous polymorphism, the arrows at the bottom of Fig. 6 would seem not to have equal strength in their effects. The expected decrease in synonymous polymorphism caused by codon usage bias is not great enough to overcome the expected increase in such polymorphisms due to lessening effects of selective sweeps.
Figure 6.
Schematic hypothesis of why, in D. melanogaster, synonymous polymorphisms are not less in genes with higher codon usage bias.
Conclusions
While the information available on codon usage bias of both microorganisms and Drosophila provides good evidence that selection can act on what had been considered prime candidates for neutral mutations, are all synonymous substitutions detected at all times? This is highly unlikely, and we argue elsewhere that there is likely a continuum in Drosophila (and other organisms) with codon usage in highly biased genes being primarily affected by selection, whereas other genes may have codon usage controlled primarily by mutation and drift along the lines of models previously proposed (33, 34). This is in agreement with Akashi’s (35) observation that the selection for optimal codons in D. simulans has been more effective than in D. melanogaster. D. simulans is thought to have an effective population size greater than D. melanogaster, so the selection coefficients on synonymous mutations (at least on some genes) are sufficiently small as to be sensitive to population size differences among species of Drosophila. This implies the selection coefficients on synonymous mutations are on the order of Nes equal to 1 (Ne is the effective population size and s is the selection coefficient), consistent with previous studies (35, 36). Further, we note that when selection is ineffective due to reduced recombination, codon usage may well reflect mutation/drift (Table 2). Nevertheless, at times selection for codon usage must be very effective, as exemplified by the phylogenetic persistence of avoidance of specific codons in specific genes for very long evolutionary periods (Table 1).
Acknowledgments
We thank the organizers of this colloquium, Francisco J. Ayala and Walter M. Fitch, for the opportunity to commemorate Theodosius Dobzhansky and the publication of arguably the most important book on evolution in the 20th Century. We appreciate the helpful review provided by Charles F. Aquadro. This work was supported by National Science Foundation Grant DEB9318836.
ABBREVIATION
- ENC
effective number of codons
References
- 1.King J L, Jukes T H. Science. 1969;164:788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- 2.Richmond R. Nature (London) 1970;225:1025–1028. doi: 10.1038/2251025a0. [DOI] [PubMed] [Google Scholar]
- 3.Shields D C, Sharp P M, Higgins D G, Wright F. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 4.Wright F. Gene. 1990;87:23–39. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 5.Ikemura T. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 6.Sharp P M, Li W-H. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powell J R, DeSalle R. Evol Biol. 1995;28:87–138. [Google Scholar]
- 8.Ibnsouda S, Schweisguth F, de Billy G, Vincent A. Development (Cambridge, UK) 1993;119:471–483. doi: 10.1242/dev.119.2.471. [DOI] [PubMed] [Google Scholar]
- 9.Moriyama E N, Hartl D L. Genetics. 1993;134:847–858. doi: 10.1093/genetics/134.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharp P M, Lloyd A T. In: An Atlas of Drosophila Genes. Moroni G, editor. New York: Oxford Univ. Press; 1993. pp. 378–397. [Google Scholar]
- 11.Kliman R M, Hey J. Mol Biol Evol. 1993;10:1239–1258. doi: 10.1093/oxfordjournals.molbev.a040074. [DOI] [PubMed] [Google Scholar]
- 12.Hill W G, Robertson A. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- 13.Bernardi G, Olofsson B, Filipski J, Zerial M, Salinas J, Cuny G, Meunier-Rotival M, Fodier F. Science. 1985;228:953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- 14.Filipski J. FEBS Lett. 1987;217:184–186. doi: 10.1016/0014-5793(87)80660-9. [DOI] [PubMed] [Google Scholar]
- 15.Wolfe K H, Sharp P M, Li W-H. Nature (London) 1989;337:283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- 16.Bernardi G, Bernardi G. J Mol Evol. 1986;24:1–11. doi: 10.1007/BF02099946. [DOI] [PubMed] [Google Scholar]
- 17.Aota S, Ikemura T. Nucleic Acids Res. 1986;14:6345–6355. doi: 10.1093/nar/14.16.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiery J-P, Macaya G, Bernardi G. J Mol Biol. 1976;108:219–235. doi: 10.1016/s0022-2836(76)80104-0. [DOI] [PubMed] [Google Scholar]
- 19.Kliman R M, Hey J. Genetics. 1994;137:1049–1056. doi: 10.1093/genetics/137.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosjean H, Fiers W. Gene. 1982;18:199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- 21.Ikemura T. In: Transfer RNA in Protein Synthesis. Hatfield D L, Lee B J, Pirtle R M, editors. Boca Raton, FL: CRC; 1992. pp. 87–111. [Google Scholar]
- 22.White B N, Tener G M, Holden J, Suzuki D T. Dev Biol. 1973;33:185–195. doi: 10.1016/0012-1606(73)90173-5. [DOI] [PubMed] [Google Scholar]
- 23.Sprinzl M, Steegborn, Bübel F, Steinber S. Nucleic Acids Res. 1996;24:68–72. doi: 10.1093/nar/24.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Precup J, Parker J. J Biol Chem. 1987;262:11351–11356. [PubMed] [Google Scholar]
- 25.Dix D B, Thompson R C. Proc Natl Acad Sci USA. 1989;86:6888–6892. doi: 10.1073/pnas.86.18.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akashi H. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharp P M, Li W-H. Mol Biol Evol. 1987;4:222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- 28.Sharp P M, Li W-H. J Mol Evol. 1989;29:398–402. doi: 10.1007/BF02603075. [DOI] [PubMed] [Google Scholar]
- 29.Moriyama E N, Powell J R. Mol Biol Evol. 1996;13:261–277. doi: 10.1093/oxfordjournals.molbev.a025563. [DOI] [PubMed] [Google Scholar]
- 30.Begun D J, Aquadro C F. Nature (London) 1993;365:548–550. doi: 10.1038/365548a0. [DOI] [PubMed] [Google Scholar]
- 31.Hudson R R. Proc Natl Acad Sci USA. 1994;91:6815–6818. doi: 10.1073/pnas.91.15.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlesworth B, Morgan M T, Charlesworth D. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp P M, Li W-H. J Mol Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 34.Bulmer M. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akashi H. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartl D L, Moriyama E N, Sawyer S A. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriyama, E. N. & Powell, J. R. J. Mol. Evol., in press.