Abstract
Our manipulation of the nonsense-mediated decay pathway in microsatellite unstable colon cancer cell lines identified the p300 gene as a potential tumor suppressor in this subtype of cancer. Here, we have demonstrated that not only the p300 gene but also the highly homologous cAMP-response element-binding protein (CREB) binding protein (CBP) gene together are mutated in >85% of microsatellite instability (MSI)+ colon cancer cell lines. A limited survey of primary tumors with MSI+ shows that p300 is also frequently mutated in these cancers, demonstrating that these mutations are not consequences of in vitro growth. The mutations in both genes occur frequently in mononucleotide repeats that generate premature stop codons. Reintroduction of p300 into MSI colon cancer cells could only be supported in the presence of an inactivated CBP gene, suggesting the idea that one or the other function must be inactivated for cancer cell viability. p300 is known to acetylate p53 in response to DNA damage, and when MSI+ cells null for p300 activity are forced to reexpress exogenous p300 cells show slower growth and a flatter morphology. p53 acetylation is increased upon reexpression of p300, suggesting that MSI+ cells constitutively activate the DNA damage response pathway in the absence of DNA-damaging agents. In support of this hypothesis, c-ABL kinase, which is also activated in response to DNA damage, shows higher levels of basal kinase activity in MSI+ cells. These observations suggest that there is a selective growth/survival advantage to mutational inactivation of p300/CBP in cells with inactivated mismatch repair capabilities.
Tumors from patients with hereditary nonpolyposis colon cancer and ≈15% of sporadic cancers of the gastrointestinal tract demonstrate microsatellite instability (MSI) that results in elevated mutation rates and especially high rates of insertion/deletion mutations at microsatellite sequences (1–3). MSI has been shown to be a result of mutations in the DNA mismatch repair (MMR) gene family (4–6), whose function is to correct DNA replication errors. Tumors with MSI have distinctive phenotypic characteristics: they are located at the right side of the colon, are poorly differentiated, and have a near-diploid chromosome number. Defects in MMR are also associated with resistance to some DNA-damaging drugs such as methylating agents and cisplatin (7).
Tumors with MSI progress through a distinctive genetic pathway, because the genes mutated in these cancers are generally different from those in cancers without MSI. For example, genes that are frequently mutated in MSI-negative colorectal cancer, such as p53, K-ras, and APC, are less frequently mutated in MSI+ tumors (1). Mutations in MSI+ tumors almost invariably occur in mononucleotide repeats within the exons. Deletions of single nucleotides within these repeats cause frameshift mutations in these coding microsatellites, which result in the generation of premature stop codons. Frameshift mutations in coding microsatellites have been reported in genes such as TGFβRII, IGFIIR, and PTEN, which are related to growth control (8–10), BAX and Caspase 5, which are involved in apoptosis (11, 12), hMSH6, hMSH3, and MBD4, which are involved in DNA repair (13, 14), and the transcriptional regulator TCF-4 (15). The frequency of mutations depends on the length of the microsatellite repeat and the extent of the growth advantage provided to the cells by the mutations (16). A shift in the translational reading frame caused by insertion/deletion mutations inevitably results in the inactivation of the normal function of the encoded proteins. This observation, therefore, suggests that where a high frequency of frameshift mutations occurs in a gene it may have a tumor suppressor function. Indeed, except for DNA repair genes, all other genes found frequently mutated in cancers with MSI are negative regulators of cell growth, either as tumor suppressor genes, such as TGFβRII and PTEN, or proapoptotic genes such as BAX and Caspase 5.
Frameshift mutations inevitably result in the appearance of the stop codons from the generation of an alternative reading frame, which frequently leads to the rapid degradation of the mutant mRNA through a mechanism known as nonsense-mediated decay (NMD) (17–19). Recently, a strategy named gene identification by NMD inhibition has been proposed for the identification of genes carrying nonsense mutations (20). This procedure uses drug combinations to inhibit translation that is normally essential for NMD to occur. As a result, mRNA molecules avoid degradation and can then be detected with expression microarrays. Using a modification of this approach we recently found that in the MSI+ colorectal cell line RKO two frameshift mutations were present in the coding microsatellite repeats in exons 3 and 27 of the p300 gene (21). p300 is a histone acetyltransferase that regulates transcription via histone acetylation and is known to acetylate p53 in response to DNA damage (22). DNA damage-induced p53 acetylation is thought to stimulate its ability to bind to DNA in a sequence-specific manner and enhance its transcription, resulting in growth arrest and/or apoptosis. Here, we describe the frequent mutation of p300 in MSI+ colon cancer cells and the homologous cAMP-response element-binding protein (CREB) binding protein (CBP) gene. Reintroduction of p300 into cells null for its activity results in flattening of the cells, a reduction in growth rate, and increased p53 acetylation. From these data we now suggest the significance of the mutational inactivation of p300 for colon cancer cell lines with MSI.
Materials and Methods
Cell Culture, Transfections, and Western Blotting. Colon cancer cell lines were grown in DMEM supplemented with 10% FBS and antibiotics. Stable transfection of HCT15 and RKO cells were performed in 30-mm plates. p300 expression plasmid (0.8 μg; Upstate Biotechnology, Lake Placid, NY) linearized by treatment with PvuI endonuclease was mixed with 0.1 μg of pCDNA3.1 plasmid (Invitrogen) and transfected with Effectene reagent (Qiagen, Chatsworth, CA). Stable clones were selected by growing in medium containing 1 mg/ml G418 (GIBCO). For Western blot analysis of p300 and CBP expression, nuclear extracts were prepared by using NE-PER nuclear and cytoplasmic extraction reagents (Pierce). Protein concentration was measured by using Bio-Rad dye-binding assays, and 20 μg of nuclear extracts was run on 8% SDS/PAGE. The separated proteins were transferred onto poly(vinylidene difluoride) membranes (Immobilon P, Millipore), blocked with skim milk, and incubated with anti-N-terminal anti-p300 or anti-CBP antibodies (Santa Cruz Biotechnology) for 2 h at room temperature. Antigen–antibody complexes were detected by secondary anti-rabbit antibody followed by enhanced chemiluminescence.
CBP Mutation Analysis. PCR primers flanking the overlapping fragments of exon 31 of CBP were used to amplify this exon from genomic DNA isolated from cell lines. The amplified fragments of exon 31 were sequenced by using an Applied Biosystems Prism Sequencer. The sequences of primers that flank the frameshift mutation in the SW48 and Lovo cells are 5′-TCTGCCTTCTCCTACCTCAGCAC (forward) and 5′-ATTCAGGCTCACGGGGGCCATC (reverse).
c-Abl Kinase Assay. c-Abl tyrosine kinase activity was measured in an immune complex by using as a substrate CTD-CRK fusion protein generously provided by J. Y. J. Wang (University of California at San Diego). The HCT116+Ch3 and HCT116+Ch2 cells generously provided by C. R. Boland (University of California at San Diego) were lysed in RIPA buffer containing 25 mM β-glycerophosphate, 1 mM orthovanadate, 10 mM sodium fluoride, and 1 mM DTT, and the lysates were incubated with ant-c-Abl K-12 agarose (Santa Cruz Biotechnology) for 2 h at 4°C. Immune complexes were washed three times in the lysis buffer and twice in kinase buffer (50 mM Tris·HCl, pH 7.5 with 10 mM MgCl2 25 mM β-glycerophosphate, 1 mM orthovanadate, 10 mM sodium fluoride, and 1 mM DTT). The washed beads were resuspended in kinase buffer with 20 μCi of [γ-32P]ATP, 10 μM cold ATP, and 3 μg of CTD-CRK substrate and incubated 30 min at room temperature. The phosphorylated proteins were visualized by using a PhosphorImager (Molecular Dynamics). Equal amounts of the c-Abl protein in immunoprecipitates were confirmed by Western blot analysis using K-12 anti-c-Abl antibodies (Santa Cruz Biotechnology).
Results
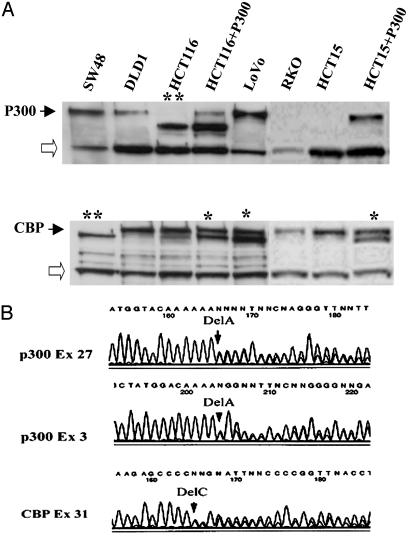
A High Frequency of p300/CBP Frameshift Mutations in Colon Cancer Cell Lines with MSI. Our previous studies used inhibition of NMD to detect genes carrying premature stop codons. In the MSI+ colorectal cancer cell line, RKO, we found two frameshift mutations in the p300 gene in the (A)5 and (A)7 coding microsatellite repeats of exons 3 and 27, respectively (21). When we analyzed this cell line by using Western blotting, no evidence of the p300 protein could be found in nuclear extracts (Fig. 1A), which demonstrated that these mutations inactivated both alleles of the p300 gene. To determine whether mutations in p300 are common events in MSI+ colon cancer cells, we performed sequence analysis of all of the individual exons of p300 in seven other MSI+ colon cancer cell lines (LS180, SW48, LoVo, DLD1, HCT15, RKO, and HCT116). Of these, four cell lines (HCT15, DLD1, RKO, and HCT116) were also shown to carry truncating mutations in the p300 gene (57%). An identical G4239T mutation in exon 16 was found in DLD1 and HCT15, which results in an E1039X premature stop codon. This mutation was homozygous in HCT15 but heterozygous in DLD1. No mutation was found in the other allele in DLD1. In HCT116, the p300 mutation constituted a deletion of nucleotide 6294A in an A(5) repeat in exon 31. When these cells were similarly subjected to Western blot analysis (Fig. 1 A), HCT15, like RKO, showed no protein, whereas HCT116 demonstrated a smaller protein product. The mutant mRNA escapes NMD because the mutation occurs in the last exon of the gene. The presence of the normal-sized protein in DLD1 demonstrates that one normal copy of the gene is present.
Fig. 1.
(A) Western blot analysis of nuclear extracts from MSI+ colorectal cancer cell lines probed with anti-N-terminal p300 and CBP antibodies (Santa Cruz Biotechnology). RKO and HCT15 do not express the p300 protein. Open arrows show nonspecific bands. A double star indicates truncated protein caused by homozygous frameshift mutations in the last exons of p300 and CBP; a single star indicates double protein bands caused by heterozygous mutation. (B) Sequence chromatograms of exons 3 and 27 of the p300 gene from genomic DNA derived from RKO cells and exon 31 of CBP gene from LoVo cells show heterozygous deletions of one adenine in (A)5 and (A)7 repeats and one cytosine in (C)5 repeat, respectively.
CBP is another protein that possesses intrinsic acetylase activity and shares significant homology with p300 in its functional domains. Because the functions of CBP often overlap with those of p300 (23), we determined whether CBP was also mutated in MSI+ colon cancer cell lines. First, using Western blotting, we analyzed the presence of CBP in colon cancer cell lines with MSI. A truncated form of CBP was seen in LoVo, which also contained a normal-sized product. In SW48 cells (Fig. 1 A) there was also a single smaller-sized protein. The presence of a truncated protein suggested that the nonsense mutation occurred within the last exon of the gene, which again avoids NMD. Sequencing this exon identified a heterozygous deletion of one cytosine in the (C)5 repeat (nucleotides 5911–5915 of the CBP cDNA) in LoVo and a homozygous (or hemizygous) deletion at the same site in SW48 cells. No mutations in either p300 or CBP were detected by using either sequencing or Western blotting in the LS180 cell line. Taken together, therefore, mutations in either the CBP or p300 genes were present in 86% (6/7) of the MSI+ colon cancer cell lines analyzed. Importantly, the mutations in these two genes were frequently mutually exclusive, demonstrating that the function of these genes is compromised in all but LS180 cells.
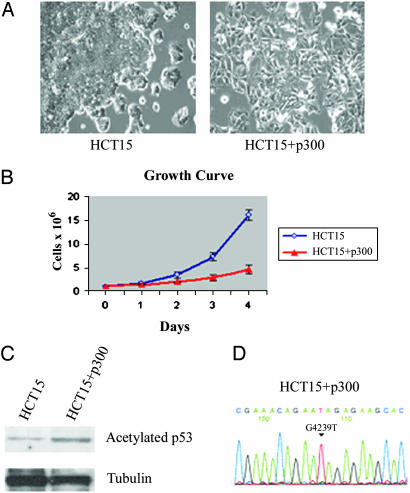
Reexpression of p300 in Colon Cancer Cells Results in Slower Growth and Higher Level of Acetylated p53. To determine the functional significance of the homozygous inactivation of p300 in RKO and HCT15 cells, we transfected a WT copy of p300 into these cells. More than 100 G418-resistant colonies were generated from RKO compared with only 2 colonies from HCT15. Western blot analysis of nuclear extracts from 12 of the RKO clones revealed that none of them expressed exogenous p300 (data not shown). This result suggests that mutational inactivation of p300 in RKO cells provides a growth advantage, such that only G418-resistant RKO cells that are unable to express p300 can grow. In contrast, both HCT15 clones expressed p300 (Fig. 1 A and data not shown). Interestingly, these HCT15 clones also carried a truncated form of CBP, which was caused by the heterozygous deletion of one cytosine in a (C)5 repeat in the last exon of the CBP gene, which is identical to the mutation seen in LoVo. It is possible that truncated CBP could have a dominant negative function, making cells less sensitive to the restoration of p300 activity. To exclude the possibility that these clones arose from a small subpopulation within the HCT15 parental cells that expressed endogenous p300, or from contamination of the HCT15 cell line with LoVo cells for example, we analyzed the genomic DNA of the p300-expressing HCT15 clones for the previously reported G4239T homozygous mutation in p300 in this cell line (24). The sequence of PCR-amplified exon 16 demonstrated the presence of the homozygous G>T mutation in all cases (Fig. 2D), which confirmed the ectopic expression of p300 in the transfected HCT15 clones. Sequencing exons 4 and 6 of the MSH6 gene from the HCT15 cells that expressed the ectopic p300 protein identified the previously reported mutations in this gene in HCT15 cells (data not shown), which confirmed that the p300-expressing clones were not derived from contaminating cells. Phenotypically, the HCT15 clones expressing WT p300 grew more slowly (Fig. 2B) and were morphologically different from the cells transfected with the vector alone, in that they were larger in size and had a more flat appearance (Fig. 2 A).
Fig. 2.
Ectopic expression of the p300 protein in HCT15 cells results in changes in cell size and shape, slower growth, and a higher level of acetylated p53. (A) The HCT15 cells (Right) cotransfected with the p300 expression plasmid (Upstate Biotechnology) together with the pCDNA3.1 vector have a larger size and a more flat appearance than the same cells transfected with the pCDNA3.1 vector alone (Left) (see Materials and Methods). (B) Cells (105) of the mixtures of the p300-expressing HCT15 clones (HCT15+p300) or HCT15 cells transfected with the pCDNA3.1 vector were seeded in 60-mm plates in triplicate, and the cell numbers were counted at the indicated times. Standard errors were calculated on the basis of triplicate measurements. The reduction in growth rate with the addition of p300 is clearly seen. (C) Western blot analysis of nuclear extracts from the HCT15+p300 cells and the HCT15 cells transfected with the empty vector probed with antiacetylated-p53 antibodies (Upper). The blot was then stripped of antibodies by using Pierce stripping reagent and reprobed with antitubulin antibodies. An ≈3-fold increase in acetylated p53 is seen (Lower). (D) Sequence chromatogram of exon 16 from the HCT15+p300 genomic DNA shows the homozygous G4239T substitution resulting in E1013X nonsense mutation.
Because HCT116 cells also express a truncated P300 protein lacking the carboxyl end, we analyzed the effect of reintroduction of WT p300 into these cells. Western blot analysis of nuclear extracts of nine G418-resistant clones of HCT116 cells after stable transfection of the p300 expression vector identified one clone that expressed the p300 protein. The phenotype of this clone was the same as that of the parental HCT116 cells. However, the level of expression of the WT p300 protein was much lower than that of the truncated mutant p300 protein (Fig. 1 A), which might explain why the phenotype was unchanged and again suggests that MSI+ cells resist the full restoration of p300 activity. In addition, this clone had a heterozygous truncating mutation in the last exon of the CBP gene (Fig. 1 A). The absence of this mutation in the remaining eight clones from HCT116 cells, which did not express exogenous p300 (data not shown), implies that only a minor fraction of the HCT116 cells that harbor the mutation in the CBP gene tolerates the introduction of a functional p300 gene. This observation provides further support that reduction of p300/CBP activity is beneficial for cells with MSI. A minor band corresponding to the truncated CBP, which can be seen on the Western blot of the nuclear extract from HCT116 cells (Fig. 1 A), indicates the presence of a CBP frameshift mutation in a small fraction of HCT116 cells.
Increased Acetylation of p53 in the Presence of p300. p300 is known to acetylate p53 in response to DNA damage (25), which is thought to result in p53 stabilization. We, therefore, compared the endogenous level of p53 acetylation in the HCT15 cells transfected with the empty vector with the level seen in the p300-transfected cells. Those cells ectopically expressing the p300 protein had a higher level of acetylated p53 than the HCT15 cells transfected with the empty vector (Fig. 2C). A higher acetylation of p53 in the p300-expressing cells, in the absence of the genotoxic stress, possibly suggests that, in cell lines with MSI, the DNA damage response pathway may be constitutively activated in the absence of DNA damage-inducing treatment. Such activation could be caused by multiple insertion/deletion DNA loops that occur at the sites of microsatellite sequences during DNA replication and remain unrepaired in cell lines with MSI. These insertion/deletion mismatches may imitate DNA damage and induce the DNA damage response, as suggested for DNA lesions produced by cisplatin or alkylating agent treatments.
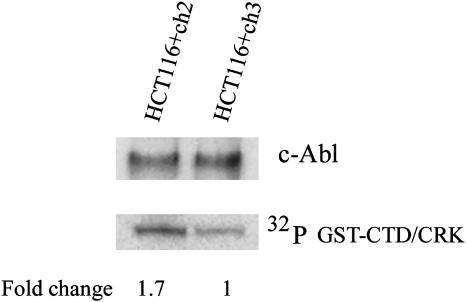
Analysis of c-Abl Kinase Activity in HCT116 Cells. The ubiquitously expressed c-Abl tyrosine kinase is known to be involved in the stress response to DNA-damaging agents (26). Ionizing radiation and alkylating agents such as cis-platinum and mitomycin C have been shown to activate c-Abl (27). To test the hypothesis that MSI+ cells have constitutively activated the DNA damage response pathway, we analyzed whether MSI+ cells elevated c-abl kinase activity in the absence of genotoxic stress. We compared the c-Abl kinase activity in the absence of DNA damage in an MMR-deficient clone of HCT116 cells with that in a clone of HCT116 cells where MMR had been restored by introduction of a WT copy of the hMLH1 gene (28). An in vitro kinase assay using immunoprecipitated c-Abl showed that, in the absence of DNA damage, c-Abl kinase activity was higher in MMR-deficient HCT116 cells (Fig. 3). This result shows that the inactivation of MMR may induce a DNA damage response pathway and that mutational inactivation of genes involved in the DNA damage response, including the p300 gene, could provide selective advantages to the cells with MSI.
Fig. 3.
A higher c-Abl activity in MMR-deficient HCT116 cells. MMR-proficient HCT116+Ch3 cells, which have a functional hMLH1 gene as a result of the introduction of a normal chromosome 3, and control MMR-deficient HCT116+Ch2 cells, which contain transferred chromosome 2, were lysed and c-Abl kinase was immunoprecipitated with an anti-c-Abl K-12 antibody (Santa Cruz Biotechnology). (Lower) Tyrosine kinase activity in c-Abl immunoprecipitates was measured in an immune complex kinase asssay by using a GST-CTD/CRK fusion protein as a substrate. The experiment was repeated three times, and similar results were obtained in each case; differences in activity were quantitated with a PhosphorImager (Molecular Dynamics). (Upper) Equal amounts of c-Abl protein in immunoprecipitates were confirmed by Western blot analysis using K-12 anti-c-Abl antibodies.
Discussion
In this article we have demonstrated that the vast majority of MSI+ colon cancer cell lines carry mutations in either the p300 or CBP genes. This observation strongly suggests that it is important for the progression of these MSI+ cells to inactivate the pathways that are regulated by these highly similar proteins. The high frequency of mutations of the p300/CBP genes can be explained in several ways. First, it may simply be caused by the generalized increase of insertion/deletion mutations in microsatellites in MSI+ cells because of the presence of mononucleotide repeats in the coding DNA of p300 and CBP. However, the frameshift mutations in RKO, SW48, and LoVo cells occurred in relatively short (A)5 and (C)5 repeats. This finding is unusual, because frameshift mutations in mononucleotide repeats <8 nt have been rarely reported in MSI+ cells in the absence of a strong selective pressure (11, 14, 29). It seems more likely, therefore, that this high frequency of p300/CBP inactivation in MSI+ cells is caused by a strong selective pressure for loss of their function during tumor progression.
It has been shown that one of the functions of p300/CBP is to regulate the transcriptional activity of p53 (22, 30). Thus, the low frequency of p53 mutations seen in cancers with MSI can be compensated for by mutations in p300/CBP or other genes involved in the p53 pathway. This hypothesis is supported by the observation that mutations in the p300 and CBP genes are absent in LS180 cells that are known not to express p53 (31). Furthermore, HCT15 cells, which carry a mutation in the p53 gene (31), can tolerate the restoration of p300 activity, but RKO cells, which have a functional p53, cannot. It is also noteworthy that frameshift mutations in the CBP gene in LoVo, SW48, and the mutation in the p300 gene in HCT116 cells all occurred at the 5′ end of the very large, last exon of the p300/CBP genes. In this way these mRNAs avoid degradation of the mutant mRNA through the NMD pathway. Thus, even though a partially functional protein is generated, they lack the p53 binding site (32) as a result of the truncations. In our study we have specifically concentrated on MSI+ cell lines that show a high frequency of p300/CBP mutations. In a large survey for mutations in a cohort of tumors that were not selected for MSI+, Gayther and colleagues (33) also described truncating p300 mutations in breast, colon, and pancreas cancer cell lines and primary tumors. Another report demonstrated p300 truncating mutations in only 6 of 107 (5.6%) cell lines regardless of their MSI status (24). It is clear, therefore, that p300 is more frequently mutated in MSI+ colon cancer cell lines than in cell lines without MSI (P < 0.001 by χ test).
The high frequency of p300/CBP mutations could be a result of an involvement of p300 and CBP in the transforming growth factor (TGF)-β pathway (34, 35), which is frequently impaired in MSI+ cancers through the mutational inactivation of the TGFβRII gene (8). However, we did not observe a reverse correlation between p300/CBP and TGFβRII mutations. For example, HCT116 and SW48 both have biallelic inactivation of TGFβRII (16) and the homozygous mutations in p300 and CBP, respectively. This finding implies that inactivation of TGFβRII and p300/CBP affect different pathways in MSI+ cells.
The higher level of p53 acetylation, in the absence of DNA damage in HCT115 cells with restored p300 activity, suggests another possible explanation for the increased frequency of mutations. Thus, in the absence of a functional MMR capability, per se, there is a selective advantage for the mutational inactivation of the p300/CBP proteins. p300 and CBP are known to be involved in the DNA damage response and maintenance of genomic stability (22, 30, 36, 37). Multiple mispairings that occur during DNA replication at the sites of microsatellite DNA, if they remain unrepaired in MSI+ cells, may imitate DNA damage, thus activating the DNA damage response that leads to growth arrest or apoptosis. Mutational inactivation of genes involved in the DNA damage response pathway, therefore, could then provide the growth advantage to cells with inactivated MMR capabilities. The higher level of the c-Abl kinase activity in the HCT116 cells with inactivated MMR, compared with the same cells with functional MMR, in the absence of exogenous DNA damaging agents supports the hypothesis that the inactivation of MMR may induce the DNA damage response. This hypothesis offers an alternative explanation for the link between the inactivation of the MMR and the tolerance to alkylating agents such as MNNG or to cisplatin. Currently, there are two models to explain this tolerance. According to one model the cells with functional MMR are more sensitive to DNA damage because MMR proteins represent an integral part of a DNA damage-inflicted apoptotic pathway (38). According to a second model, the higher sensitivity of the cells with functional MMR to the DNA adducts produced by MNNG or cisplatin is explained by the structural similarity of DNA adducts produced by MNNG or cisplatin to the DNA mismatches that occur during DNA replication. The MMR system recognizes MNNG or cisplatin adducts as a substrate, excises the DNA strand opposite the DNA adduct, and replaces a new DNA strand without repairing the DNA damage. Multiple futile cycles of DNA repair then finally result in double-stranded DNA breaks that induce apoptosis (39). Our hypothesis, which considers the function of the microsatellite DNA to be a sensor of the integrity of the MMR, suggests that inactivation of MMR results in multiple unrepaired insertion/deletion mismatches that occur at microsatellite sequences during new strand DNA synthesis. These mismatches induce a DNA damage response in the same way proposed for MNNG or cisplatin adducts. The reported binding of the p53 protein to insertion/deletion misalignments (40, 41) supports this hypothesis. Only those cells that acquire mutations in the DNA damage response pathway genes gain a selective advantage and finally develop into cancer cells. Cell lines with MSI, therefore, are resistant to DNA damaging agents because of the prior selection of clones with mutations in genes associated with DNA damage response pathways. Fig. 4 represents a model describing the tumorigenic pathway for cancers with MSI. The frequent mutations of the p300/CBP genes in cell lines with MSI may be explained by the involvement of these genes in the DNA damage response pathway. Although the introduction of the p300 cDNA into HCT15 cells did not resulted in the higher sensitivity to cisplatin (data not shown) this could be caused by the involvement of p300 in the mediation of the DNA damage response induced by genotoxic agents other then cisplatin.
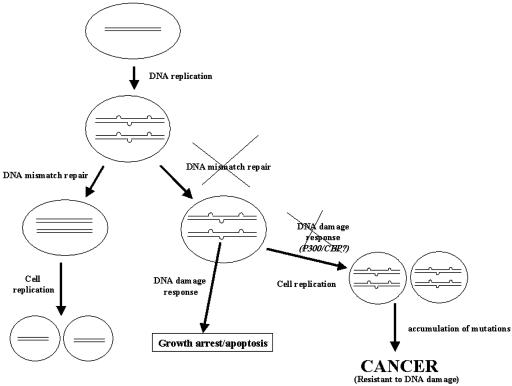
Fig. 4.
A model for tumorigenic pathway for cancers with MSI, a possible role for microsatellite DNA. Because of intrinsic instability of microsatellite DNA sequences, multiple single-strand loops arise by strand slippage at the sites of microsatellite DNA after each cycle of DNA replication. In cells with inactivated MMR, such single-strand loops remain unrepaired, and, by imitating DNA damage lesions, may induce DNA damage response, leading to growth arrest or apoptosis. The cells that acquire mutations in the DNA damage response genes, such as p300 or CBP, for example, then gain a growth advantage and eventually become tumorigenic because of the accumulation of mutations in other growth-controlling genes.
In summary, we have demonstrated frequent mutational inactivation of p300/CBP in colon cancer cell lines with MSI and have shown that p300 has growth suppressor activity when expressed in cells null for its activity.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CBP, cAMP-response element-binding protein (CREB) binding protein; MSI, microsatellite instability; NMD, nonsense-mediated decay; MMR, mismatch repair; TGF, transforming growth factor.
References
- 1.Ionov, Y., Peinado, M. A., Malkhosyan, S., Shibata, D. & Perucho, M. (1993) Nature 363, 558–561. [DOI] [PubMed] [Google Scholar]
- 2.Aaltonen, L. A., Peltomaki, P., Leach, F. S., Sistonen, P., Pylkkanen, L., Mecklin, J. P., Jarvinen, H., Powell, S. M., Jen, J., Hamilton, S. R., et al. (1993) Science 260, 812–816. [DOI] [PubMed] [Google Scholar]
- 3.Thibodeau, S. N., Bren, G. & Schaid, D. (1993) Science 260, 816–819. [DOI] [PubMed] [Google Scholar]
- 4.Fishel, R., Lescoe, M. K., Rao, M. R., Copeland, N. G., Jenkins, N. A., Garber, J., Kane, M. & Kolodner, R. (1993) Cell 75, 1027–1038. [DOI] [PubMed] [Google Scholar]
- 5.Parsons, R., Li, G. M., Longley, M. J., Fang, W. H., Papadopoulos, N., Jen, J., de la Chapelle, A., Kinzler, K. W., Vogelstein, B. & Modrich, P. (1993) Cell 75, 1227–1236. [DOI] [PubMed] [Google Scholar]
- 6.Kolodner, R. D. & Marsischky, G. T. (1999) Curr. Opin. Genet. Dev. 9, 89–96. [DOI] [PubMed] [Google Scholar]
- 7.Karran, P. (2001) Carcinogenesis 22, 1931–1937. [DOI] [PubMed] [Google Scholar]
- 8.Markowitz, S., Wang, J., Myeroff, L., Parsons, R., Sun, L., Lutterbaugh, J., Fan, R. S., Zborowska, E., Kinzler, K. W., Vogelstein, B., et al. (1995) Science 268, 1336–1338. [DOI] [PubMed] [Google Scholar]
- 9.Souza, R. F., Appel, R., Yin, J., Wang, S., Smolinski, K. N., Abraham, J. M., Zou, T. T., Shi, Y. Q., Lei, J., Cottrell, J., et al. (1996) Nat. Genet. 14, 255–257. [DOI] [PubMed] [Google Scholar]
- 10.Guanti, G., Resta, N., Simone, C., Cariola, F., Demma, I., Fiorente, P. & Gentile, M. (2000) Hum. Mol. Genet. 9, 283–287. [DOI] [PubMed] [Google Scholar]
- 11.Rampino, N., Yamamoto, H., Ionov, Y., Li, Y., Sawai, H., Reed, J. C. & Perucho, M. (1997) Science 275, 967–969. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz, S., Jr., Yamamoto, H., Navarro, M., Maestro, M., Reventos, J. & Perucho, M. (1999) Cancer Res. 59, 2995–3002. [PubMed] [Google Scholar]
- 13.Malkhosyan, S., Rampino, N., Yamamoto, H. & Perucho, M. (1996) Nature 382, 499–500. [DOI] [PubMed] [Google Scholar]
- 14.Riccio, A., Aaltonen, L. A., Godwin, A. K., Loukola, A., Percesepe, A., Salovaara, R., Masciullo, V., Genuardi, M., Paravatou-Petsotas, M., Bassi, D. E., et al. (1999) Nat. Genet. 23, 266–268. [DOI] [PubMed] [Google Scholar]
- 15.Duval, A., Gayet, J., Zhou, X. P., Iacopetta, B., Thomas, G. & Hamelin, R. (1999) Cancer Res. 59, 4213–4215. [PubMed] [Google Scholar]
- 16.Woerner, S. M., Gebert, J., Yuan, Y. P., Sutter, C., Ridder, R., Bork, P. & von Knebel Doeberitz, M. (2001) Int. J. Cancer 93, 12–19. [DOI] [PubMed] [Google Scholar]
- 17.Losson, R. & Lacroute, F. (1979) Proc. Natl. Acad. Sci. USA 76, 5134–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Culbertson, M. R. (1999) Trends Genet. 15, 74–80. [DOI] [PubMed] [Google Scholar]
- 19.Frischmeyer, P. A. & Dietz, H. C. (1999) Hum. Mol. Genet. 8, 1893–1900. [DOI] [PubMed] [Google Scholar]
- 20.Noensie, E. N. & Dietz, H. C. (2001) Nat. Biotechnol. 19, 434–439. [DOI] [PubMed] [Google Scholar]
- 21.Ionov, Y., Nowak, N., Perucho, M., Markowitz, S. & Cowell, J. K. (2004) Oncogene, in press. [DOI] [PubMed]
- 22.Grossman, S. R. (2001) Eur. J. Biochem. 268, 2773–2778. [DOI] [PubMed] [Google Scholar]
- 23.Goodman, R. H. & Smolik, S. (2000) Genes Dev. 14, 1553–1577. [PubMed] [Google Scholar]
- 24.Ozdag, H., Batley, S. J., Forsti, A., Iyer, N. G., Daigo, Y., Boutell, J., Arends, M. J., Ponder, B. A., Kouzarides, T. & Caldas, C. (2002) Br. J. Cancer 87, 1162–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan, Z. M., Huang, Y., Ishiko, T., Nakada, S., Utsugisawa, T., Shioya, H., Utsugisawa, Y., Yokoyama, K., Weichselbaum, R., Shi, Y. & Kufe, D. (1999) J. Biol. Chem. 274, 1883–1886. [DOI] [PubMed] [Google Scholar]
- 26.Gong, J. G., Costanzo, A., Yang, H. Q., Melino, G., Kaelin, W. G., Jr., Levrero, M. & Wang, J. Y. (1999) Nature 399, 806–809. [DOI] [PubMed] [Google Scholar]
- 27.Kharbanda, S., Ren, R., Pandey, P., Shafman, T. D., Feller, S. M., Weichselbaum, R. R. & Kufe, D. W. (1995) Nature 376, 785–788. [DOI] [PubMed] [Google Scholar]
- 28.Koi, M., Umar, A., Chauhan, D. P., Cherian, S. P., Carethers, J. M., Kunkel, T. A. & Boland, C. R. (1994) Cancer Res. 54, 4308–4312. [PubMed] [Google Scholar]
- 29.Suzuki, K., Dai, T., Suzuki, I., Dai, Y., Yamashita, K. & Perucho, M. (2002) Cancer Res. 62, 1961–1965. [PubMed] [Google Scholar]
- 30.Costanzo, A., Merlo, P., Pediconi, N., Fulco, M., Sartorelli, V., Cole, P. A., Fontemaggi, G., Fanciulli, M., Schiltz, L., Blandino, G., et al. (2002) Mol. Cell 9, 175–186. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues, N. R., Rowan, A., Smith, M. E., Kerr, I. B., Bodmer, W. F., Gannon, J. V. & Lane, D. P. (1990) Proc. Natl. Acad. Sci. USA 87, 7555–7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan, H. M. & La Thangue, N. B. (2001) J. Cell. Sci. 114, 2363–2373. [DOI] [PubMed] [Google Scholar]
- 33.Gayther, S. A., Batley, S. J., Linger, L., Bannister, A., Thorpe, K., Chin, S. F., Daigo, Y., Russell, P., Wilson, A., Sowter, H. M., et al. (2000) Nat. Genet. 24, 300–303. [DOI] [PubMed] [Google Scholar]
- 34.Massague, J., Blain, S. W. & Lo, R. S. (2000) Cell 103, 295–309. [DOI] [PubMed] [Google Scholar]
- 35.Suganuma, T., Kawabata, M., Ohshima, T. & Ikeda, M. A. (2002) Proc. Natl. Acad. Sci. USA 99, 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tini, M., Benecke, A., Um, S. J., Torchia, J., Evans, R. M. & Chambon, P. (2002) Mol. Cell 9, 265–277. [DOI] [PubMed] [Google Scholar]
- 37.Snowden, A. W. & Perkins, N. D. (1998) Biochem. Pharmacol. 55, 1947–1954. [DOI] [PubMed] [Google Scholar]
- 38.Kat, A., Thilly, W. G., Fang, W. H., Longley, M. J., Li, G. M. & Modrich, P. (1993) Proc. Natl. Acad. Sci. USA 90, 6424–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karran, P. & Bignami, M. (1994) BioEssays 16, 833–839. [DOI] [PubMed] [Google Scholar]
- 40.Szak, S. T. & Pietenpol, J. A. (1999) J. Biol. Chem. 274, 3904–3909. [DOI] [PubMed] [Google Scholar]
- 41.Lee, S., Elenbaas, B., Levine, A. & Griffith, J. (1995) Cell 81, 1013–1020. [DOI] [PubMed] [Google Scholar]