Abstract
Johne's disease in ruminants is caused by Mycobacterium avium subsp. paratuberculosis. Diagnosis of M. avium subsp. paratuberculosis infection is difficult, especially in the early stages. To date, ideal antigen candidates are not available for efficient immunization or immunodiagnosis. This study reports the in silico selection and subsequent analysis of epitopes of M. avium subsp. paratuberculosis proteins that were found to be upregulated under stress conditions as a means to identify immunogenic candidate proteins. Previous studies have reported differential regulation of proteins when M. avium subsp. paratuberculosis is exposed to stressors which induce a response similar to dormancy. Dormancy may be involved in evading host defense mechanisms, and the host may also mount an immune response against these proteins. Twenty-five M. avium subsp. paratuberculosis proteins that were previously identified as being upregulated under in vitro stress conditions were analyzed for B and T cell epitopes by use of the prediction tools at the Immune Epitope Database and Analysis Resource. Major histocompatibility complex class I T cell epitopes were predicted using an artificial neural network method, and class II T cell epitopes were predicted using the consensus method. Conformational B cell epitopes were predicted from the relevant three-dimensional structure template for each protein. Based on the greatest number of predicted epitopes, eight proteins (MAP2698c [encoded by desA2], MAP2312c [encoded by fadE19], MAP3651c [encoded by fadE3_2], MAP2872c [encoded by fabG5_2], MAP3523c [encoded by oxcA], MAP0187c [encoded by sodA], and the hypothetical proteins MAP3567 and MAP1168c) were identified as potential candidates for study of antibody- and cell-mediated immune responses within infected hosts.
INTRODUCTION
Mycobacterium avium subsp. paratuberculosis is an intracellular, Gram-positive, facultative, and slow-growing acid-fast bacillus that causes Johne's disease (JD), primarily in ruminants. The predominant ruminant hosts are cattle and sheep, and bacilli are known to be shed in the feces and milk of an infected host. The disease is typically observed as a subclinical or clinical form, but the largest proportion of animals is found to remain subclinically infected (34). The detection of infection becomes efficient as the disease progresses to its clinical stage, but there are no sensitive tests available for detection of subclinical or early-stage infection.
Outside the host system, M. avium subsp. paratuberculosis is known to survive in food, water, and soil (26). In an external environment, typically on soil, M. avium subsp. paratuberculosis is known to undergo a state of dormancy (40). In vitro experiments have reported differential regulation of certain M. avium subsp. paratuberculosis proteins when the organism is exposed to stressors similar to dormancy (13, 15, 21). It is believed that the in vivo regulation of dormancy genes and associated proteins by M. avium subsp. paratuberculosis may play a role in the ability of the organism to evade host defense mechanisms, and in addition, the host may mount an immune response against these proteins.
Previous evaluation of M. avium subsp. paratuberculosis proteins identified as differentially regulated under stress conditions found only a few to be immunogenic (14), suggesting that not all proteins differentially regulated under in vitro stress conditions are expressed in the host and/or are immunogenic. Knowledge of the proteomic response alone is not a sufficient classifier of M. avium subsp. paratuberculosis protein immunogenicity. Therefore, there is a need for a robust selection method to identify possibly immunogenic M. avium subsp. paratuberculosis proteins.
In recent years, immunoinformatics (a subdiscipline of bioinformatics), an integration of immunology and informatics, has become the driving force in understanding the networks regulating the immune system. Such approaches have been used to predict immune epitopes in pathogens to identify potential vaccine candidates in an approach called reverse vaccinology (11, 36). This process has been used successfully to identify proteins for use in vaccines against pathogens such as Neisseria meningitidis and Mycobacterium tuberculosis (28, 32). The availability of the M. avium subsp. paratuberculosis K-10 genome (24) and bioinformatics tools makes an in silico approach to identifying potentially immunogenic antigens from M. avium subsp. paratuberculosis proteins possible. Such an approach allows for systematic antigen selection and provides a basis for laboratory experiments that may reduce the high cost of cloning and subsequent evaluation.
The aim of this study was to identify immunogenic candidate antigens which can be evaluated later in M. avium subsp. paratuberculosis diagnostic assays. This study utilized the in silico approach to analyze 25 M. avium subsp. paratuberculosis proteins that had previously been identified as upregulated when the organism was exposed to in vitro stressors (13, 15, 21).
MATERIALS AND METHODS
Selection of stress-regulated M. avium subsp. paratuberculosis proteins.
The M. avium subsp. paratuberculosis proteins were selected based on the prior demonstration of an upregulated proteomic response to at least one in vitro stress condition (13, 15, 21), leading to a set of 25 M. avium subsp. paratuberculosis proteins. Each protein was submitted to a basic local alignment search (BLASTp) at the National Center for Biotechnology and Information (NCBI) database to confirm the M. avium subsp. paratuberculosis K-10 sequence identity. Proteins which showed 100% sequence identity to M. avium subsp. paratuberculosis K-10 were retained, while those with 100% sequence identity to other mycobacterial taxa were excluded. Eighteen such proteins unique to M. avium subsp. paratuberculosis K-10 were identified for epitope prediction.
T cell epitope prediction.
T cell epitopes were identified using prediction tools located at the Immune Epitope Database and Analysis Resource (IEDB-AR), a database of experimentally characterized immune epitopes (B and T cell epitopes) for humans, nonhuman primates, rodents, and other animal species (http://tools.immuneepitope.org/analyze/html/mhc_binding.html). T cell epitopes were classified based on their binding affinity for mouse major histocompatibility complex (MHC) alleles, using the half-maximal inhibitory concentration of a biological substance (IC50) as the unit of measure, as follows: high-affinity binding, IC50s of <50 nM; intermediate-affinity binding, IC50s of <500 nM; and low-affinity binding, IC50s of <5,000 nM. A lower IC50 is indicative of a higher affinity of binding to host MHC alleles. The mouse MHC alleles used for antigen peptide binding were H-2Db, H-2Dd, H-2Kb, H-2Kd, H-2Kk, and H-2Ld for MHC class I epitopes and H-2IAb, H-2IAd, and H-2IEd for MHC class II epitopes. T cell epitopes binding to mouse MHC molecules were identified by submitting the FAST-All (FASTA) format M. avium subsp. paratuberculosis protein sequence to IEDB-AR. Nine-mer MHC class I T cell epitope prediction was performed using the artificial neural network (ANN) method as described previously (29, 35), and 15-mer MHC class II T cell epitope prediction was performed using the consensus method (38), a combination of the average relative binding (ARB) matrix method and the stabilization matrix alignment method (SMM_align).
3D structure template modeling for selected M. avium subsp. paratuberculosis proteins.
The SWISS-MODEL Workspace (2) (http://swissmodel.expasy.org) and ElliPro (31) (http://tools.immuneepitope.org/tools) were used for three-dimensional (3D) structure template/scaffold modeling. The modeling results were compared for agreement between SWISS-MODEL Workspace and ElliPro. A template model was obtained for each M. avium subsp. paratuberculosis protein by submitting FASTA format protein sequences and modeling them.
Conformational B cell epitope prediction.
ElliPro was used to predict conformational B cell epitopes from M. avium subsp. paratuberculosis proteins, using a modeled 3D structure template for each protein. The 3D structure template was selected based on best-fitting scaffold criteria. The best-fitting template was defined as having a long alignment length, a large number of identities, and a small number of gaps. Prediction at a minimum level of 0.5 was considered the most lenient, with a level of 1.0 as the most stringent, and the maximum distance for residue clustering was defined as 6.0 Å. Selection of M. avium subsp. paratuberculosis proteins as B cell epitope candidates was based on the number of epitopes predicted with a minimum cutoff score of 0.8. Each predicted epitope formed by a group of amino acid residues was viewed with a Java viewer for chemical structures in 3D (Jmol) to illustrate its 3D structure and relative orientation to the protein molecule. The Jmol viewer was also used to confirm the positions of residues of each predicted epitope.
Prediction of protein secretion.
Prediction of the secretory or nonsecretory nature of stress-regulated M. avium subsp. paratuberculosis proteins was performed. Protein secretion was categorized as classical or nonclassical. Prediction of classical secretion and detection of the presence of signal sequences were performed using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP) as described previously (6). SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP) was used to predict nonclassical secretion as described previously (5). Both types of prediction were performed at a default setting score of 0.5. The proteins with predicted scores of 0.5 and above were considered to be secreted.
RESULTS
Upregulated M. avium subsp. paratuberculosis proteins exposed to stress conditions.
Twenty-five proteins from the S and C strains of M. avium subsp. paratuberculosis which had not been evaluated for immunogenicity and had an upregulated proteomic response to different stress conditions were identified (Table 1). Each protein identified was upregulated under at least one stress condition.
Table 1.
Sequence identity mapping of proteins that were upregulated in response to stress conditions
| Protein | Mass (kDa) | M. avium subsp. paratuberculosis strain(s)a | Regulation in response to in vitro stress conditionsb |
Sequence identity (%)c |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T | H | S | ROI | RNI | M. avium subsp. paratuberculosis K-10 | M. avium subsp. hominissuis 104 | M. avium subsp. avium ATCC 25291 | |||
| MAP0187c | 23.0 | S | ↑ | 100 | 99 | 99 | ||||
| MAP0540 | 17.6 | C | ↑ | 100 | 100 | 100 | ||||
| MAP1017c | 27.8 | C | ↑ | 100 | 99 | 99 | ||||
| MAP1168c | 31.9 | C | ↑ | 100 | 99 | 99 | ||||
| MAP1297 | 25.3 | S | ↑ | 100 | 99 | 99 | ||||
| MAP1560 | 15.2 | C | ↑ | 100 | 100 | 99 | ||||
| MAP1588c | 18.8 | C | ↑ | ↑ | ↑ | 100 | 99 | 100 | ||
| MAP1589c | 21.6 | C, S | ↑ | ↑ | ↑ | 100 | 99 | 99 | ||
| MAP1653 | 16.4 | C | ↑ | ↑ | ↑ | 100 | 99 | 99 | ||
| MAP1698c | 16.3 | C | ↑ | 100 | 99 | 99 | ||||
| MAP1834c | 27.9 | S | ↑ | 100 | 100 | 100 | ||||
| MAP2058c | 25.4 | C | ↑ | 100 | 99 | 100 | ||||
| MAP2312c | 41.4 | C, S | ↑ | 100 | 99 | 99 | ||||
| MAP2487c | 17.8 | C | ↑ | 100 | 99 | 99 | ||||
| MAP2698c | 31.4 | C | ↑ | 100 | 99 | 99 | ||||
| MAP2872c | 26.7 | C | ↑ | 100 | 99 | 99 | ||||
| MAP3268 | 16.4 | C | ↑ | 100 | 99 | 99 | ||||
| MAP3393c | 17.5 | C | ↑ | ↑ | 100 | 99 | 99 | |||
| MAP3523c | 62.3 | S | ↑ | 100 | NA | 99 | ||||
| MAP3538 | 15.9 | C | ↑ | 100 | 100 | 96 | ||||
| MAP3567 | 30.1 | C | ↑ | 100 | 99 | 99 | ||||
| MAP3651c | 43.9 | C | ↑ | 100 | 99 | 99 | ||||
| MAP3974c | 18.6 | S | ↑ | 100 | 39 | * | ||||
| MAP4098 | 18.7 | C | ↑ | ↑ | 100 | 100 | 100 | |||
| MAP4340 | 12.4 | C | ↑ | 100 | 99 | NA | ||||
C, cattle strain; S, sheep strain.
T, temperature flux (18); H, hypoxia; S, nutrient starvation (16); ROI, oxidative stress; RNI, nitrosative stress (22); ↑, upregulated.
Values in bold indicate 100% sequence identity only to M. avium subsp. paratuberculosis K-10, and the corresponding proteins were selected for epitope prediction. NA, sequence identity data not available at NCBI; *, the MAP3974c protein revealed 39% and 99% sequence identities to the hypothetical proteins MaviaA2_17319 and MaviaA2_20904, respectively, both from Mycobacterium avium subsp. avium ATCC 25291.
Sequence identity mapping of stress-regulated M. avium subsp. paratuberculosis proteins.
Protein sequence identity analysis at the NCBI GenBank database showed that the amino acid sequences of all 25 proteins were 100% identical to those in M. avium subsp. paratuberculosis K-10, with an equal or lesser identity to closely related taxa, e.g., Mycobacterium avium subsp. hominissuis 104 and Mycobacterium avium subsp. avium ATCC 25291 (Table 1). Eighteen M. avium subsp. paratuberculosis proteins were 100% identical only to those in M. avium subsp. paratuberculosis K-10. Seven proteins had 100% sequence identity with either M. avium subsp. hominissuis 104, M. avium subsp. avium, or both. Thus, the 18 proteins that were 100% identical only to proteins in M. avium subsp. paratuberculosis K-10 were selected for epitope prediction. The MAP3974c protein revealed 39% and 99% sequence identity with the hypothetical proteins MaviaA2_17319 and MaviaA2_20904 of Mycobacterium avium subsp. avium ATCC 25291, respectively, indicating comparison with two different proteins in the database.
Predicted MHC class I T cell epitopes.
Each protein was predicted to have a large number of low-affinity and a relatively small number of intermediate- and high-affinity epitopes (Table 2). The number of predicted epitopes was lowest in the high-affinity category. Four proteins (MAP1297, MAP1653, MAP4340, and MAP2487c) failed to identify with any epitopes in the high-affinity category. The remaining 14 proteins were consistently predicted to have high-affinity epitopes in addition to a large number of epitopes of intermediate affinity. The MHC class I T cell epitopes with high-affinity binding are presented in Table 3. Five proteins (MAP2698c, MAP3567, MAP3651c, MAP3523c, and MAP2312c) were found to carry the largest numbers of MHC class I T cell epitopes of intermediate- and high-affinity binding and were selected as potential candidates. Details of the epitopes of these five proteins are shown in Table 3. Intermediate- and high-affinity nine-mer MHC class I T cell epitopes for all 18 M. avium subsp. paratuberculosis proteins are presented in Table S1 in the supplemental material.
Table 2.
Numbers of conformational B cell and T cell (MHC class I and II) epitopes
| Proteina | PDB IDb | No. of B cell epitopes at prediction cutoff score |
No. of T cell epitopesc |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MHC class I T cell epitopes |
MHC class II T cell epitopes |
|||||||||||
| 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | L | I | H | L | I | H | ||
| MAP1297 | 2VEP | 5 | 7 | 9 | 3 | 0 | 234 | 1 | 225 | 4 | ||
| MAP1653 | 1Y25 | 4 | 4 | 2 | 2 | 2 | 155 | 1 | 138 | 12 | ||
| MAP3393c | 1O4V | 5 | 4 | 4 | 3 | 2 | 158 | 6 | 1 | 149 | 10 | |
| MAP2872cc | 2ZAT | 4 | 9 | 5 | 5 | 4 | 241 | 7 | 3 | 213 | 13 | |
| MAP1589c | 2BMX | 8 | 7 | 6 | 3 | 3 | 183 | 3 | 1 | 181 | ||
| MAP4340 | 1NW2 | 8 | 7 | 2 | 2 | 1 | 106 | 3 | 103 | |||
| MAP2698ca | 1ZA0 | 7 | 7 | 7 | 4 | 3 | 249 | 9 | 9 | 256 | 5 | |
| MAP1168cb | 1EZW | 9 | 7 | 6 | 4 | 2 | 288 | 7 | 2 | 267 | 20 | 4 |
| MAP3567ac | 1ZBQ | 10 | 12 | 9 | 5 | 2 | 268 | 7 | 4 | 268 | 5 | |
| MAP1017c | 3H81 | 7 | 7 | 6 | 4 | 3 | 249 | 4 | 2 | 249 | ||
| MAP2487c | 1YLK | 5 | 6 | 4 | 2 | 2 | 148 | 6 | 140 | 9 | ||
| MAP3268 | 1SHS | 4 | 4 | 5 | 4 | 2 | 138 | 1 | 1 | 133 | ||
| MAP1698c | 1SHS | 8 | 8 | 6 | 4 | 2 | 133 | 4 | 1 | 131 | 1 | |
| MAP3651cac | 2PGO | 10 | 10 | 10 | 10 | 6 | 381 | 10 | 6 | 381 | 10 | |
| MAP3523cabc | 2Q27 | 9 | 7 | 9 | 9 | 5 | 570 | 13 | 3 | 539 | 41 | |
| MAP0187cb | 1GN3 | 6 | 7 | 4 | 2 | 1 | 188 | 8 | 3 | 175 | 18 | |
| MAP2312cac | 1JQI | 6 | 8 | 7 | 8 | 5 | 364 | 11 | 5 | 373 | 1 | |
| MAP3974c | 163 | 2 | 1 | 160 | ||||||||
Proteins are marked as follows. a, proteins with the largest numbers of MHC class I T cell epitopes; b, proteins with the largest numbers of MHC class II T cell epitopes; c, proteins with the largest numbers of conformational B cell epitopes.
PDB ID of the 3D structural template for each M. avium subsp. paratuberculosis protein. A detailed description of the source of each template is presented in Table S4 in the supplemental material.
L, low-affinity binding; I, intermediate-affinity binding; H, high-affinity binding.
Table 3.
High-affinity (IC50 of <50 nM) MHC class I and II T cell epitopes
| Protein | Epitope sequence (positions) |
|---|---|
| Proteins with MHC class I T cell epitopes (9–mer) | |
| MAP2698c | RPVANALTL (4–12) |
| HRELVEHFI (83–91) | |
| IALRNYLVV (112–120) | |
| LEEPILSGL (173–181) | |
| EEPILSGLV (174–182) | |
| HELFFSNLV (192–200) | |
| LEYTRDETI (204–212) | |
| TENDLRQVI (246–254) | |
| DEPALKPFV (265–273) | |
| MAP3567 | RAVANYDSV (65–73) |
| VGLINSLAL (172–180) | |
| KYNIHANAI (184–192) | |
| NPNSASVFV (230–238) | |
| MAP3651c | SPVSMPCYV (81–89) |
| VSMPCYVRV (83–91) | |
| CYVRVTEQL (87–95) | |
| IEQMKRIGI (61–69) | |
| SNARRSGLI (178–186) | |
| MEAGMAKLF (336–344) | |
| MAP3523c | NATTNCFPM (115–123) |
| AYAQADNEI (247–255) | |
| SEFDSNQQI (331–339) | |
| MAP2312c | YGFMNEYPV (349–357) |
| SYQAISFKI (285–293) | |
| NEEQKRKWL (108–116) | |
| KEAAIAKMI (322–330) | |
| NEYPVARHY (353–361) | |
| MHC class II T cell epitopes (15–mer) | |
| MAP1168c | TPIPVYVAAMGPKAL (155–169) |
| PIPVYVAAMGPKALQ (156–170) | |
| IPVYVAAMGPKALQV (157–171) | |
| PVYVAAMGPKALQVT (158–172) |
Predicted MHC class II T cell epitopes.
Thirteen proteins were predicted to have at least one MHC class II T cell epitope of intermediate-affinity binding. Five proteins (MAP1589c, MAP4340, MAP1017c, MAP3974c, and MAP3268) did not reveal any epitopes with intermediate- or high-affinity binding. The MAP3523c protein showed the largest number of epitopes in the intermediate-affinity category. The number of predicted MHC class II T cell epitopes is shown in Table 2. Only one protein (MAP1168c) was found to have epitopes with high-affinity binding in addition to a large number of epitopes of low- and intermediate-affinity binding. High-affinity MHC class II T cell epitopes of the MAP1168c protein are shown in Table 3. Three proteins (MAP1168c, MAP3523c, and MAP0187c) with the largest numbers of MHC class II epitopes were selected as potential candidates. Intermediate- and high-affinity 15-mer MHC class II T cell epitopes for all 13 M. avium subsp. paratuberculosis proteins are presented in Table S2 in the supplemental material.
Overlapping residues in MHC class I and II T cell epitopes.
Eight M. avium subsp. paratuberculosis proteins (MAP1297, MAP3393c, MAP3567, MAP3651c, MAP3523c, MAP1168c, MAP0187c, and MAP2487c) were found to have at least one T cell epitope with binding affinity for both MHC class I and MHC class II molecules (Table 4). The 9-mer MHC class I T cell epitope was found to overlap with the 15-mer MHC class II T cell epitope, offset at either the C-terminal or N-terminal end. The MAP2487c protein had the largest number of overlapping epitopes.
Table 4.
Overlapping MHC class I and II T cell epitopes
| Protein | MHC class II T cell epitope |
MHC class I T cell epitope |
||
|---|---|---|---|---|
| Sequencea | Positions | Sequence | Positions | |
| MAP1297 | ALYAGRFTLPQALAA | 226–240 | ALYAGRFTL | 226–234 |
| MAP3393c | LPGMVASATPLPVIG | 75–89 | GMVASATPL | 77–85 |
| MAP3567 | KYNIHANAIAPIAAT | 184–198 | KYNIHANAI | 184–192 |
| MAP3651c | DQKRAYLPRMASGEI | 123–137 | LPRMASGEI | 129–137 |
| MAP3523c | HPMRFYNALGAIRAV | 393–407 | HPMRFYNAL | 393–401 |
| MAP1168c | SELSAAPSFPVRVPG | 139–153 | SELSAAPSF | 139–147 |
| AAPSFPVRV | 143–151 | |||
| MAP0187c | WDYAALEPHISGQIN | 11–25 | DYAALEPHI | 12–20 |
| LEPHISGQI | 16–24 | |||
| MAP2487c | YASSFEGPLPMPPSK | 14–28 | YASSFEGPL | 14–22 |
| SSFEGPLPM | 16–24 | |||
| EETGIKPPWAAEAFA | 107–121 | KPPWAAEAF | 112–120 | |
| PPWAAEAFA | 113–121 | |||
The underlined residues are the 9-mer MHC class I T cell epitopes overlapping the 15-mer MHC class II T cell epitopes, offset at either the N-terminal or C-terminal end.
3D structure modeling for selected proteins.
Seventeen 3D structure templates were successfully modeled by two modeling programs—ElliPro and SWISS-MODEL Workspace. Neither program could find a suitable model for the MAP3974c protein. The Protein Data Bank (PDB) ID for each 3D structure template was generated in 4-character identifier format, i.e., (0 to 9)(a to z, 0 to 9)(a to z, 0 to 9)(a to z, 0 to 9). Two proteins (MAP1698c and MAP3268) were identified by the same 3D structure template—that for a small heat shock protein from Methanococcus jannaschii (PDB accession no. 1SHS) (22). All of the modeled templates showed various lengths of sequence alignment and sequence identity with M. avium subsp. paratuberculosis proteins.
The modeled 3D structure templates for 14 proteins were the same after modeling by both the ElliPro and the SWISS-MODEL Workspace programs. However, the programs identified different templates for three proteins, MAP4340, MAP3651c, and MAP3523c. For MAP4340, the template 2ILU, the crystal structure of lactaldehyde dehydrogenase from Escherichia coli (a binary complex with NADPH aldehyde dehydrogenase A), was identified by SWISS-MODEL Workspace, while the template 1NW2, the crystal structure of the mutant R82e of thioredoxin from Alicyclobacillus acidocaldarius (30), was identified by ElliPro. Similarly, for MAP3651c, template 2CX9 was modeled by SWISS-MODEL Workspace and template 2PGO (L. Chen et al., unpublished data) was modeled by ElliPro. The MAP3523c protein was also modeled with two different templates, 2Q29 (39) by SWISS-MODEL Workspace and 2Q27 (39) by ElliPro, from the crystal structure of oxalyl-coenzyme A (oxalyl-CoA) decarboxylase from E. coli. The details of the 3D structure template IDs and the descriptions of the source molecules are shown in Table S3 in the supplemental material.
Conformational B cell epitopes.
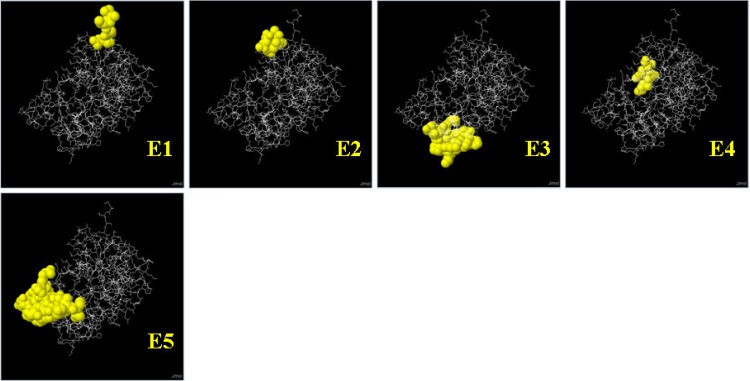
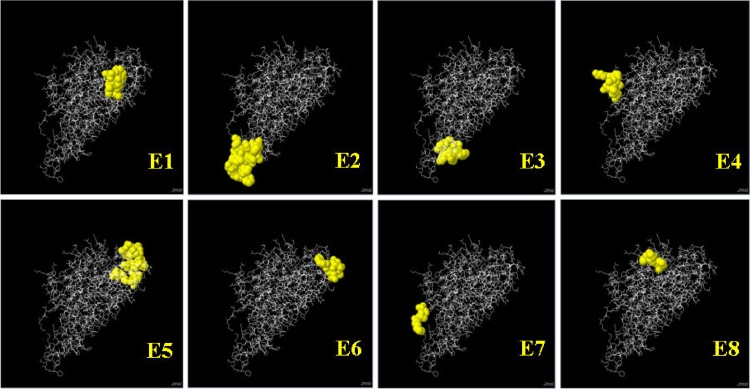
The number of conformational B cell epitopes was found to be reduced as the prediction parameter was made more stringent by gradually increasing the cutoff prediction score (Table 2). However, a few proteins were found to have more epitopes between cutoff prediction scores of 0.5 and 0.9. The MAP1297 protein did not reveal any epitopes at a cutoff or minimum prediction score of 0.9, whereas the other 16 M. avium subsp. paratuberculosis proteins revealed at least one epitope at the same cutoff score. A comparison of the numbers of epitopes and the cutoff scores illustrated the effect of stringent prediction parameters. The effect indicated that the stringent prediction identified fewer epitopes with higher scores and vice versa. With the default prediction parameter, all M. avium subsp. paratuberculosis proteins showed a minimum of 4 and a maximum of 10 epitopes. Eight M. avium subsp. paratuberculosis proteins (MAP2872c, MAP1589c, MAP3567, MAP1017c, MAP1698c, MAP3651c, MAP3523c, and MAP2312c) were identified as having relatively large numbers of conformational B cell epitopes that were consistently predicted even with more stringent cutoffs of 0.5 to 0.9. However, the number of residues in each epitope was greatly reduced as the stringency of the prediction parameters was increased, and thus the size of the epitope became smaller. A list of conformational B cell epitopes for all 17 M. avium subsp. paratuberculosis proteins is shown in Table S4 in the supplemental material. Five M. avium subsp. paratuberculosis proteins (MAP2872c, MAP3567, MAP3651c, MAP3523c, and MAP2312c) were consistently predicted to have a large number of conformational B cell epitopes at a cutoff prediction score of 0.8 (Fig. 1 to 5). For the purpose of B cell immune response studies, these proteins would be selected. The epitopes of the MAP2872c protein selected as potential B cell candidates are shown in Fig. 1. The 3D images of the predicted epitopes are shown as ball-and-stick models (viewed with Jmol), which illustrate their spatial locations relative to the protein molecule. The predicted residues of the epitopes were identified from the complete length of the MAP2872c protein sequence. The residues that formed the epitopes were scattered over the surface of the protein molecule. The predicted epitopes had scores of between 0.816 and 0.921, as the minimum prediction cutoff score was set at 0.80. The predicted epitopes were of the conformational type. The details of epitopes and Jmol views of MAP3567, MAP3651c, MAP3523c, and MAP2312c are shown in Fig. 2 to 4.
Fig 1.
Conformational B cell epitopes of MAP2872c (fabG4_2) predicted from the 3D structure template under PDB ID 2ZAT (a crystal structure of a mammalian reductase). In the ball-and-stick model, yellow balls are the residues of predicted epitopes (E1 to E5) and white sticks are the structures for nonepitope and core residues. Each epitope is shown with predicted residues (abbreviated amino acids) and residue positions (superscript numerals): E1, MTSQ1–4; E2, A50-GPQAL53–57; E3, PAYGPLIEQDHARF94–107-A151; E4, KQE41–43-D46; and E5, LWKDHEDP195–202-SSS204–206-ALGR208–211-GT243–244-APTPS255–259.
Fig 5.
Conformational B cell epitopes of MAP2312c (fadE19) predicted from the 3D structure template under PDB ID 1JQI (a crystal structure of rat short-chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA). In the ball-and-stick model, yellow balls are the residues of predicted epitopes (E1 to E8) and white sticks are the structures for nonepitope and core residues. Each epitope is shown with predicted residues (abbreviated amino acids) and residue positions (superscript numerals): E1, ARSLGLS382–388; E2, YAKERQSFGQPIGSYQAISF272–291; E3, GGY347–349-FMNEY351–355; E4, AGTLPK7–12-QD15–16; E5, RLEDGE147–152-V154-N156-TPG198–200-SD223–224-R226-E229; E6, GTVGAD180–185-K187-G237; E7, MTT1–3; and E8, AGKP316–319.
Fig 2.
Conformational B cell epitopes of MAP3567 predicted from the 3D structure template under PDB ID 1ZBQ (a crystal structure of human 17-beta-hydroxysteroid dehydrogenase type 4 in complex with NAD). In the ball-and-stick model, yellow balls are the residues of predicted epitopes (E1 to E5) and white sticks are the structures for nonepitope and core residues. Each epitope is shown with predicted residues (abbreviated amino acids) and residue positions (superscript numerals): E1, QNAGVTYDKPPTVQDVAARWDEITDLSAA251–279; E2, F108-MLF111–113; E3, KLG185–187; E4, LPKEV205–209-AKL211–213; and E5, DGT105–107-HK109–110-NFG158–160.
Fig 4.
Conformational B cell epitopes of MAP3523c (oxcA) predicted from the 3D structure template under PDB ID 2Q27 (a crystal structure of oxalyl-CoA decarboxylase from E. coli). In the ball-and-stick model, yellow balls are the residues of predicted epitopes (E1 to E9) and white sticks are the structures for nonepitope and core residues. Each epitope is shown with predicted residues (abbreviated amino acids) and residue positions (superscript numerals): E1, MAGQ1–4; E2, YPHPHGR5–11; E3, SRHMSATSASAGPAPESAR12–30; E4, ASAASPAAVSAGA582–594; E5, PAPEAVDRALDVLADARRP220–238-STGI261–264-AD295–296-DALAARPITVPA356–367-E371; E6, RGDEAPHGDDPAPTV504–518-P567; E7, SD130–131-ALVDLRRGDY133–142-DL144–145; E8, LSARAR519–524-L527-EAF530–532-H538; and E9, SAGVK318–322.
Fig 3.
Conformational B cell epitopes of MAP3651c (fadE2_2) predicted from the 3D structure template under PDB ID 2PGO (a crystal structure of acyl-CoA dehydrogenase from Geobacillus kaustophilus). In the ball-and-stick model, yellow balls are the residues of predicted epitopes (E1 to E10) and white sticks are the structures for nonepitope and core residues. Each epitope is shown with predicted residues (abbreviated amino acids) and residue positions (superscript numerals): E1, RESVR16–20; E2, A396-LVTRGGI399–405; E3, YAQQRESFGQPIWQHQSVGNY286–306; E4, LPDPDSDG160–167; E5, TDPKATP192–198; E6, GGY361–363-YSQEY365–369; E7, V7-AQQVDV9–14-WAQ21–23; E8, G213-GN236–237; E9, MGANS1–5; and E10, NDE25–27.
Protein secretion.
SignalP 3.0 identified a single protein (MAP2872c) as being secreted, with a score of 0.56, whereas all other protein sequences submitted had low scores suggesting the absence of a signal sequence. The MAP2872c protein was predicted to be secreted with a 33-amino-acid signal sequence at the N-terminal end and, most likely, with a cleavage site for peptidase I at positions 33 and 34, between alanine and asparagine residues, with the first residue of the mature protein being asparagine. The signal sequence with the cleavage site was MTSQDLTGRTAIITGASRGIGLAIAQQLAAAGA|NV. Prediction of nonclassical secretion showed SecP scores ranging from 0.028 to 0.300 for all 18 proteins. These values were below the cutoff score (0.50), indicating that none were secreted via a nonclassical secretory pathway.
In summary, from 18 proteins with 100% sequence identity to M. avium subsp. paratuberculosis K-10 that were upregulated under stress conditions, 8 were identified as potentially immunogenic based on the number of epitopes predicted from their protein sequence/structure. Three proteins (MAP0187, MAP1168c, and MAP2698c) were identified as having epitopes that could be potential candidates for cell-mediated immune responses, while MAP2872c was identified as a candidate protein for an antibody-mediated immune response. Four proteins (MAP3523c, MAP3651c, MAP2312c, and MAP3567) had large numbers of epitopes for both cell- and antibody-mediated immune responses. The MAP2872c protein was found to be secretory in nature, while the other 17 proteins were nonsecretory. The proteins identified as potential candidates for immunogenicity evaluation are shown in Table 5.
Table 5.
Candidate proteins identified for immune response studies
| Protein | Gene encoding the protein or hypothetical protein | Type of epitope | Type of immune response |
|---|---|---|---|
| MAP2698c | desA2 | MHC class I T cell | Cell mediated |
| MAP3567 | Hypothetical protein | MHC class I T cell and B cell | Cell and antibody mediated |
| MAP3651c | fadE3_2 | MHC class I T cell and B cell | Cell and antibody mediated |
| MAP2312c | fadE19 | MHC class I T cell and B cell | Cell and antibody mediated |
| MAP1168c | Hypothetical protein | MHC class II T cell | Cell mediated |
| MAP3523c | oxcA | MHC class I and II T cell and B cell | Cell and antibody mediated |
| MAP0187c | sodA | MHC class II T cell | Cell mediated |
| MAP2872ca | fabG5_2 | B cell | Antibody mediated |
Secreted protein.
DISCUSSION
Knowledge of the interactions between MHC alleles, peptides, and host immune cells has immense immunological value for identifying immune epitopes and for the development of diagnostic assays or vaccines against pathogens. The information obtained from in silico analysis in this study may be used for evaluating M. avium subsp. paratuberculosis recombinant proteins and/or selected peptides in cattle and sheep to examine their potential for use in an effective vaccine. The findings of this study may be validated by evaluating identified proteins/peptides in gamma interferon (IFN-γ) and antibody assays.
Previous proteomic analyses identified a number of M. avium subsp. paratuberculosis proteins (MAP1297, MAP2872c, MAP3567, and MAP3651c [17], as well as MAP1168c [23]). However, none of these proteins were identified by in silico analysis for the purpose of assessing immunogenic potential, and only one of these proteins (MAP1297) was evaluated by antibody assay to confirm a reaction with sera obtained from sheep infected with M. avium subsp. paratuberculosis.
In the present study, these and other potential candidate M. avium subsp. paratuberculosis proteins which were identified as immunogenic based upon the results of epitope prediction were selected. Random selection of proteins has not proven to be an effective method of antigen characterization (20). Therefore, an additional step of in silico analysis was utilized to make the antigen characterization process more efficient.
The stress-regulated proteins from M. avium subsp. paratuberculosis K-10 and other closely related taxa showed 99 to 100% amino acid sequence identity, which is in agreement with findings from other studies (3, 33).
T cell epitope prediction was performed based on the probability of MHC-peptide ligand formation and presentation to different T cell populations (35). MHC-peptide ligand formation is dependent on the generation of antigenic peptides in proteasomal and phagolysosomal complexes. The availability of MHC alleles for the generated peptides is also important for effective binding and presentation by antigen-presenting cells (APCs). This in silico analysis was performed using tools designed for mouse MHC alleles. Although there are several reports on antigen processing and presentation in a mouse MHC model (27) and on identification of immunogenic peptides (19, 42), this is the first study to investigate MHC binding of peptides by using an in silico model to identify epitopes for use in a cattle and sheep study. It is recognized that the bovine MHC profile in particular is more complex than the mouse MHC profile. However, MHC haplotype sequence homology within the NCBI database revealed more than 80% identity between the MHC alleles of mice, sheep, and cows.
There was variation in the number of predicted epitopes among the different proteins. One reason for this may simply have been variation in the sizes of the proteins. The relatively large protein molecule MAP3523c, with a molecular mass of 62.3 kDa, showed a larger total number of epitopes than that for smaller proteins. However, with an increase in the stringency of the prediction cutoffs, this did not hold true. Conversely, the MAP2698c protein, with a molecular mass of 31.4 kDa, was predicted to have 9 high-affinity epitopes with the most stringent prediction cutoff. Therefore, a possible explanation for this observation is the ability of prediction tools to consistently predict large numbers of epitopes in immunogenic proteins, irrespective of size.
Proteins with oxidoreductase and antioxidant activity were shown to have more epitopes of stronger affinity for MHC class I molecules than those of other proteins. These bacterial proteins are involved in the termination of oxidation reactions, thereby reducing the production of host reactive oxygen intermediates targeted to destroy pathogens (41, 43). The oxidoreductase class protein MAP2312c (predicted to have a large number of T cell epitopes) was previously found to be upregulated under conditions of oxidative and nitrosative stress (21). A study on the inhibition of the host defense mechanism by mycobacteria was suggested to play an important role in the pathogenesis of tuberculosis (10).
Interestingly, the only protein with predicted high-affinity MHC class II T cell epitopes was also from this oxidoreductase class. Compared to numerous proteins predicted to have large numbers of MHC class I T cell epitopes, only the MAP1168c protein was predicted to have high-affinity MHC class II T cell epitopes. This may have been the result of using the consensus prediction method.
A recent study showed that some exogenous antigens were presented efficiently via the MHC class I pathway in addition to the MHC class II pathway (1, 18). The T cell epitope prediction in this analysis revealed several overlapping peptides/epitopes, suggesting the possibility of antigen presentation to immune cells via both MHC class I and II pathways. The presence of such peptides supports findings from other studies of antigen cross-presentation (1).
The 3D structures of M. avium subsp. paratuberculosis K-10 proteins analyzed in this study were determined by matching their structural orientation with that of the most relevant template in the PDB library. Templates were available for most of the proteins in this study, although there were various degrees of sequence identity. Conformational B cell epitope prediction is influenced by the degree of sequence alignment, by sequence identity, and by the similarity of structural scaffolding between the target protein and the modeled template (12). Therefore, a template modeled with a longer range of sequence alignment and a higher percentage of sequence identity may have resulted in a more accurate epitope prediction. The majority of proteins in this study showed 40 to 90% sequence identity, which is in concurrence with findings from other studies where modeling was proved significantly accurate when sequence identity was ≥40% (37). Two proteins (MAP1698c and MAP3268) were modeled on the same 3D structure template due to their relatively high structural equivalence or unavailability of a more relevant template in the PDB library. BLASTp revealed 60% sequence identity between these two proteins.
The in silico epitope prediction in this study revealed several M. avium subsp. paratuberculosis proteins containing both B and T cell epitopes, and these proteins are potential candidates for the investigation of both cellular and humoral immune responses in an infected host. Our findings support those of a recent study which reported a combined antibody and IFN-γ response in M. avium subsp. paratuberculosis-infected sheep rather than a defined switch from a cell-mediated Th1 response to an antibody-mediated Th2 response (4). The coexistence of both B and T cell epitopes within several M. avium subsp. paratuberculosis proteins could be a reason for the development of such an immune response.
The majority of the proteins in this study were predicted to be nonsecretory in nature, suggesting that they are probably not secreted by M. avium subsp. paratuberculosis. Only one protein (MAP2872c) with oxidoreductase activity was identified as classically secreted by SignalP 3.0. Studies have identified several secreted M. avium subsp. paratuberculosis proteins in culture filtrates, but MAP2872c was not one of them (8), and its involvement in pathogenesis may be worth investigating.
SecretomeP predicted the highest score for MAP0187c but did not classify it as a non-classically secreted protein. Its homologue in M. tuberculosis was also identified as nonsecretory. However, M. avium subsp. paratuberculosis sod, with a manganese-binding cofactor, was found to be secreted into the culture supernatant without a signal sequence of its own and was translocated by the specific export machinery of mycobacteria (25). The copper- and zinc-cofactored superoxide dismutase encoded by the sodC gene from M. avium subsp. paratuberculosis was also found to be a secreted protein (9). The protein encoded by the same gene in Mycobacterium tuberculosis was found in the host cell cytoplasm, which suggested that the proteins encoded by sod genes are secretory (7, 16). However, neither tool (SignalP 3.0 or SecretomeP 2.0) predicted the MAP0187c protein to be a secretory protein, suggesting that the default prediction threshold of 0.5 in these tools may be too stringent.
The selection of proteins was based on three main criteria: an upregulated response to physiological stress similar to the case with dormancy, a large number of predicted epitopes, and 100% sequence identity to M. avium subsp. paratuberculosis K-10. However, due to the high levels of genetic similarity of the M. avium subsp. paratuberculosis genome and those of other mycobacteria, selection of candidate proteins based only on 100% identity to M. avium subsp. paratuberculosis K-10 was difficult and may not be accurate, and species excluded due to 1 to 2% differences in genetic composition may not actually be different. Therefore, to increase specificity, most immunological assays for detection of M. avium subsp. paratuberculosis infection involve a step dedicated to eliminating cross-reactions encountered as a result of such genetic similarity.
The in silico analysis in this study led to identification of eight M. avium subsp. paratuberculosis proteins as potential immunogens, based on the presence of relatively large numbers of B and T cell epitopes in their amino acid sequences. T cell epitope prediction was performed on a recently created data set in IEDB-AR, and conformational B cell epitope prediction was performed to overcome the limitations of predicting immunogenicity based on linear epitopes. These proteins may be tested further for their immunoreactivity in sheep or cattle to support the in silico findings. Immunogenicity testing of these proteins may prove their value as diagnostic antigens. The performance of prediction tools depends on the quality of the databases, which are dynamic and evolving as new information is submitted. Therefore, it is likely that epitope prediction results obtained for the same set of proteins at later time points may vary.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Meat and Livestock Australia and by the Cattle Council of Australia, Sheepmeat Council of Australia, and WoolProducers Australia through Animal Health Australia. The first author of this work was awarded an Endeavor Postgraduate Award by the Department of Education, Employment and Workplace Relations (DEEWR), Australia.
We thank Julia Ponomarenko, the author of “ElliPro: a New Structure-Based Tool for the Prediction of Antibody Epitopes,” at San Diego Supercomputer Center, University of California, San Diego, CA, for her guidance in using the ElliPro tool to predict conformational B cell epitopes.
Footnotes
Published ahead of print 11 April 2012
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1. Amigorena S, Savina A. 2010. Intracellular mechanisms of antigen cross presentation in dendritic cells. Curr. Opin. Immunol. 22: 109–117 [DOI] [PubMed] [Google Scholar]
- 2. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- 3. Bannantine JP, Baechler E, Zhang Q, Li L, Kapur V. 2002. Genome scale comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium reveals potential diagnostic sequences. J. Clin. Microbiol. 40: 1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Begg DJ, et al. 2011. Does a Th1 over Th2 dominancy really exist in the early stages of Mycobacterium avium subspecies paratuberculosis infections? Immunobiology 216: 840–846 [DOI] [PubMed] [Google Scholar]
- 5. Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. 2004. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17: 349–356 [DOI] [PubMed] [Google Scholar]
- 6. Bendtsen JD, Nielsen H, Von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783–795 [DOI] [PubMed] [Google Scholar]
- 7. Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR., Jr 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48: 453–464 [DOI] [PubMed] [Google Scholar]
- 8. Cho D, Sung N, Collins MT. 2006. Identification of proteins of potential diagnostic value for bovine paratuberculosis. Proteomics 6: 5785–5794 [DOI] [PubMed] [Google Scholar]
- 9. Dupont C, Murray A. 2001. Identification, cloning and expression of sodC from an alkaline phosphatase gene fusion library of Mycobacterium avium subspecies paratuberculosis. Microbios 106: 7–19 [PubMed] [Google Scholar]
- 10. Ehrt S, et al. 1997. A novel antioxidant gene from Mycobacterium tuberculosis. J. Exp. Med. 186: 1885–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guerfali FZ, et al. 2009. An in silico immunological approach for prediction of CD8+ T cell epitopes of Leishmania major proteins in susceptible BALB/c and resistant C57BL/6 murine models of infection. Infect. Genet. Evol. 9: 344–350 [DOI] [PubMed] [Google Scholar]
- 12. Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- 13. Gumber S, Taylor DL, Marsh IB, Whittington RJ. 2009. Growth pattern and partial proteome of Mycobacterium avium subsp. paratuberculosis during the stress response to hypoxia and nutrient starvation. Vet. Microbiol. 133: 344–357 [DOI] [PubMed] [Google Scholar]
- 14. Gumber S, Taylor DL, Whittington RJ. 2009. Evaluation of the immunogenicity of recombinant stress-associated proteins during Mycobacterium avium subsp. paratuberculosis infection: implications for pathogenesis and diagnosis. Vet. Microbiol. 137: 290–296 [DOI] [PubMed] [Google Scholar]
- 15. Gumber S, Whittington RJ. 2009. Analysis of the growth pattern, survival and proteome of Mycobacterium avium subsp. paratuberculosis following exposure to heat. Vet. Microbiol. 136: 82–90 [DOI] [PubMed] [Google Scholar]
- 16. Harth G, Horwitz MA. 1999. Export of recombinant Mycobacterium tuberculosis superoxide dismutase is dependent upon both information in the protein and mycobacterial export machinery: a model for studying export of leaderless proteins by pathogenic mycobacteria. J. Biol. Chem. 274: 4281–4292 [DOI] [PubMed] [Google Scholar]
- 17. Hughes V, et al. 2008. Immunogenicity of proteome-determined Mycobacterium avium subsp. paratuberculosis-specific proteins in sheep with paratuberculosis. Clin. Vaccine Immunol. 15: 1824–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ikeuchi N, et al. 2010. Efficient cross-presentation of soluble exogenous antigens introduced into dendritic cells using a weak-based amphiphilic peptide. Biochem. Biophys. Res. Commun. 392: 217–222 [DOI] [PubMed] [Google Scholar]
- 19. Jones GJ, Bagaini F, Hewinson RG, Vordermeier HM. 2011. The use of binding-prediction models to identify M. bovis-specific antigenic peptides for screening assays in bovine tuberculosis. Vet. Immunol. Immunopathol. 141: 239–245 [DOI] [PubMed] [Google Scholar]
- 20. Kawaji S, Gumber S, Whittington RJ. 2012. Evaluation of the immunogenicity of Mycobacterium avium subsp. paratuberculosis (MAP) stress-associated recombinant proteins. Vet. Microbiol. 155: 298–309 doi:10.1016/j.vetmic.2011.08.021 [DOI] [PubMed] [Google Scholar]
- 21. Kawaji S, Zhong L, Whittington RJ. 2010. Partial proteome of Mycobacterium avium subsp. paratuberculosis under oxidative and nitrosative stress. Vet. Microbiol. 145: 252–264 [DOI] [PubMed] [Google Scholar]
- 22. Kim KK, Kim R, Kim SH. 1998. Crystal structure of a small heat-shock protein. Nature 394: 595–599 [DOI] [PubMed] [Google Scholar]
- 23. Leroy B, et al. 2007. Antigen discovery: a postgenomic approach to paratuberculosis diagnosis. Proteomics 7: 1164–1176 [DOI] [PubMed] [Google Scholar]
- 24. Li L, et al. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102: 12344–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu XF, et al. 2001. Identification of a secreted superoxide dismutase in Mycobacterium avium subspecies paratuberculosis. FEMS Microbiol. Lett. 202: 233–238 [DOI] [PubMed] [Google Scholar]
- 26. Mackintosh CG, De Lisle GW, Collins DM, Griffin JFT. 2004. Mycobacterial diseases of deer. N. Z. Vet. J. 52: 163–174 [DOI] [PubMed] [Google Scholar]
- 27. Men Y, et al. 1999. MHC class I- and class II-restricted processing and presentation of microencapsulated antigens. Vaccine 17: 1047–1056 [DOI] [PubMed] [Google Scholar]
- 28. Mustafa AS, Shaban FA. 2006. ProPred analysis and experimental evaluation of promiscuous T-cell epitopes of three major secreted antigens of Mycobacterium tuberculosis. Tuberculosis 86: 115–124 [DOI] [PubMed] [Google Scholar]
- 29. Nielsen M, et al. 2003. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 12: 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pedone E, Cannio R, Saviano M, Rossi M, Bartolucci S. 1999. Prediction and experimental testing of Bacillus acidocaldarius thioredoxin stability. Biochem. J. 339: 309–317 [PMC free article] [PubMed] [Google Scholar]
- 31. Ponomarenko J, et al. 2008. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics 9: 514 doi:10.1186/1471-2105-9-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sardinas G, et al. 2009. Assessment of vaccine potential of the Neisseria-specific protein NMB0938. Vaccine 27: 6910–6917 [DOI] [PubMed] [Google Scholar]
- 33. Saxegaard F, Baess I. 1988. Relationship between Mycobacterium avium, Mycobacterium paratuberculosis and ‘wood pigeon mycobacteria.’ Determinations by DNA-DNA hybridization. APMIS 96: 37–42 [PubMed] [Google Scholar]
- 34. Stevenson K. 2010. Diagnosis of Johne's disease: current limitations and prospects. Cattle Pract. 18: 104–109 [Google Scholar]
- 35. Tenzer S, et al. 2005. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell. Mol. Life Sci. 62: 1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vani J, Shaila MS, Chandra NR, Nayak R. 2006. A combined immuno-informatics and structure-based modeling approach for prediction of T cell epitopes of secretory proteins of Mycobacterium tuberculosis. Microbes Infect. 8: 738–746 [DOI] [PubMed] [Google Scholar]
- 37. Vijayasri S, Agrawal S. 2005. Domain-based homology modeling and mapping of the conformational epitopes of envelope glycoprotein of West Nile virus. J. Mol. Model. 11: 248–255 [DOI] [PubMed] [Google Scholar]
- 38. Wang P, et al. 2008. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 4: e1000048 doi:10.1371/journal.pcbi.1000048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Werther T, et al. 2006. New insights into structure-function relationships of oxalyl CoA decarboxylase from Escherichia coli. FEBS J. 277: 2628–2640 [DOI] [PubMed] [Google Scholar]
- 40. Whittington RJ, Marshall DJ, Nicholls PJ, Marsh IB, Reddacliff LA. 2004. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl. Environ. Microbiol. 70: 2989–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zahrt TC, Deretic V. 2002. Reactive nitrogen and oxygen intermediates and bacterial defenses: unusual adaptations in Mycobacterium tuberculosis. Antioxid. Redox Signal. 4: 141–159 [DOI] [PubMed] [Google Scholar]
- 42. Zhao BP, et al. 2011. In silico prediction of binding of promiscuous peptides to multiple MHC class-II molecules identifies the Th1 cell epitopes from secreted and transmembrane proteins of Schistosoma japonicum in BALB/c mice. Microbes Infect. 13: 709–719 [DOI] [PubMed] [Google Scholar]
- 43. Zhu X, et al. 2008. Transcriptional analysis of diverse strains of Mycobacterium avium subspecies paratuberculosis in primary bovine monocyte derived macrophages. Microbes Infect. 10: 1274–1282 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.