Abstract
Ywp1 is a prominent glycosylphosphatidylinositol (GPI)-anchored glycoprotein of the cell wall of Candida albicans; it is present in the yeast form of this opportunistic fungal pathogen but absent from filamentous forms and chlamydospores. Yeast cells that lack Ywp1 are more adhesive and form thicker biofilms, implying an antiadhesive activity for Ywp1, with a possible role in yeast dispersal. The antiadhesive effect of Ywp1 is transplantable from yeast to hyphae, as hyphae that are forced to express YWP1 lose adhesion in an in vitro assay. Deletion of the GPI anchor results in loss of Ywp1 to the surrounding medium and reduction of the antiadhesive effect, implying an importance of time-dependent residency in the cell wall. Anchor-negative versions of Ywp1 possessing or lacking a C-terminal green fluorescent protein (GFP) tag were created in C. albicans and harvested from culture supernatants; in addition to serving as quantifiable markers for Ywp1 secretion, they revealed that the cleaved 11-kDa propeptide of Ywp1 remains strongly but noncovalently associated with the Ywp1 core. This association is resistant to highly acidic and basic solutions, 8 M urea, and 1% SDS (below 45°C). Above 50°C, SDS dissociates the isolated complex, but even higher temperatures are required to dissociate the propeptide from native Ywp1 that is anchored in a cell wall. This property has permitted detection, for the first time, of orthologs of Ywp1 in other members of the Candida clade. The cleaved propeptide, which carries the sole N-glycan of Ywp1, must participate in the antiadhesive effect of Ywp1.
INTRODUCTION
Candida albicans is the most prevalent fungal pathogen of humans, often existing as a commensal in the alimentary tract but being capable of dispersing and causing disease in diverse anatomical locations when immune defenses are compromised. Superficial infections of skin and mucosae are usually not life-threatening, but systemic infections have a high mortality rate (46). Adhesion and biofilm formation are critical components of the persistence and pathogenesis of this fungus in the human population. Biofilms typically start as adherent yeast forms (also termed blastospores or blastoconidia) that develop into filamentous (hyphal and pseudohyphal) forms as they proliferate; the cells become enmeshed in a loose polymeric matrix and develop enhanced drug resistance (3, 8, 13) in a complex and dynamic process that is influenced by a diverse array of regulatory and surface molecules (14). Cells may adhere to tissue surfaces as well as catheters and medical prostheses in vivo and to various substrates in vitro; the adhesion properties of yeast forms are relevant to both initial adhesion and later dispersal and thus have an important role in the commensalism and pathogenesis of this organism.
Just outside the plasma membrane of each Candida cell is an enclosing wall that provides physical protection from hypo-osmosis and other environmental perils and is also the point of contact between the fungus and the host. It is composed of a durable network of β-1,3-glucans cross-linked by β-1,6-glucans and reinforced with chitin (32). Glycoproteins embedded in the plasma membrane or covalently attached to the glucans make up the major protein component of the wall. Rich in mannose-containing glycans, these mannoproteins are prevalent at the outer surface of the wall and are responsible for functions such as recognition, adhesion, and biofilm formation (7). Many of these proteins acquire a permanent or temporary glycosylphosphatidylinositol (GPI) anchor that dictates their wall association (11). This paper focuses on one such protein that has a structure similar to that of the known cell wall adhesins of C. albicans but, unexpectedly, has an antiadhesive effect.
Ywp1 (Pga24) is a GPI-anchored glycoprotein of the C. albicans cell wall, based on predictions from its sequence (11, 35, 62) and on investigations of the protein itself (10, 23, 52). The 533-amino-acid (aa) polypeptide encoded by YWP1 has the following features: (i) an N-terminal signal peptide of 21 aa that directs the nascent polypeptide into the secretory pathway but is then cleaved and lost; (ii) a tribasic sequence (RRR) followed closely by a dibasic sequence (KR), both of which are cleaved to create an N-terminal propeptide of about 100 aa (at least one of these cleavages is predicted to occur intracellularly, prior to secretion); (iii) a segment of about 360 aa that is especially rich in threonine, with a third of the residues in this segment being either threonine or serine (this segment contains three tandem copies of a 42- to 51-aa motif that is also found in several other wall proteins [23, 39, 54]); and (iv) a hydrophobic C terminus and adjacent signal that result in replacement of the C-terminal 22 aa with a GPI moiety anchored in the plasma membrane. The GPI anchor is subsequently cleaved, and the 378-aa core of Ywp1 becomes attached covalently to the glucan network of the cell wall through a lipid-free remnant of the GPI anchor. A single N-glycan attachment site is present in the Ywp1 polypeptide, near the C terminus of the cleaved propeptide. The Ser/Thr-rich central region of Ywp1 is predicted to be heavily O-glycosylated, consistent with mass spectroscopic analysis of two tryptic peptides from this region (10).
YWP1, the gene encoding Ywp1, is located on chromosome 2 of C. albicans (2, 30), about 6 kb downstream of a nonexpressed pseudogene paralog (2, 5, 23, 64). YWP1 is highly expressed during exponential-phase growth of yeast forms (23) and is strongly suppressed under conditions that promote filamentation (23, 41, 56). It continues to be regarded as a specific marker for the yeast form of C. albicans (18). Deletion of both alleles of YWP1 results in strains with normal growth rates, morphologies, and physicochemical sensitivities (23, 43, 47) but increased adhesiveness of the yeast forms (23). Deletion of just one allele of YWP1 results in a slight increase in adhesiveness, suggesting a nonlinear dosage effect (23). Ywp1 is thus abundant in the yeast cell wall but not the hyphal cell wall, correlating with its demonstrated effect on yeast cell adhesion but not hyphal cell adhesion. In systemically infected mice, YWP1 deletion has little effect on kidney colonization (43) or mouse survival times (23), but numerous other aspects of infection and pathogenicity have yet to be examined.
The molecular mechanism of the antiadhesive effect of Ywp1 is currently unknown. The amino acid sequence of Ywp1 reveals no obvious binding or enzymatic activities that might be important for this effect. Orthologs of YWP1 are present in other members of the Candida clade (6), both pathogenic and nonpathogenic, but not in the Saccharomyces clade or more distant genera; since Ywp1 has so far been identified and studied only in C. albicans, there is currently no information from homologs that might provide clues about its activities. YWP1 expression patterns have been revealed by dozens of published transcript profiling and mass spectroscopy experiments with C. albicans (see File S1 in the supplemental material), but the conditions that have been tested to date represent a small fraction of those that might be encountered by C. albicans in vitro and in vivo; collectively, clear correlations of YWP1 expression have not yet emerged for phenotypes other than the proportion of actively growing yeast forms (as opposed to filamentous or filamenting forms) in the populations that have been sampled, and none of the findings are inconsistent with an antiadhesive role for Ywp1. This paper focuses on the Ywp1 protein itself and reveals several functional and structural features that promote a better understanding of how Ywp1 might effect its adhesion-inhibiting activities.
MATERIALS AND METHODS
Strains and media.
Candida strains (detailed in Table S1 in the supplemental material) were routinely grown as yeast cells in well-aerated shaking flask cultures at 30°C in a defined minimal medium containing 100 mM glucose and 80 mM ammonium chloride as the carbon and nitrogen sources (medium 13 in reference 23). Supplemental arginine, histidine, and uridine were added as needed, and phosphate was limited in some experiments by reducing its concentration from 5 mM to 0.2 to 0.3 mM. These and other modifications are specified below or in individual figure legends.
Mycelial growth of C. albicans for adhesion assays was performed in multiwell polystyrene tissue culture plates containing RPMI medium at 37 to 38°C. Similar results were observed for Thermanox and polyvinyl chloride substrates. A version of RPMI 1640 (Sigma) that contains 11 mM glucose but no bicarbonate or phenol red was supplemented with 25 mM HEPES, 10 to 15 mM NaOH (to raise the pH to 7.5 to 7.7), 0.1 to 0.5 mM uridine, and 0.02% bovine serum albumin (BSA). For growth of mycelia that were utilized for protein analyses, the RPMI medium was supplemented as described above, except with an extra 14 mM glucose (to bring the total to 25 mM) and no BSA.
Mammalian cell lines were grown in Dulbecco's modified Eagle's medium containing 6% heat-inactivated fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2. Lines included CV-1 (African green monkey kidney), NRK (normal rat kidney), HeLa (human cervical carcinoma), and CHO (Chinese hamster ovary) cells.
Genetic transformations.
For the ectopic (hyphal) expression of YWP1, the initial coding sequence of one allele of HWP1 was replaced, by homologous recombination (67), with one of four YWP1-HIS1 cassettes (detailed in Table S2 in the supplemental material). Lithium acetate-polyethylene glycol (PEG) transfections (20) of the PCR amplicons into C. albicans strains BWP17 (YWP1/YWP1) and Ca#12 (ywp1/ywp1), as well as subsequent selections and genetic analyses, were performed as described previously (23). Selected transformants of strain Ca#12, which previously had YWP1 coding nucleotides +64 to +815 replaced by ARG4 in one allele and by a recyclable URA3-dpl cassette in the other allele (23), were grown in the presence of 5-fluoroorotic acid (5-FOA) to select progeny that had lost the inserted URA3 cassette; this allowed disruption of the ectopic YWP1 gene with a new URA3 cassette designed to replace coding nucleotides +173 to +435 of the ectopic YWP1 gene (see Table S2 in the supplemental material).
To create an anchor-negative version of Ywp1, a HIS1 cassette was designed to place a stop codon just upstream of the GPI attachment site codon of YWP1 (detailed in Table S2 in the supplemental material). This cassette was transfected into three strains of C. albicans that were derivatives of strain BWP17, with either two wild-type YWP1 alleles or codons 22 to 271 of one YWP1 allele replaced with either ARG4 or URA3. The STOP/HIS1 cassette was targeted to (and homologously recombined with) the wild-type YWP1 alleles as well as the previously disrupted (Δ) YWP1 alleles, giving transformants (histidine prototrophs) with all combinations of wild-type (WT) and anchor-negative (anchor-minus [AM]) Ywp1: WT/AM, WT/Δ, and Δ/AM. Along with the parental strains (WT/WT and WT/Δ), this made comparison of several independent transformants possessing various auxotrophies possible, thus strengthening the attribution of phenotypic changes to anchor-negative Ywp1. The engineered genetic changes were confirmed by numerous PCR analyses; for one transformant, the genomic DNA that encompassed the introduced stop codon was amplified and confirmed by sequencing.
To create YWP1-GFP chimeras, GFP-HIS1 amplicons (23) were used to transform C. albicans strain BWP17 and a URA3 derivative of BWP17. The GFP portion of these constructs was the yeast enhanced green fluorescent protein (EGFP) construct (yEGFP3; encodes S65G and S72A enhancements and is optimized for Candida codon usage) of Cormack et al. (9) and was targeted to codons 119, 165, 520, and 533 of YWP1 (see Table S2 in the supplemental material). Transformants (histidine prototrophs) were screened for correct insertion into YWP1 by PCR analyses and by fluorescence detection of secreted GFP; the success rate was 55% (10/18 transformants) for Ywp165-GFP and 69% (9/13 transformants) for Ywp520-GFP; these values are much higher than those typically seen with only 58 to 63 bp of matched flanking sequence (21).
Protein analysis.
Mannoproteins that were present in culture supernatants or extracted from live cells by use of agents such as SDS and disulfide-reducing agents (dithiothreitol [DTT] and Tris-carboxyethyl phosphine [TCEP]) were concentrated by precipitation with 1 to 2 volumes of ethanol (23); secreted Ywp1-GFP chimeras were harvested from culture supernatants by centrifugal ultrafiltration in devices with nominal cutoffs of 10 to 50 kDa. Unlike yeast cells, mycelia could not be centrifuged into compact pellets, so they were separated from surrounding media and from subsequent washes and extracts by filtration in small, disposable columns with porous frits. β-1,3-Glucanase (Quantazyme ylg) digestions of cell walls previously extracted with DTT, hot SDS, and saturated urea were performed as described previously (23), as were mannoprotein digestions with peptide N-glycanase F (PNGase F; New England BioLabs).
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) utilized the discontinuous Tris-glycine-chloride system of Laemmli (34) or the Tris-Tricine-chloride modification (for Fig. 4 and 7A) of Schägger and von Jagow (51); samples in Fig. 1 and 3 were reduced with DTT prior to electrophoresis. For the Ywp1-GFP fusion proteins, the Laemmli system was modified as described in File S2 in the supplemental material. Western blotting was performed as described previously (23), using polyvinylidene difluoride (PVDF) filters and chromogenic detection for Fig. 3. For Fig. 1, secondary goat anti-mouse IgG was conjugated to horseradish peroxidase, and detection involved enhanced chemiluminescence capture by X-ray film. In both cases, mouse antisera elicited by DNA vaccination against Ywp1 amino acids 51 to 197 and 105 to 161 were used (23). The brightness and contrast of all images were adjusted uniformly with Adobe Photoshop.
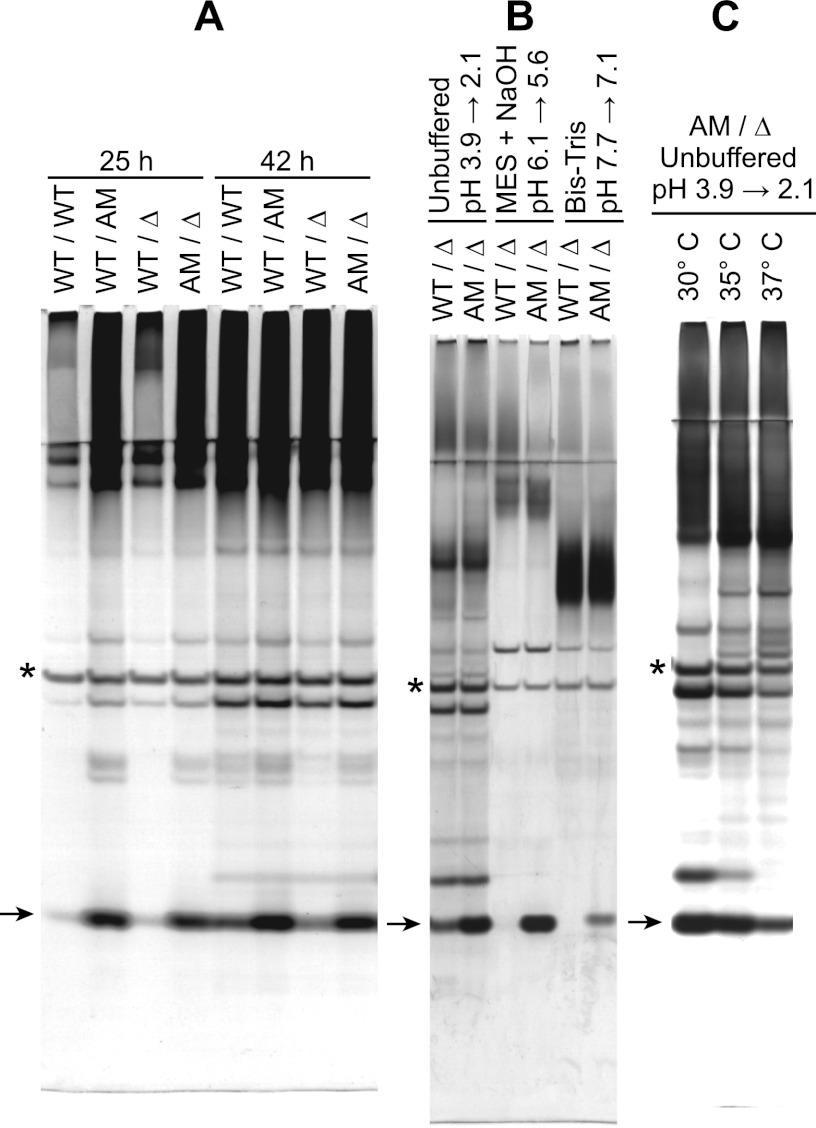
Fig 4.
Ywp1 propeptide quantity monitored by SDS-PAGE and silver staining. Four independent strains possessing wild-type (WT), anchor-negative (AM), and/or deleted (Δ) versions of Ywp1 were compared. All cultures were grown in medium 13 at 30°C (A and B) or as indicated (C); for panel B, the medium was unbuffered, buffered with 100 mM morpholineethanesulfonic acid (MES) plus 50 mM NaOH, or buffered with 100 mM Bis-Tris, with the indicated initial and final pH values for each culture. After growth for 88 h (B), 48 h (C), or as indicated (A), mannoproteins were precipitated from the culture supernatants with ethanol and digested with PNGase F. All samples were heated in SDS without disulfide reduction and resolved in Tris-Tricine gels. Each lane in panel A represents the same number of cells. Arrows point to the deglycosylated Ywp1 propeptide (11 kDa), and asterisks are adjacent to PNGase F (36 kDa).
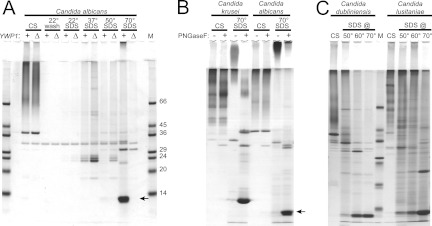
Fig 7.
Extraction of the Ywp1 propeptide from intact cells by use of SDS. Yeast cell culture supernatants (CS) were harvested and compared with sequential SDS extracts of whole cells. SDS-PAGE was followed by protein staining with Coomassie blue R-250. (A) Wild-type C. albicans strain SC5314 (+) compared with ywp1/ywp1 knockout strain 4L1 (Δ). All samples were ethanol precipitated and digested with PNGase F prior to electrophoresis. The Ywp1 propeptide (arrow) was extracted from intact cells by SDS at 70°C but not at 50°C or below. (B) The “C. albicans 70° SDS” lanes show that the migration of the band at 11 kDa depended upon removal of its N-glycan with PNGase F; otherwise, it migrated only into the stacking gel. A similar pattern was seen for C. krusei. (C) PNGase F digests of similar samples from C. dubliniensis and C. lusitaniae; again, most of the putative Ywp1 propeptide was extracted from intact cells by SDS only at temperatures above 50°C. M, markers with the indicated masses (in kDa).
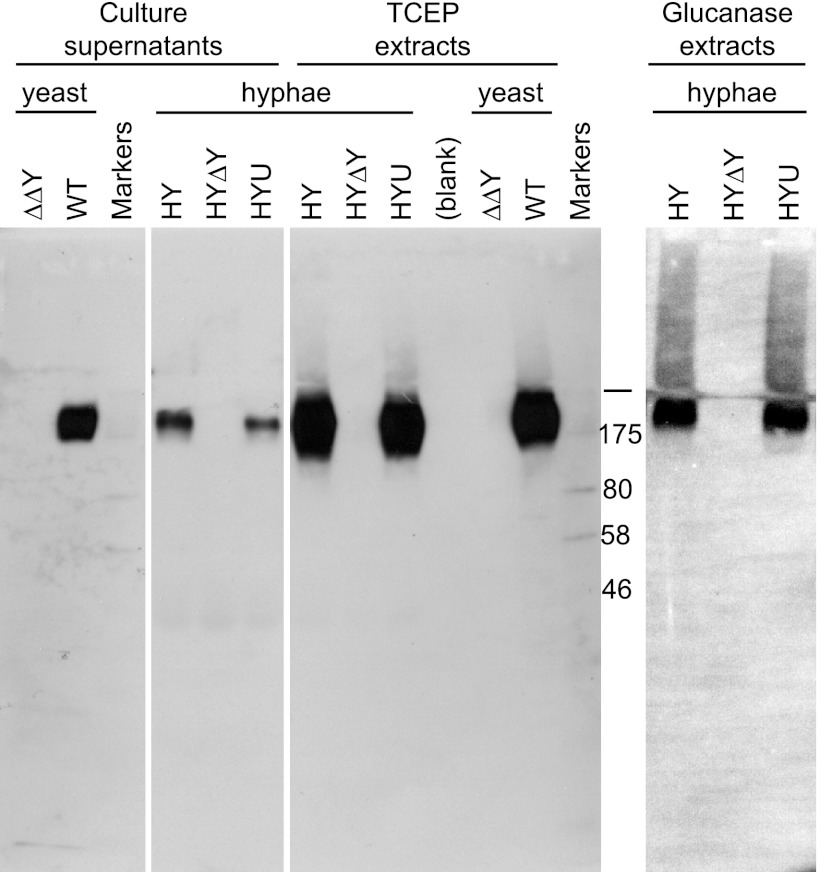
Fig 1.
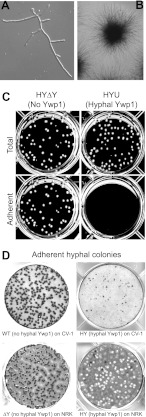
Normal (yeast) and ectopic (hyphal) production of Ywp1. Western blotting reveals the presence of Ywp1 in culture supernatants, in disulfide-reduced (TCEP) extracts of intact cells, and in β-1,3-glucanase extracts of cell walls, as indicated. Strains (all derived from C. albicans SC5314) included the wild type (WT; DAY185), a ywp1/ywp1 knockout (ΔΔY; 4L1), and a ywp1/ywp1 knockout that subsequently had YWP1 placed under the control of the HWP1 promoter (HY); a transfected URA3 cassette then either disrupted the ectopic YWP1 gene in HY (giving HYΔY) or was inserted randomly into the genome (giving HYU). Cells were grown as yeast or hyphae, as indicated. Nonspecifically labeled marker bands provide size references of 46, 58, 80, and 175 kDa. The bar denotes the junction of stacking and resolving gels.
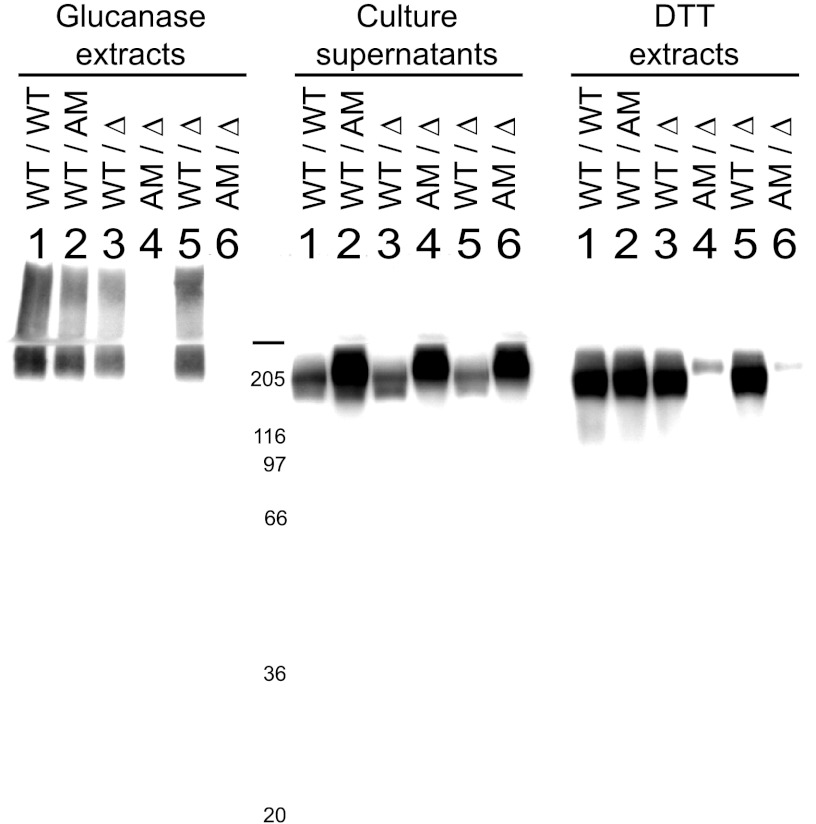
Fig 3.
Anchor-negative Ywp1 visualized by Western blotting. Strains possessing wild-type (WT), anchor-negative (AM, anchor-minus), and deleted (Δ) versions of Ywp1 (as indicated for each allele of YWP1) were grown as yeast in low-phosphate medium 13 at 30°C for 95 h. Culture supernatants, DTT extracts of the intact cells, and β-1,3-glucanase extracts of cell walls were ethanol precipitated, resolved by SDS-PAGE, electroblotted onto a PVDF filter, and probed with anti-Ywp1. The DTT extracts represent 3.3 times as many cells as the other two samples. The six lanes represent independent strains with auxotrophies for arginine (lanes 1, 2, 5, and 6), uridine (lanes 3 and 4), and/or histidine (lanes 1). Marker proteins migrated as indicated (masses in kDa), and the horizontal bar marks the top of the resolving gel.
N-terminal sequencing by Edman degradation of a polypeptide electroblotted onto a PVDF filter was performed at the Iowa State University Protein Facility. Mass spectrometry was performed at the Montana State University Mass Spectrometry and Proteomics Facility, as follows. Gel-eluted polypeptides were bound to an Agilent 1100 reverse-phase high-performance liquid chromatography (HPLC) column in 0.1% formic acid, eluted with a gradient of acetonitrile, and fed into a Bruker micrOTOF instrument with electrospray ionization (set at 100 V) and a time-of-flight detector.
RESULTS
YWP1 expression correlates with cellular morphology.
C. albicans YWP1 is expressed abundantly in yeast but is downregulated upon filamentation; this has been shown by transcript profiling (31, 40, 41, 56) and by using GFP as a reporter of YWP1 gene expression (23). The latter technique has also revealed that YWP1 is not expressed by suspensor cells or chlamydospores (see Fig. S1 in the supplemental material). Even though the rate of YWP1 downregulation and functional mRNA loss upon filamentation can be high (31, 40, 41, 56), contributions from the diffusion of existing mRNA molecules or from incompletely shut-down YWP1 in early daughter filaments are likely low but have not been ruled out.
Ectopic expression of YWP1.
Deletion of both alleles of YWP1 was previously found to increase C. albicans yeast adhesiveness, implying that the presence of Ywp1 in the yeast cell wall results in decreased adhesiveness (23). If this is indeed due to an effect of Ywp1 in the cell wall, then ectopic Ywp1 might similarly decrease the adhesiveness of hyphae, which normally lack this protein. To test this, YWP1 was placed under the control of the endogenous hyphal wall protein 1 gene (HWP1) promoter so that it would be expressed in filamentous forms of C. albicans rather than in yeast forms (61); the adhesiveness of the filamentous forms was then examined.
Strain Ca#12, which previously had both of its YWP1 alleles disrupted (23), was transformed with a series of constructs that targeted cloned versions of YWP1 to the HWP1 locus. The start codon of one allele of HWP1, along with 32 or 412 downstream HWP1 codons, was replaced with the coding sequence of YWP1, resulting in strains that drove expression of YWP1 exclusively with the natural HWP1 promoter. The other HWP1 allele remained unaltered. Six independent strains were created, representing both alleles of YWP1 and different extents of HWP1 replacement; all gave similar results (described below), lessening the possibility that the observed changes were due to unforeseen mutations. For further confirmation and to ensure that the observed effects were not due simply to loss of one of the two HWP1 alleles, the ectopic YWP1 gene was disrupted with URA3 in two of the strains. The data presented here are from one representative set of three strains, denoted HY (hyphal YWP1 strain), HYΔY (hyphal YWP1 strain in which the ectopic YWP1 gene was disrupted with URA3), and HYU (hyphal YWP1 strain in which the URA3 cassette was inserted elsewhere in the genome, leaving the ectopic YWP1 gene intact).
Ectopic hyphal Ywp1 was found to have properties similar to those of yeast Ywp1. (The term “hyphal” is used here to include hyphae and pseudohyphae found in the filamentous growth of mycelia.) Figure 1 shows that ectopic Ywp1 is detectable by Western blotting of hyphal culture supernatants and that Ywp1 is extractable from hyphae by use of disulfide-reducing agents. Similar samples from wild-type and ywp1 knockout yeast are shown for comparison. The deglycosylated Ywp1 propeptide was also detected readily in these samples by silver staining (data not shown). In addition, β-1,3-glucanase digestion liberated Ywp1 from hyphal cell walls. Thus, ectopic Ywp1 appears to be incorporated covalently into the hyphal cell wall, as it is in the yeast cell wall, with a small fraction being lost to the surrounding medium.
The effect of ectopic Ywp1 on hyphal adhesion was demonstrated most easily in a simple microcolony assay. Yeast cells were seeded sparsely in tissue culture plates containing RPMI 1640 (a mammalian cell culture medium) and incubated undisturbed at 37 to 38°C. Virtually all of the cells initiated filamentous growth, with no obvious morphological differences between strains possessing and lacking hyphal Ywp1 (Fig. 2A). The developing mycelial colonies remained adherent to the substrate, forming radially stellate colonies of ∼0.5 to 3 mm in diameter after 1 to 3 days of growth (Fig. 2B). Physical disruption of these microcolonies, which resembled hemispherical Koosh balls, revealed few blastoconidia. After 15 to 25 h of growth, and for several days thereafter, gentle agitation of the culture dislodged those colonies that expressed ectopic Ywp1 but left the other colonies adherent (Fig. 2C). The dislodged colonies retained their hemispherical shapes. Thus, the antiadhesive effect of Ywp1 appears to be transplantable from yeast to hyphae.
Fig 2.
Hyphal microcolony growth and adhesion. Sparsely plated yeast cells grown under filament-inducing conditions generated adherent hyphae regardless of YWP1 expression. (A) Strain HY after 8 h of growth. (B) Within 2 days, macroscopic, radially stellate colonies developed, and they remained adherent unless YWP1 was expressed ectopically. (C) Gentle agitation of a 6-well plate, followed by removal of nonadherent colonies, demonstrated the release of strains expressing hyphal YWP1 (strain HYU in this case) and the continued adherence of both wild-type strains and those in which the ectopic YWP1 gene had been disrupted (strain HYΔY in this case). (D) Growth of colonies on monolayers of mammalian cells (monkey CV-1 and rat NRK cells in this case) resulted in similar adhesion properties, with hyphal expression of YWP1 resulting in loss of adherence. The wells were stained with crystal violet after gentle removal of nonadherent colonies, resulting in dark fungal colonies and lighter thinning of the NRK monolayer under dislodged colonies.
Under identical conditions, two independent strains lacking Als3 (42, 69) had unaltered colony morphology and initial attachment to the substrate, but the stellate colonies showed eventual loss of adhesion, typically several hours sooner than the colonies expressing ectopic hyphal Ywp1 (data not shown). This detachment was not observed in a parental control strain (69), and detachment was inhibited when ALS3 was restored (68). In contrast, strains lacking Hyr1, Cht2, Ece1, Rbt5, Ecm331 (42), or Als1 (16) all maintained adhesion of their stellate colonies (data not shown). Thus, among the few GPI-anchored proteins tested, only the absence of Als3 and the ectopic presence of Ywp1 resulted in loss of hyphal adhesion under these conditions.
Interestingly, none of the strains formed adherent microcolonies unless exogenous protein was present, either in the culture medium or preadsorbed to the plastic. BSA was typically included in the culture medium at 0.02% for these assays, and gelatin and fetal bovine serum could substitute for the BSA. One of four tested brands of gelatin, however, as well as heating of the BSA to 80°C, negated this effect (resulted in loss of adhesion for all strains) (data not shown). Even though the in vitro growth assays that have been devised to document the antiadhesive effect of Ywp1 are not identical to any growth condition in vivo, they are nevertheless robust and convincingly document a role for Ywp1 in reduced cellular adhesion.
Similar antiadhesion effects of ectopic hyphal Ywp1 were observed on a more complex substrate, namely, live monolayers of various mammalian cell lines (Fig. 2D). To cultures of confluent or nearly confluent monolayers of CV-1, NRK, HeLa, and CHO cells were added a small number of wild-type or HY C. albicans yeast cells, which were allowed to grow undisturbed for 1 to 2 days. Poorly adherent microcolonies were dislodged with gentle agitation, and the adherent cells were fixed with ethanol and stained with aqueous crystal violet (shown in Fig. 2D for CV-1 and NRK cell monolayers). As observed for microcolony growth on bare plastic, hyphal expression of YWP1 inhibited adhesion to the mammalian cells. The mammalian cells continued to look healthy throughout the culture period, but there was diminished cell mass directly beneath the fungal microcolonies, visualized most readily by diminished staining where microcolonies had detached from the NRK cell monolayer (Fig. 2D). Similar culture conditions have resulted in minimal disturbance to a variety of primary human cell monolayers (60). The extent of fungal penetration through the mammalian cell monolayers to the plastic substrate was not determined; details of this adhesion are therefore uncertain, but it nonetheless appears to be inhibited by ectopic Ywp1.
Hyphal expression of YWP1 was also engineered into strain BWP17, which has two unaltered YWP1 alleles, resulting in strains that exhibited both normal yeast expression of YWP1 and ectopic hyphal expression of YWP1. Phenotypically, the adhesion properties of the hyphae were the same as those described above, i.e., apparently unaffected by the wild-type YWP1 alleles (data not shown). Since YWP1 and HWP1 have largely complementary and exclusive expression patterns (in yeast and hyphae, respectively), the adhesion properties of the two morphologies appear to be independently assayable. Nevertheless, since mycelial cultures can also generate blastoconidia under various conditions, the strains possessing just hyphal Ywp1 ruled out the possibility that yeast Ywp1 or its effects might have influenced the results of these experiments.
Anchor-negative Ywp1.
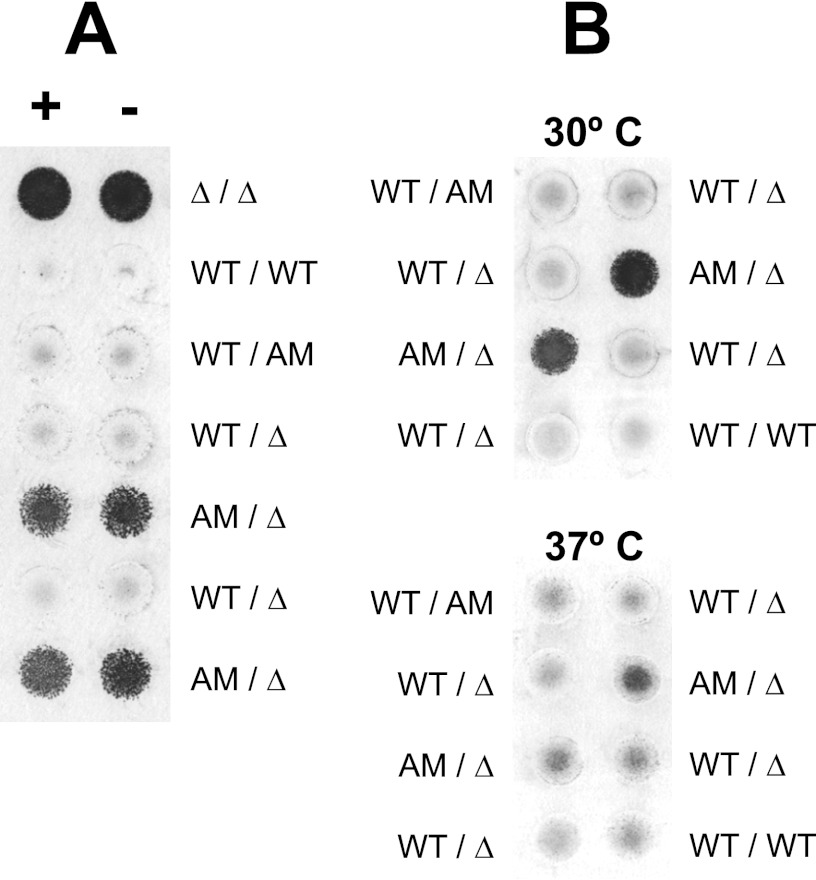
An anchor-negative version of Ywp1 was created by inserting a stop codon immediately upstream of the “omega” (glycine 511) codon of the GPI attachment site of YWP1, thus eliminating the C-terminal 24 amino acids of Ywp1. Multiple strains were engineered for comparison, differing in their auxotrophies and in the numbers of WT, AM (anchor-minus), and ΔYWP1 alleles. As shown below, the observed phenotypes were found to be independent of the auxotrophies.
Western blotting with anti-Ywp1 revealed that virtually all of the anchor-negative Ywp1 was secreted into the medium, with little or none remaining associated with the cell wall (Fig. 3). Culture supernatants showed immunoreactive Ywp1 near the top of the resolving gel; the low mobility is presumably attributable to extensive O-glycosylation of the 38-kDa core polypeptide. In these stationary-phase supernatants, strains with anchor-negative Ywp1 showed the greatest amount of immunoreactivity, with a slightly diminished mobility compared to that for wild-type Ywp1 (some of which is released from the cells by unknown mechanisms). DTT extracts of intact cells showed immunoreactive Ywp1 only if those cells possessed a wild-type allele of YWP1. (The faint signal in the lanes for the anchor-negative allele only [lanes 4 and 6] is likely due to supernatant carryover from the unwashed cells.) β-1,3-Glucanase extracts of cell wall matrices also revealed immunoreactive Ywp1 only in those strains possessing a wild-type allele of YWP1, confirming that none of the anchor-negative Ywp1 was covalently linked to wall glucan. Much of that signal was found in the stacking gel, presumably because heterogeneous, covalently linked β-1,6-glucans and β-1,3-glucan remnants increased the size and diminished the mobility of this form of Ywp1. No Ywp1 immunoreactivity has been seen in any sample from ywp1 Δ/Δ strains (data not shown).
The appearance and accumulation of wild-type and anchor-negative Ywp1 in the culture medium were monitored semiquantitatively over time through visualization of the deglycosylated propeptide of Ywp1. (Propeptide cleavage is predicted to occur intracellularly, giving a 1:1 stoichiometry for the Ywp1 core and the propeptide.) Figure 4A shows that anchor-negative Ywp1 was prevalent in the culture medium at the end of exponential-phase growth (25 h), whereas wild-type Ywp1 was not. After 42 h, both forms had increased in quantity, but there was a greater fold increase in wild-type Ywp1. (Aside from the case for Ywp1, which can be identified in protein-stained gels only after deglycosylation, no consistent differences in band patterns have been observed between WT/WT and Δ/Δ strains [23; our unpublished observations].) Numerous analyses such as these have suggested that a substantial fraction (half or more) of the wild-type Ywp1 that is initially anchored to the cell eventually ends up in the medium in 3- to 4-day-old stationary-phase yeast cultures. However, this loss (“shedding”) appears to be pH dependent, as shown in Fig. 4B. Using the anchor-negative form of Ywp1 as a quantitative standard, it was apparent that the wild-type form accumulated in the medium when the culture was unbuffered, attaining a stationary-phase pH of about 2, but was nearly absent from the medium when the buffer kept the pH close to 6. Parallel cultures buffered at pH 7 to 8 also showed little or no wild-type Ywp1 in the medium. The quantity of the anchor-negative form was diminished at pH 7 to 8; this was not because the anchor-negative Ywp1 protein was partially incorporated into the cell wall at alkaline pH (as revealed by silver staining of DTT extracts and by Western blotting of glucanase extracts [data not shown]), however, suggesting that it was due to downregulation of YWP1 expression under a condition (alkaline pH) that promotes filamentation. As assessed by light microscopy, the cells from the acidic cultures in Fig. 4B were virtually all blastoconidia (yeast forms), while about 20% of the cells in the alkaline (pH 7 to 8) cultures consisted of short pseudohyphae. Notably, these alkaline cultures showed as much YWP1 expression as the acidic cultures did (as estimated by propeptide abundance in the medium) when phosphate was limiting or when exogenous farnesol was added to the medium, conditions that each resulted in all yeast forms (data not shown).
Another condition that promotes a filamentation pathway in C. albicans is temperature elevation to 37°C. Figure 4C shows acidic cultures grown at 30°C, 35°C, and 37°C, with the propeptide of secreted anchor-negative Ywp1 indirectly quantifying expression of YWP1; there was a significant drop-off in YWP1 expression at 37°C compared to 35°C. The same trend was seen in cultures buffered at around pH 5 and, to a lesser extent, in phosphate-limited cultures (data not shown). The cells in these cultures were virtually all yeast forms, yet growth at 37°C still appears to have downregulated YWP1 expression.
Strains that lack Ywp1 show increased blastoconidial adhesiveness, as demonstrated previously in a simple yeast biofilm assay (23). Yeast microcultures were arrayed on a flat surface, allowed to grow, rinsed of nonadherent cells, and stained with crystal violet. Strains that made wild-type Ywp1 had significantly sparser adherent biofilms (23). Figure 5A reveals that anchor-negative Ywp1 poorly mimics wild-type Ywp1 in this antiadhesive effect. Several independent strains with various auxotrophies, making WT and/or AM Ywp1, were compared using this assay. Strains making wild-type Ywp1 showed little yeast biofilm adhesion, while strains making only anchor-negative Ywp1 showed nearly as much adhesion as strains that made no Ywp1 of either sort. Results were independent of requirements for arginine, histidine, and uridine for growth. Enhanced adhesion in the absence of any Ywp1 was found to largely disappear when the growth temperature was raised from 30°C to 37°C, indicating that other (as yet unidentified) antiadhesion mechanisms must become operational in yeast cells at 37°C (23). Figure 5B shows that the same holds true for strains making only anchor-negative Ywp1. Thus, it appears that the antiadhesive effect of Ywp1 in yeast cells is largely dependent upon anchorage of Ywp1 at the cell surface or in the cell wall; mere extracellular presence and accumulation in the medium are not sufficient to markedly reduce yeast biofilm adhesion.
Fig 5.
Yeast biofilm adhesion assay. Microcultures arrayed on a polystyrene plate were grown in medium 13 with (A+ and B) or without (A−) supplemental arginine, histidine, and uridine for 92 h (A) or 63 h (B) at 30°C (A) or as indicated (B). Nonadherent cells were rinsed away, and the adherent cells were stained with crystal violet. Independent strains possessing wild-type (WT), anchor-negative (AM), and/or deleted (Δ) versions of Ywp1 were compared. The upper and lower portions of panel B show cultures that were identical except for their growth temperatures.
Ywp1-GFP chimeras.
Four Ywp1-GFP chimeras were created by transfection of C. albicans with selectable GFP-HIS1 PCR amplicons targeted to different positions in YWP1. The resulting chimeras included the N-terminal 119, 165, 520, or 533 amino acids of Ywp1 fused to C-terminal GFP. The Ywp119-GFP and Ywp533-GFP chimeras were barely detectable and were not investigated further. The Ywp165-GFP and Ywp520-GFP chimeras were more abundant and were secreted into the yeast culture medium, where they were easily harvested by ultrafiltration. Significantly, these chimeras represented the predominant protein species in these samples. By microscopy, the fluorescence of both chimeras could be visualized in transit through the secretory pathway, but neither showed detectable association with the cell wall after secretion (data not shown).
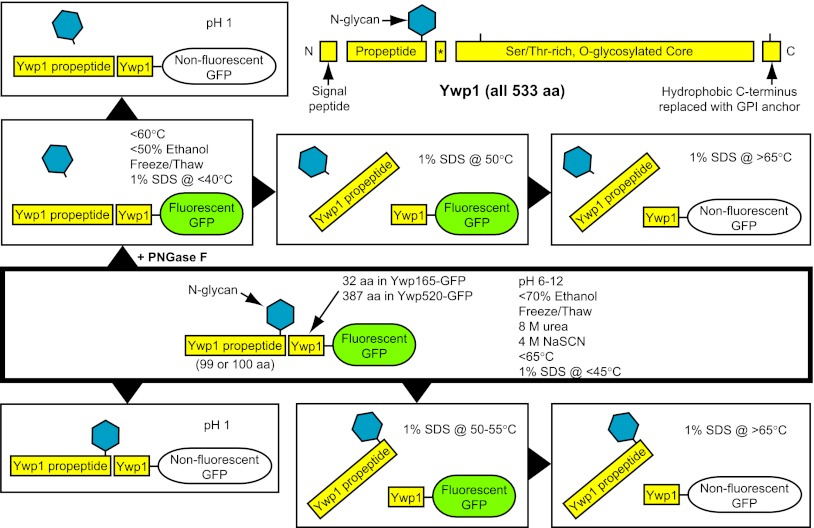
Ywp165-GFP included 32 amino acids downstream from the dibasic site of Ywp1 (…KRDQIDDFIASIENTEGTALEGSTLEVVDYVPGS-GFP; dibasic site is underlined); N-terminal sequence analysis of the gel-purified GFP moiety gave the sequence “DQIDD…,” confirming that cleavage had taken place immediately downstream from the dibasic site. The Ywp520-GFP chimera consisted of a replacement of the C-terminal 13 amino acids of Ywp1 (VLALALIPLAYFI), encompassing the hydrophobic GPI signal anchor, with GFP, giving a splice site sequence of …FEGAAAASAGAS-GFP, where the underlined G is the “omega” site for GPI attachment. This substitution thus disrupted the normal GPI attachment and anchor cleavage process. The resulting chimera included the repetitive and heavily O-glycosylated core stalk region of Ywp1.
Perhaps the most striking feature of the secreted Ywp1-GFP chimeras was that the cleaved Ywp1 propeptide remained strongly but noncovalently associated with the downstream (core) Ywp1 sequences. In the case of Ywp165-GFP, a segment of only 32 downstream Ywp1 amino acids evidently maintained this association. Remarkably, this association persisted at pH values between 1 and 12, at temperatures up to 65°C, in 8 M urea, and in 1% SDS at temperatures up to 45°C (see Fig. S2 and S3 and File S2 in the supplemental material). The properties of the Ywp1-GFP chimeras are summarized schematically in Fig. 6.
Fig 6.
Ywp1 schematic and summary of the known properties of the secreted Ywp1-GFP chimeras. Schematic natural fragments of the entire Ywp1 polypeptide are drawn to scale in the upper right, with ticks indicating the two points of GFP fusion. The 10- to 12-aa peptide (asterisk) between the cleaved tribasic and dibasic sites has not been detected experimentally and is presumably lost from mature Ywp1. In the other areas, the text within each box indicates conditions under which the indicated protein scenario predominates for the secreted Ywp1-GFP chimeras (at pH 8 and 20°C, unless specified otherwise), highlighting removal of the N-glycan with PNGase F, dissociation of the propeptide from the Ywp1 core, and maintenance of GFP fluorescence.
Independent analyses using mass spectrometry, carboxypeptidase B digestion, and electrophoretic mobility shifts all revealed that the Ywp165-GFP preparations always possessed similar amounts of two versions of the propeptide, differing by a C-terminal arginine (the first arginine of the tribasic site). Upon prolonged storage, limited end nibbling of the propeptide by endogenous proteases also became evident (see Fig. S2 in the supplemental material). The Ywp1 core region of Ywp520-GFP (387 aa) was more prone to cleavage upon storage than that of Ywp165-GFP (32 aa), as it afforded either specific sites or more opportunities for cleavage by endogenous proteases (see Fig. S3 in the supplemental material).
Ywp1 in the walls of wild-type cells.
The Ywp1-GFP chimera data suggested that in wild-type C. albicans, the Ywp1 propeptide might be dissociable from the cell wall by use of SDS at 50°C, leaving the downstream Ywp1 core covalently anchored within the wall matrix. To test this, whole wild-type and ywp1/ywp1 knockout yeast cells were washed and sequentially treated with 1% SDS at increasing temperatures. The original culture supernatant, wash, and SDS extracts were precipitated with ethanol, digested with PNGase F, and resolved by SDS-PAGE. Strikingly, there was little evidence of any cytosolic protein being extracted by the SDS; the predominant SDS-extracted protein was the Ywp1 propeptide (as evidenced by its deglycosylated form, which was absent from the knockout strain) (Fig. 7A). Instead of dissociating from the wall-anchored Ywp1 core at 50°C, however, as expected from the Ywp1-GFP chimera results, it dissociated at 70°C. Additional experiments (not shown) revealed that a minor proportion was extractable at 60°C, while most was released at 70°C. Identical results were obtained for five laboratory strains of C. albicans (SC5314, 3153A, 64550, CA-1, and A9), as well as a recent oral isolate (JG1). A strain lacking cell wall phosphomannan (CDH5 [MNN4Δ]) (27) gave the same result in these experiments as a wild-type strain (SC5314) and a strain with MNN4 restored (CDH13) (data not shown), indicating that simple electrostatic repulsion of the anionic dodecyl sulfate by anionic wall phosphodiesters cannot explain the resistance of the Ywp1 propeptide to dissociation. In addition, when hyphae that expressed ectopic YWP1 were tested (namely, strains HY and HYU versus the Ywp1-free strain HYΔY), the Ywp1 propeptide was liberated by SDS at 70°C but not at 50°C (data not shown).
Finally, regardless of their growth pH, when intact SC5314 yeast cells were subsequently treated with HCl at pH 2, no Ywp1 propeptide was extracted (data not shown); this is consistent with the inability of HCl at pH 1 to dissociate the propeptide from isolated Ywp165-GFP (see Fig. S2 in the supplemental material) and confirms that the low pH attained by unbuffered yeast cultures does not liberate the Ywp1 propeptide.
Ywp1 orthologs.
Orthologs of YWP1 can be identified in the genome sequences of other species of Candida and in closely related genera (6), and the N-terminal domains of the corresponding translations are highly conserved (see Fig. S4 in the supplemental material). In contrast to the genes, none of the non-C. albicans protein orthologs has yet been shown to exist. Our antisera that specifically bind to the C. albicans Ywp1 polypeptide (23) have not shown cross-reactivity against equivalent samples from non-C. albicans species, but candidate Ywp1 propeptide bands have been observed on gels containing samples similar to those that originally revealed C. albicans Ywp1 (unpublished observations). In light of the results shown in Fig. 7A, we tested non-C. albicans Candida species for the presence of candidate propeptides extractable from whole cells by use of hot SDS. We found that all of the tested species indeed exhibited such a protein; most of it was liberated from the cells by SDS at 60 to 70°C (but not at 50°C), and it appeared as a sharp band with a size similar to that of the C. albicans Ywp1 propeptide only after N-glycan removal by PNGase F. Figure 7B and C show a selection of the data for C. dubliniensis MYA-578, C. lusitaniae 64125, and C. krusei M-103 in comparison to C. albicans SC5314; C. parapsilosis 90018 and C. tropicalis 750 showed similar patterns, but with lesser amounts of these putative Ywp1 propeptide bands (data not shown). Except for C. krusei, whose genome has not been sequenced, the Candida Ywp1 propeptides are all predicted to have unglycosylated masses of 10.7 to 11.5 kDa, consistent with the observed bands.
DISCUSSION
The YWP1 gene is expressed primarily, if not exclusively, by the budding yeast form of C. albicans. The product of this gene, Ywp1, is a prominent glycoprotein of the yeast cell wall. Deletion of YWP1 results in greater adhesiveness and biofilm accumulation of the yeast morphotype in vitro, implying an antiadhesive effect of Ywp1. Evidence is presented here to show that this effect is attributable to the sustained presence of Ywp1 in the yeast cell wall. The effect is largely but not completely lost if Ywp1 lacks an anchor and passes through the wall without becoming incorporated, suggesting a time-dependent component of its antiadhesive activity. Furthermore, the antiadhesive effect is transplantable, as shown by the ectopic expression of YWP1 in hyphae. Detachment of such hyphae was observed only after many hours, again suggesting a requirement for a prolonged presence of Ywp1 in the wall. The molecular mechanism of this effect remains unknown but conceivably involves binding or enzymatic modification of other components of the cell wall. A cleaved propeptide that carries the sole N-glycan of Ywp1 must have a role in these effects, as it remains strongly but noncovalently associated with the Ywp1 core polypeptide in the cell wall.
The antiadhesive effect of Ywp1 presumably has an advantageous role in the growth or survival of C. albicans in mammalian hosts. Yeast forms and YWP1 expression are prominent during alimentary commensalism (50, 53, 66), and yeast forms usually coexist with filamentous forms at sites of pathogenic infection (44). Certain routes of dissemination to new growth sites may be favored by the yeast form, due to differences in size, shape, and interactions with host cells (22, 24, 29). The immune system responds differentially to yeast and hyphae (22), but no evidence has yet been presented to show that Ywp1 is a component of that recognition or response. Biofilms that grow on tissue surfaces or implanted medical devices have features that are similar to those of in vitro models, which are typically composed of basal yeast layers and overlaid filamentous layers; as such, they may depend on subsequent production of low-adherence yeast to effect dispersal (65). A recent report (49) suggested that phosphate limitation, which increases YWP1 expression in yeast cells (23), attenuates C. albicans SC5314 biofilm formation in vitro and in vivo and may lead to greater dissemination from the mouse cecum to the kidney, liver, and lung. The yeast cell adhesion that Ywp1 inhibits appears to be a highly complex process, considering that 30 transcription factors have been identified as promoting yeast adhesion to a silicone substrate (14). The postulated role for Ywp1 in yeast dispersal does not rule out other phenotypic traits that have not yet been discerned experimentally, however.
Ywp1 is prominent as a shed, thiol-extractable, SDS-extractable, and unanchored truncated component of the yeast secretome (23; this study) and is prominent in yeast secreted samples and wall samples that have been prepared for mass spectroscopy (26, 37, 57–59). This abundance appears to be an essential feature of the function of Ywp1, as reductions by half or more result in yeast cells that are clearly more adhesive (23). Loss of Ywp1 has no obvious effect on wall resistance properties (23, 47), suggesting that Ywp1 does not have a major structural role in the wall. The extensive shedding of Ywp1 and its accumulation in stationary-phase yeast cultures (23) appear to be largely a function of the low pH attained by these cultures and may be mediated by acid hydrolases (17, 33). GPI-anchored Sap9 may have a role in this shedding (52; our unpublished observations). Compared to anchor-negative Ywp1, shed wild-type Ywp1 has increased mobility during SDS-PAGE, suggesting that one or more proteolytic cleavage events simultaneously liberate and shorten it. Shedding of GPI-anchored proteins may also result from cell wall remodeling or from washout during the membrane-to-glucan transfer process (12, 33, 37, 52, 58). Notably, the extractability of Ywp1 with disulfide-reducing agents increases as yeast cultures age (23), suggesting that a subfraction of the shed (as well as unshed) molecules may become disulfide bonded to other polypeptides within the wall and thiol extractable only if the link to β-1,6-glucan (through the GPI remnant) has been severed. Ywp1 has nine cysteines in its 378-aa core, at least one of which is not involved in an intramolecular disulfide bond, and similar cysteines in another GPI-anchored protein have been shown to mediate wall anchorage (4). Formation of such disulfides may require prolonged residence times of the polypeptides in the wall, as the anchor-negative version of Ywp1 and the Ywp520-GFP chimera pass through the wall without obvious trapping by disulfide bond formation. Based on the results presented here, extensive shedding of Ywp1 may be largely an artifact of highly acidic cultures in vitro and is likely less prevalent in more moderate environments in vivo.
Cleaved propeptides often serve as intramolecular chaperones, folding catalysts, signaling molecules, or regulated inactivators. The current data do not reveal the function of the Ywp1 propeptide but do reveal the surprising persistence with which it remains associated with the Ywp1 core. As few as 32 aa of the N terminus of the Ywp1 core are required for this noncovalent association. Whether there are nondenaturing, nonproteolytic conditions that allow dissociation of the Ywp1 propeptide from the core as part of the normal functioning of Ywp1 remains to be determined. Nevertheless, no physical condition that C. albicans is likely to encounter in the human body, including passage through the stomach, is likely to dissociate the Ywp1 propeptide from the cell wall.
Formation of the 11-kDa propeptide by cleavage of Ywp1 at its tribasic site requires Kex2 and is predicted to occur intracellularly (23). The resulting propeptide exists predominantly as two versions that are present in similar amounts, retaining either one or none of the three arginines of the tribasic site (23; this study). Interestingly, Saccharomyces cerevisiae Kex2 itself has a cleaved propeptide, and C-terminal basic residues on this propeptide are critical for Kex2 activity (36); by extension, the C-terminal arginine on a subpopulation of the Ywp1 propeptides conceivably has an important regulatory function.
In C. albicans, Sap9 and Sap10 are surface GPI-anchored proteins whose proteolytic functions may overlap those of intracellular Kex2 (1, 52). The proteolytic activities of Sap9 and Sap10 span a broad pH range (pH 5 to 8), and although these proteins show a preference for basic and dibasic sites, their cleavage targets include a wide range of sequences (52). When applied to SDS-stripped cell walls, soluble recombinant Sap9 is capable of cleaving the Ywp1 core just 23 aa from its N terminus; Ywp1 that is shed into the culture medium from mutants lacking Sap9 and Sap10 shows no such cleavage upon incubation with recombinant Sap9 or Sap10, presumably because of conformational differences or carbohydrate masking (52). Similarly, no such cleavage appears to occur in intact cells, as no differences have been found for the 11-kDa Ywp1 propeptide or the 378-aa Ywp1 core in comparing wild-type cells with mutants lacking Sap9 and Sap10 (52). The possibility that Sap9 or Sap10 cleaves the Ywp1 core near its C terminus, possibly contributing to some of the observed shedding of Ywp1, has not been ruled out, however; such a process would resemble the liberation of GPI-anchored proteins (such as the adhesin Epa1) from the cell wall of C. glabrata by the GPI-anchored Yps proteases (33). Still other proteases might be responsible for such cleavage, however, as shown for shedding of the extracellular domain of C. albicans Msb2 (63). Cleavage of Ywp1 at its tribasic site (but not its dibasic site) depends on Kex2 (23), but whether Sap9 or Sap10 has a role in cleavage at the dibasic site remains to be determined.
Many C. albicans wall adhesins contain short (4 to 9 aa) segments with high β-aggregation potential; these are thought to form amyloids that are important for cell adhesion (19, 45, 48). These segments are rich in the amino acids Val, Thr, and Ile (average of 77%) and poor in aromatic amino acids, and they have high scores for β-aggregation potential (34 to 100%). Analysis of the Ywp1 sequence reveals five 6- to 8-aa segments with β-aggregation potentials of 94 to 98%, two located in the propeptide and three located in the core region; two of those in the core region are devoid of aromatics and have Val-plus-Thr-plus-Ile contents of 83% and 100%. Reconciling this with the antiadhesive effects of Ywp1 may require that these sequences serve some other function, not be positioned to form amyloids, or, more interestingly, interfere with the adhesive functions of amyloid-forming adhesins.
Cell surface hydrophobicity can promote adhesiveness of C. albicans yeast under certain conditions (25). It is thus conceivable that Ywp1 decreases yeast adhesiveness simply by decreasing cellular hydrophobicity. Various assays have suggested that deletion of YWP1 increases the surface hydrophobicity of yeast cells (unpublished observations), but efforts to reliably quantify this effect have been unsuccessful, in part because wild-type parental strain SC5314 is already relatively hydrophobic (38). Such an antihydrophobicity effect might be due to the abundance of Ywp1 if Ywp1 is indeed hydrophilic, but the mechanistic relationship between glycans, hydrophobicity, and adhesion remains complex and enigmatic at this time (38, 55). Alternatively, the effect may be more specific. For example, adhesins of the Als family possess tandem repeats of a 36-aa threonine-rich segment (28) that exhibits hydrophobic patches and mediates binding to similar domains in other proteins as well as to hydrophobic substrates (15); although these repeats have no obvious homology to Ywp1, it is conceivable that Ywp1 might normally block hydrophobic components of the wall and thus result in a more hydrophobic surface when absent.
In conclusion, the existing body of knowledge about Ywp1 suggests that it is made predominantly by rapidly proliferating yeast forms and that it exerts an antiadhesive effect as a resident of the C. albicans cell wall. It will be of interest to determine whether Ywp1 orthologs in other members of the Candida clade share these features. Knowledge of the complex biosynthetic pathway and structural properties of Ywp1 provides a firm foundation for further exploration of the molecular mechanisms of its antiadhesive effect in C. albicans.
Supplementary Material
ACKNOWLEDGMENTS
I am indebted and grateful to Jim Cutler for provision of a laboratory and for support during the initial stages of this work. I also thank Heini Miettinen for material support; Al Jesaitis, Jim Burritt, and Heini Miettinen for equipment usage; Jonathan Hilmer (for mass spectrometry) and Dan Siemsen (for laser scanning) at the Montana State University Proteomics and Mass Spectrometry Facility, established and run by Ed Dratz and Brian Bothner; Joel Nott for N-terminal sequencing; Jim Cutler, Lois Hoyer, Heini Miettinen, and John Mills for illuminating discussions; Jim Cutler, Diane Brawner, Lois Hoyer, Aaron Mitchell, Frank Smith, Yue Fu, and Jack Edwards for Candida strains; and Heini Miettinen, Jim Cutler, and the anonymous reviewers for critical reviews of the manuscript.
Footnotes
Published ahead of print 13 April 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Albrecht A, et al. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281:688–694 [DOI] [PubMed] [Google Scholar]
- 2. Arnaud MB, et al. 2005. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 33:D358–D363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blankenship JR, Mitchell AP. 2006. How to build a biofilm: a fungal perspective. Curr. Opin. Microbiol. 9:1–7 [DOI] [PubMed] [Google Scholar]
- 4. Boisramé A, Cornu A, Da Costa G, Richard ML. 2011. Unexpected role for a serine/threonine-rich domain in the Candida albicans Iff protein family. Eukaryot. Cell 10:1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruno VM, et al. 2010. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 20:1451–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Butler G, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaffin WL. 2008. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72:495–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chandra J, et al. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cormack BP, et al. 1997. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143:303–311 [DOI] [PubMed] [Google Scholar]
- 10. de Groot PWJ, et al. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3:955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Groot PWJ, Hellingwerf KJ, Klis FM. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20:781–796 [DOI] [PubMed] [Google Scholar]
- 12. de Nobel H, Lipke PN. 1994. Is there a role for GPIs in yeast cell-wall assembly? Trends Cell Biol. 4:42–45 [DOI] [PubMed] [Google Scholar]
- 13. Douglas LJ. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30–36 [DOI] [PubMed] [Google Scholar]
- 14. Finkel JS, et al. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog. 82:e1002525 doi:10.1371/journal.ppat.1002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frank AT, et al. 2010. Structure and function of glycosylated tandem repeats from Candida albicans Als adhesins. Eukaryot. Cell 9:405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fu Y, et al. 2002. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 44:61–72 [DOI] [PubMed] [Google Scholar]
- 17. Gagnon-Arsenault I, Parisé L, Tremblay J, Bourbonnais Y. 2008. Activation mechanism, functional role and shedding of glycosylphosphatidylinositol-anchored Yps1p at the Saccharomyces cerevisiae cell surface. Mol. Microbiol. 69:982–993 [DOI] [PubMed] [Google Scholar]
- 18. Ganguly S, et al. 2011. Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryot. Cell 10:1448–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garcia MC, et al. 2011. A role for amyloid in cell aggregation and biofilm formation. PLoS One 6:e17632 doi:10.1371/journal.pone.0017632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87–96 [DOI] [PubMed] [Google Scholar]
- 21. Gola S, Martin R, Walther A, Dünkler A, Wendland J. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20:1339–1347 [DOI] [PubMed] [Google Scholar]
- 22. Gow NAR, van de Veerdonk FL, Brown AJP, Netea MG. 2012. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat. Rev. Microbiol. 10:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Granger BL, Flenniken ML, Davis DA, Mitchell AP, Cutler JE. 2005. Yeast wall protein 1 of Candida albicans. Microbiology 151:1631–1644 [DOI] [PubMed] [Google Scholar]
- 24. Grubb SEW, et al. 2008. Candida albicans-endothelial cell interactions: a key step in the pathogenesis of systemic candidiasis. Infect. Immun. 76:4370–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hazen KC. 1989. Participation of yeast cell surface hydrophobicity in adherence of Candida albicans to human epithelial cells. Infect. Immun. 57:1894–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heilmann CJ, et al. 2011. Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 157:2297–2307 [DOI] [PubMed] [Google Scholar]
- 27. Hobson RP, et al. 2004. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 279:39628–39635 [DOI] [PubMed] [Google Scholar]
- 28. Hoyer LL, Green CB, Oh Zhao S-HX. 2008. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med. Mycol. 46:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jacobsen ID, Grosse K, Berndt A, Hube B. 2011. Pathogenesis of Candida albicans infections in the alternative chorio-allantoic membrane chicken embryo model resembles systemic murine infections. PLoS One 6:e19741 doi:10.1371/journal.pone.0019741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones T, et al. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 101:7329–7334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kadosh D, Johnson AD. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kapteyn JC, et al. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 35:601–611 [DOI] [PubMed] [Google Scholar]
- 33. Kaur R, Ma B, Cormack BP. 2007. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. U. S. A. 104:7628–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 35. Lee SA, et al. 2003. An analysis of the Candida albicans genome database for soluble secreted proteins using computer-based prediction algorithms. Yeast 20:595–610 [DOI] [PubMed] [Google Scholar]
- 36. Lesage G, et al. 2000. The Kex2p proregion is essential for the biosynthesis of an active enzyme and requires a C-terminal basic residue for its function. Mol. Biol. Cell 11:1947–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maddi A, Bowman SM, Free SJ. 2009. Trifluoromethanesulfonic acid-based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans. Fungal Genet. Biol. 46:768–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Masuoka J, Hazen KC. 2004. Cell wall mannan and cell surface hydrophobicity in Candida albicans serotype A and B strains. Infect. Immun. 72:6230–6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moreno-Ruiz E, et al. 2009. The GPI-modified proteins Pga59 and Pga62 of Candida albicans are required for cell wall integrity. Microbiology 155:2004–2020 [DOI] [PubMed] [Google Scholar]
- 40. Murillo LA, Newport G, Lan Habelitz C-YS, Dungan J, Agabian NM. 2005. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot. Cell 4:1562–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nantel A, et al. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nobile CJ, et al. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63 doi:10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Odds FC, Van Nuffel L, Gow NAR. 2000. Survival in experimental Candida albicans infections depends on inoculum growth conditions as well as animal host. Microbiology 146:1881–1889 [DOI] [PubMed] [Google Scholar]
- 45. Otoo HN, Lee GK, Qiu W, Lipke PN. 2008. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot. Cell 7:776–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53 [DOI] [PubMed] [Google Scholar]
- 47. Plaine A, et al. 2008. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 45:1404–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramsook CB, et al. 2010. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot. Cell 9:393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Romanowski K, et al. 2012. Candida albicans isolates from the gut of critically ill patients respond to phosphate limitation by expressing filaments and a lethal phenotype. PLoS One 7:e30119 doi:10.1371/journal.pone.0030119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rosenbach A, Dignard D, Pierce JV, Whiteway M, Kumamoto CA. 2010. Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot. Cell 9:1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379 [DOI] [PubMed] [Google Scholar]
- 52. Schild L, et al. 2011. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot. Cell 10:98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sellam A, et al. 2010. Experimental annotation of the human pathogen Candida albicans coding and noncoding transcribed regions using high-resolution tiling arrays. Genome Biol. 11:R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sharkey LL, McNemar MD, Saporito-Irwin SM, Sypherd PS, Fonzi WA. 1999. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 181:5273–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Singleton DR, Masuoka J, Hazen KC. 2005. Surface hydrophobicity changes of two Candida albicans serotype B mnn4Δ mutants. Eukaryot. Cell 4:639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sohn K, Urban C, Brunner H, Rupp S. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89–102 [DOI] [PubMed] [Google Scholar]
- 57. Sorgo AG, et al. 2011. Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryot. Cell 10:1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sorgo AG, et al. 2010. Mass spectrometric analysis of the secretome of Candida albicans. Yeast 27:661–672 [DOI] [PubMed] [Google Scholar]
- 59. Sosinska GJ, et al. 2011. Mass spectrometric quantification of the adaptations in the wall proteome of Candida albicans in response to ambient pH. Microbiology 157:136–146 [DOI] [PubMed] [Google Scholar]
- 60. Southern P, Horbul J, Maher D, Davis DA. 2008. C. albicans colonization of human mucosal surfaces. PLoS One 3:e2067 doi:10.1371/journal.pone.0002067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Staab JF, Sundstrom P. 1998. Genetic organization and sequence analysis of the hypha-specific cell wall protein gene HWP1 of Candida albicans. Yeast 14:681–686 [DOI] [PubMed] [Google Scholar]
- 62. Sundstrom P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461–469 [DOI] [PubMed] [Google Scholar]
- 63. Szafranski-Schneider E, et al. 2012. Msb2 shedding protects Candida albicans against antimicrobial peptides. PLoS Pathog. 8:e1002501 doi:10.1371/journal.ppat.1002501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tuch BB, et al. 2010. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 6:e1001070 doi:10.1371/journal.pgen.1001070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Uppuluri P, et al. 2010. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 6:e1000828 doi:10.1371/journal.ppat.1000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. White SJ, et al. 2007. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog. 3:e184 doi:10.1371/journal.ppat.0030184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao X, et al. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152:2287–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhao X, et al. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150:2415–2428 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.