Abstract
The human basidiomycetous fungal pathogen Cryptococcus neoformans serves as a model fungus to study sexual development and produces infectious propagules, basidiospores, via the sexual cycle. Karyogamy is the process of nuclear fusion and an essential step to complete mating. Therefore, regulation of nuclear fusion is central to understanding sexual development of C. neoformans. However, our knowledge of karyogamy genes was limited. In this study, using a BLAST search with the Saccharomyces cerevisiae KAR genes, we identified five C. neoformans karyogamy gene orthologs: CnKAR2, CnKAR3, CnKAR4, CnKAR7 (or CnSEC66), and CnKAR8. There are no apparent orthologs of the S. cerevisiae genes ScKAR1, ScKAR5, and ScKar9 in C. neoformans. Karyogamy involves the congression of two nuclei followed by nuclear membrane fusion, which results in diploidization. ScKar7 (or ScSec66) is known to be involved in nuclear membrane fusion. In C. neoformans, kar7 mutants display significant defects in hyphal growth and basidiospore chain formation during both a-α opposite and α-α unisexual reproduction. Fluorescent nuclear imaging revealed that during kar7 × kar7 bilateral mutant matings, the nuclei congress but fail to fuse in the basidia. These results demonstrate that the KAR7 gene plays an integral role in both opposite-sex and unisexual mating, indicating that proper control of nuclear dynamics is important. CnKAR2 was found to be essential for viability, and its function in mating is not known. No apparent phenotypes were observed during mating of kar3, kar4, or kar8 mutants, suggesting that the role of these genes may be dispensable for C. neoformans mating, which demonstrates a different evolutionary trajectory for the KAR genes in C. neoformans compared to those in S. cerevisiae.
INTRODUCTION
Cryptococcus neoformans belongs to the Basidiomycota and is an opportunistic fungal pathogen causing meningoencephalitis in immunocompromised cohorts, especially patients with HIV/AIDS, and in some cases, in otherwise immunocompetent individuals (10). Basidiospores generated via sexual development are considered major infectious propagules (6, 18, 60). Therefore, investigation of the sexual development of this fungus is of importance to understand its global impact on human health resulting in more than 1,000,000 infections annually (47).
C. neoformans has two mating types, a and α, and opposite-sex mating occurs when a and α cells encounter and recognize each other (25–27), resulting in formation of mating hyphae and bulb-like basidia generated at the apex of aerial hyphae followed by spore chain formations (31, 63). The mating type locus of C. neoformans represents a bipolar mating system, in which the homeodomain and pheromone/pheromone receptor loci are genetically and physically linked, whereas in many other basidiomycetes, the two loci are unlinked and segregate independently to comprise tetrapolar mating systems (reviewed in reference 28). However, the mating type is unbalanced in natural populations, where α mating types are predominant over a mating type isolates; hence, the rarity of mating type a poses a barrier to opposite-sex mating (reviewed in reference 28). Indeed, C. neoformans undergoes α-α unisexual reproduction, which mimics opposite-sex mating morphologically but occurs during solo culture of α isolates.
The molecular mechanism(s) underlying the increase in ploidy during mating is of central interest; in the case of unisexual reproduction, diploidization may occur “early” before the sexual hyphal growth phase, wherein diploidization could be a necessary condition for filamentation. In contrast, as seen in opposite-sex mating, diploidization may occur “late” in the basidia just prior to meiosis and production of spore progeny (Fig. 1A). A promising avenue to understand this process involves the identification and characterization of karyogamy genes that enable nuclear fusion. In addition, although C. neoformans is predominantly a haploid organism, clinical and environmental diploid isolates are also found (12, 34). Thus, elucidation of the roles of karyogamy genes during mating will advance our knowledge about this ubiquitous fungal pathogen and its routes to diploidization and sexual reproduction.
Fig 1.
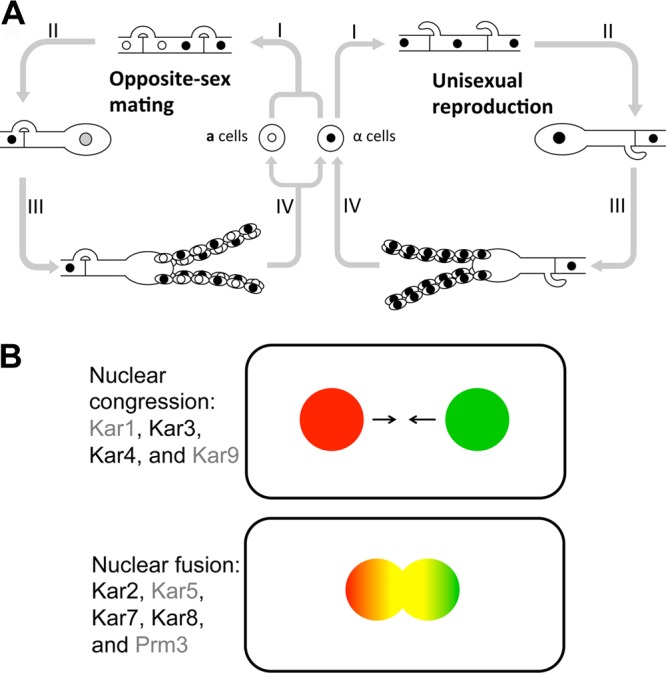
Illustration of two modes of C. neoformans sexual development and two distinct stages involved in karyogamy. (A) Opposite-sex mating. (I) Cells of two different mating types recognize each other and undergo cell-cell fusion to form dikaryotic hyphae. (II) At the apex of aerial hyphae, a basidium forms and karyogamy occurs, representing “late” diploidization. (III) Meiosis occurs, and four basidiospore chains are produced from the surface of the basidium by repeated rounds of mitosis of the postmeiotic nuclei. (IV) Progeny of two mating types disperse. (A) Unisexual reproduction. (I) Only α mating type cells undergo hyphal growth, where “early” diploidization might contribute to filamentation. (II) At the apex of aerial hyphae, a basidium forms and “late” diploidization can occur. (III) Meiosis occurs, and four basidiospore chains are formed. (IV) Haploid progeny disperse. Thus, during unisexual reproduction, two different hypothetical timings for diploidization exist. The figure depicts the hypothesis of “late” diploidization. (B) Nuclear congression and nuclear fusion. First, two nuclei migrate toward each other (nuclear congression): Kar1, Kar3, Kar4, and Kar9 are involved in this process in S. cerevisiae. Second, nuclear membrane fusion occurs and Kar2, Kar5, Kar7, and Kar8 are involved in this process. Karyogamy components that are unique to S. cerevisiae compared to C. neoformans are shown in gray.
Karyogamy (or nuclear fusion) during mating is well understood in the model yeast Saccharomyces cerevisiae (52). There are two distinct steps in karyogamy: nuclear congression and nuclear fusion. The first step involves the genes encoding Kar1, Kar3, Kar4, and Kar9 and serves to pair the nuclei close to one another (24, 40, 43, 44, 52). The second step, in which nuclear membranes fuse, involves the genes encoding Kar2, Kar5, Kar7/Sec66, and Kar8 (3, 7, 52, 54) (Fig. 1B). Kar1 is an essential protein involved in nuclear fusion during mating and also plays a role in mitosis for spindle pole body duplication. Kar1 localizes to the half-bridge of the spindle pole body (13, 53, 57). Kar3 is a minus-end-directed motor kinesin that functions during mitosis and meiosis and localizes to the spindle pole body (40, 42). Kar4 is a transcription factor induced in response to pheromone that functions in nuclear congression (23, 24). One of its targets is the KAR3 gene (24). Kar9 is a cortical protein required for proper localization of the mitotic spindle and orienting cytoplasmic microtubules (43). Kar2 is a homolog of BiP/GRP78 (54) and an ATPase that functions as a molecular chaperone involved in the transport of proteins into the endoplasmic reticulum (ER) (51, 59). Kar5 is an ER membrane protein whose expression is regulated by pheromone (3, 7). Kar7 (or Sec66) is a subunit of the Sec63 complex that is involved in protein targeting and import into the ER (23, 65). Kar8 (or Jem1) is a DnaJ-like chaperone localized to the ER membrane and is known to interact genetically with KAR2 (46, 62).
Schizosaccharomyces pombe Tht1, a Kar5 homolog, and Tht2 (meiotically upregulated gene, Mug22), are known to be involved in karyogamy (39, 58). In Neurospora, the UV-sensitive uvs-3 mutant exhibits an arrest prior to karyogamy during ascus formation in the perithecium (49). In the pathogenic fungus Candida albicans, the KAR3 gene is required for karyogamy during mating (4). C. albicans KAR8 (JEM1) is also known to complement the S. cerevisiae kar8 mutant to restore karyogamy (38). However, little is known about the roles of KAR genes in other fungi, including C. neoformans. In this study, we identified orthologs of karyogamy genes in C. neoformans and explored their functions during opposite-sex and unisexual reproduction. We found that the KAR7/SEC66 gene is required for both types of sexual reproduction. kar7 mutants exhibited defects in sexual spore formation, indicating that karyogamy occurs inside the basidium prior to meiosis and spore formation. We found that the KAR3, KAR4, and KAR8 genes are dispensable for mating in C. neoformans. KAR2 was found to be essential for viability, and its possible functions in mating remain to be studied. We also found that orthologs of KAR1, KAR5, and KAR9 are apparently missing from the C. neoformans genome, possibly reflecting an evolutionary trajectory for the mechanics of mating and nuclear fusion different from that of S. cerevisiae.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The C. neoformans strains and plasmids used in this study are listed in Table 1. The strains were grown on yeast extract-peptone-dextrose (YPD) agar containing the appropriate drugs for marker gene selections. For mating, strains were cultured in liquid YPD medium overnight, washed with distilled water, and plated on V8 medium (pH 7) for serotype D and MS medium (64) for serotype A. Plasmid preparation and cloning were performed as described previously (55).
Table 1.
Strains and plasmids used in this study
| Strain or plasmida | Serotype | Mating type | Genotype |
|---|---|---|---|
| Strains | |||
| Wild type | |||
| KN99α | A | α | Wild type |
| KN99a | A | a | Wild type |
| XL280 | D | α | Wild type |
| JEC21 | D | α | Wild type |
| JEC20 | D | a | Wild type |
| MN142.3 | D | α | Diploid, ura5::NAT/ura5::NEO |
| kar7 mutants | |||
| SL2801 | A | α | KN99α kar7::NEO |
| SL2811 | A | α | KN99α kar7::NEO |
| SL2822 | A | α | KN99α kar7::NEO |
| SL2832 | A | α | KN99α kar7::NEO |
| SL2842 | A | α | KN99α kar7::NEO |
| SL2851 | A | a | KN99akar7::NAT |
| SL2861 | A | a | KN99akar7::NAT |
| SL2871 | A | a | KN99akar7::NAT |
| SL2882 | A | a | KN99akar7::NAT |
| SL2892 | A | a | KN99akar7::NAT |
| SL2903 | A | a | KN99akar7::NEO |
| SL2913 | A | a | KN99akar7::NEO |
| SL2923 | A | a | KN99akar7::NEO |
| SL2933 | A | a | KN99akar7::NEO |
| SL2943 | A | a | KN99akar7::NEO |
| SL2761 | D | α | XL280 kar7::NEO |
| SL2771 | D | α | XL280 kar7::NEO |
| SL2782 | D | α | XL280 kar7::NEO |
| SL2792 | D | α | XL280 kar7::NEO |
| SL275 | D | α | JEC21 kar7::NEO |
| SL3551 | D | α | MN142.3 kar7::HYG/KAR7 |
| SL3561 | D | α | MN142.3 kar7::HYG/KAR7 |
| SL3572 | D | α | MN142.3 kar7::HYG/KAR7 |
| SL3582 | D | α | MN142.3 kar7::HYG/KAR7 |
| SL3592 | D | α | MN142.3 kar7::HYG/KAR7 |
| SL3602 | D | α | MN142.3 kar7::HYG/KAR7 |
| SL3611 | D | α | SL355 kar7::HYG/kar7::URA5 ura5::NAT/ura5::NEO |
| SL3622 | D | α | SL355 kar7::HYG/kar7::URA5 ura5::NAT/ura5::NEO |
| SL3632 | D | α | SL355 kar7::HYG/kar7::URA5 ura5::NAT/ura5::NEO |
| SL3643 | D | α | SL355 kar7::HYG/kar7::URA5 ura5::NAT/ura5::NEO |
| SL3653 | D | α | SL355 kar7::HYG/kar7::URA5 ura5::NAT/ura5::NEO |
| kar8 mutants | |||
| SL2701 | D | α | XL280 kar8::NAT |
| SL2742 | D | α | XL280 kar8::NAT |
| SL271 | D | a | JEC20 kar8::NAT |
| SL2721 | D | α | JEC21 kar8::NAT |
| SL2732 | D | α | JEC21 kar8::NAT |
| kar3 mutants | |||
| SL2951 | A | α | KN99α kar3::NEO |
| SL2962 | A | α | KN99α kar3::NEO |
| SL2971 | A | a | KN99akar3::NAT |
| SL2982 | A | a | KN99akar3::NAT |
| kar4 mutants | |||
| SL3661 | D | α | Diploid MN142.3 kar4::HYG/KAR4 ura5::NAT/ura5::NEO |
| SL3671 | D | α | Diploid MN142.3 kar4::HYG/KAR4 ura5::NAT/ura5::NEO |
| SL3682 | D | α | Diploid MN142.3 kar4::HYG/KAR4 ura5::NAT/ura5::NEO |
| SL3721 | D | α | Haploid progeny of SL366 kar4::HYG |
| SL3732 | D | α | Haploid progeny of SL366 kar4::HYG |
| kar2 mutants | |||
| SL3691 | D | α | Diploid MN142.3 kar2::HYG/KAR, ura5::NAT/ura5::NEO |
| SL3701 | D | α | Diploid MN142.3 kar2::HYG/KAR2 ura5::NAT/ura5::NEO |
| SL3712 | D | α | Diploid MN142.3 kar2::HYG/KAR2 ura5::NAT/ura5::NEO |
| Strains with fluorescent Nop1 | |||
| SL305 | A | α | KN99α with NOP1-mCherry (NEO) |
| SL306 | A | α | KN99α with NOP1-mCherry(NEO) |
| SL307 | A | α | KN99α with NOP1-mCherry(NEO) |
| SL321 | A | a | KN99a with GFP-NOP1(NAT) |
| SL322 | A | a | KN99a with GFP-NOP1(NAT) |
| SL323 | A | a | KN99a with GFP-NOP1(NAT) |
| SL324 | A | a | KN99a with GFP-NOP1(NAT) |
| SL308 | A | a | SL285 (kar7) NOP1-mCherry(NEO) |
| SL309 | A | a | SL285 (kar7) NOP1-mCherry(NEO) |
| SL310 | A | a | SL285 (kar7) NOP1-mCherry(NEO) |
| SL311 | A | a | SL285 (kar7) NOP1-mCherry(NEO) |
| SL312 | A | a | SL285 (kar7) NOP1-mCherry(NEO) |
| SL313 | A | a | SL285 (kar7) NOP1-mCherry(NEO) |
| SL316 | A | a | SL290 (kar7) GFP-NOP1(NAT) |
| SL317 | A | a | SL290 (kar7) GFP-NOP1(NAT) |
| SL318 | A | a | SL290 (kar7) GFP-NOP1(NAT) |
| SL319 | A | a | SL290 (kar7) GFP-NOP1(NAT) |
| SL347 | A | α | SL280 (kar7) GFP-NOP1(NAT) |
| SL348 | A | α | SL280 (kar7) GFP-NOP1 (NAT) |
| SL349 | A | α | SL280 (kar7) GFP-NOP1(NAT) |
| SL350 | A | α | SL280 (kar7) GFP-NOP1(NAT) |
| SL338 | D | α | XL280 GFP-NOP1(NAT) |
| SL339 | D | α | XL280 GFP-NOP1(NAT) |
| SL340 | D | α | XL280 GFP-NOP1(NAT) |
| SL341 | D | α | XL280 GFP-NOP1(NAT) |
| SL342 | D | α | XL280 GFP-NOP1(NAT) |
| SL380 | A | α | SL295 (kar3) GFP-NOP1(NAT) |
| SL381 | A | α | SL295 (kar3) GFP-NOP1(NAT) |
| SL383 | A | α | SL296 (kar3) GFP-NOP1(NAT) |
| SL385 | A | α | SL296 (kar3) GFP-NOP1(NAT) |
| SL374 | D | α | SL277 (kar7) GFP-NOP1(NAT) |
| SL375 | D | α | SL277 (kar7) GFP-NOP1(NAT) |
| SL376 | D | α | SL277 (kar7) GFP-NOP1(NAT) |
| SL377 | D | α | SL277 (kar7) GFP-NOP1(NAT) |
| Plasmids | |||
| pSL01 | Ampr KanrNOP1 fragment in pCR21-TOPO for N-terminal GFP tagging with pCN19 | ||
| pSL02 | Ampr KanrNOP1 fragment in pCR21-TOPO for N-terminal GFP tagging with pCN19 | ||
| pSL04-02 | Ampr BglII fragment of NOP1 from pSL01 in pCN19/BamHI, vector for GFP-Nop1 for serotype A | ||
| pSL05-1 | Ampr BglII fragment of NOP1 from pSL02 in pCN19/BamHI, vector for GFP-Nop1 for serotype D | ||
| pCN19 | Ampr plasmid harboring GFP under histone H3 promoter |
Each superscript 1, 2, or 3 indicates a group of independent transformants or progeny.
Disruption of KAR genes.
kar null mutant strains were generated using disruption cassettes with neomycin (NEO) or nourseothricin (NAT) drug resistance markers flanked by ∼1-kb sequences corresponding to the 5′ and 3′ noncoding flanking regions of each KAR gene as described previously (14). The primers used in this study are listed in Table S1 in the supplemental material. For example, to disrupt the KAR7 gene in the KN99a or KN99α strain, the 5′ region of the KAR7 gene was amplified with primers JOHE20336 and JOHE20337 from genomic DNA, the NEO or NAT drug resistance gene cassette was amplified with JOHE20338 and JOHE20339 from plasmid pNATSTM#209 or pJAF1 (17), and the 3′ region was amplified with primers JOHE20340 and JOHE20341. The three DNA fragments obtained were combined and amplified with two internal nested primers, JOHE20342 and JOHE20343, and the final overlap PCR products were purified and precipitated onto 0.6-μm gold particles (Bio-Rad, Hercules, CA). The wild-type strains then were transformed biolistically with the DNAs obtained. The bombarded cells were transferred onto YPD medium containing the appropriate selection drugs. Positive transformants were screened by PCR and Southern blotting. The disruption procedures for the serotype D KAR7 and other KAR genes are described in the Materials and Methods section in the supplemental material.
Tagging Nop1 with mCherry or GFP.
The primers used in these experiments are listed in Table S1 in the supplemental material. The nucleolar protein Nop1 was used to monitor nuclear positioning during mating. The serotype A strain NOP1 gene was replaced with NOP1-mCherry by homologous recombination with NOP1-mCherry-NEO as described previously (22). The mCherry-encoding gene was flanked by the 3′ end of the NOP1 gene open reading frame (ORF). Positive transformants were selected, and homologous recombination was confirmed by PCR. To tag the N terminus of Nop1 with green fluorescent protein (GFP), the NOP1 gene was amplified with primers JOHE23127 and JOHE23128 for serotype A or primers JOHE23129 and JOHE23130 for serotype D and the resulting fragments were cloned into the pCR21-TOPO vector (Invitrogen, Carlsbad, CA). The resulting plasmids, pSL01 and pSL02 (Table 1), respectively, were digested with BglII, and the BglII NOP1 fragments were purified and cloned into pCN19 digested with BamHI (48), producing plasmids pSL04-1 (serotype A) and pSL05-1 (serotype D), which express GFP-Nop1 under the constitutive histone H3 promoter. The wild-type and kar7 mutant strains were transformed with the plasmids as described above, and each positive transformant was screened for the GFP nuclear signal.
Microscopy.
Colony morphology and mating hyphae were observed by using a Nikon Eclipse E400 microscope equipped with a Nikon DXM1200F camera. Initial fluorescent signal screening was performed by using a Zeiss Axioskop 2 Plus with an AxioCam MRm camera (Carl Zeiss, Inc., Thornwood, NY). Further nuclear positioning analyses during mating were performed by using the Zeiss Axio Observer Z1 microscope system (Carl Zeiss, Inc., Thornwood, NY) equipped with an Opto-electronically motorized XY stage, Pecon XL S1 incubator, and Coolsnap ES2 high-resolution charge-coupled device (CCD) camera (Photometrics, Inc., Huntington Beach, CA) in the Duke University Light Microscopy Core Facility (LMCF).
For scanning electron microscopy (SEM), the edges of mating colonies were washed with 0.1 M Na cacodylate buffer (pH 6.8), and 1-mm3 blocks of mating areas were excised and incubated in fixation buffer at 4°C. Samples were then rinsed in cold 0.1 M Na cacodylate buffer three times, postfixed in 2% osmium tetroxide in 0.1 M Na cacodylate buffer for 2.5 h at 4°C, critical point dried, and sputter coated before being viewed by SEM.
RESULTS
Identification of KAR genes in Cryptococcus neoformans.
To elucidate the roles of the karyogamy genes, we identified KAR gene orthologs in the C. neoformans serotype A (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html) and D (http://www-sequence.stanford.edu/group/C.neoformans/) genomes (21, 36). S. cerevisiae KAR genes and protein sequences were used to identify C. neoformans KAR gene (CnKAR) orthologs in C. neoformans serotype A and serotype D. In the BLASTp analyses (1), proteins with reciprocal best hits were considered orthologs for each Kar protein. A summary of the C. neoformans KAR genes (KAR2, KAR3, KAR4, KAR7, and KAR8) is listed in Table 2. Interestingly, there were no orthologs of ScKAR1, ScKAR5, and ScKAR9 in either the C. neoformans serotype A or D genomes, where our analyses resulted in no apparent BLAST hits. The related C. gattii genomes (VGI strain WM276 and VGII strain R265) (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans_b/GenomeStats.html) also lack genes encoding these four Kar protein homologs (see Table S1 in the supplemental material) (16).
Table 2.
Karyogamy genes in S. cerevisiae and C. neoformans serotypes A and D
| Function | Karyogamy gene in: |
Description | |||
|---|---|---|---|---|---|
| S. cerevisiae |
C. neoformans |
||||
| Designation | Serotype A | Serotype D | |||
| Nuclear congression | KAR1 | None | None | None | Kar1 protein localizes to the half-bridge of the spindle pole body |
| KAR3 | CnKAR3 | CNAG_05752 | CNF02260 | Kar3 is a minus-end-directed microtubule motor that localizes to the spindle pole body. | |
| KAR4 | CnKAR4 | CNAG_04487 | CNI00070 | Kar4 is a transcription factor required for gene regulation in response to pheromones, and expression of KAR3 and CIK1. | |
| KAR9 | None | None | None | Kar9 is a karyogamy protein required for correct positioning of the mitotic spindle. | |
| Nuclear membrane fusion | KAR2 | CnKAR2 | CNAG_06443 | CNN01680 | Kar2 is an ATPase involved in protein import into the ER and functions as a chaperone to mediate protein folding in the ER. |
| KAR5 | None | None | None | The Kar5 protein is required for nuclear membrane fusion during karyogamy and is regulated by pheromone. | |
| KAR7 | CnKAR7 | CNAG_01647 | CNC01600 | Kar7 or Sec66 is a nonessential subunit of the Sec63 complex (Sec63p, Sec62p, Sec66p, and Sec72p) and together with the Sec61 complex, Kar2p/BiP, and Lhs1p forms a channel competent for SRP-dependent protein translocation. | |
| KAR8 | CnKAR8 | CNAG_01347 | CND04620 | Kar8 or Jem1 is a DnaJ-like chaperone required for nuclear membrane fusion during mating; a null mutant is viable. | |
The C. neoformans KAR7 ortholog is required for opposite-sex mating.
To assess the role of the KAR7 gene in opposite-sex mating, the C. neoformans KAR7 gene was replaced with the neomycin (NEO) or nourseothricin (NAT) drug resistance cassettes in strains of both opposite mating types, as described in Materials and Methods. Two congenic serotype A strains of opposite mating type, KN99a and KN99α, were employed to assess the roles of KAR7. We obtained independent KAR7 disruption mutants from separate transformations for each mating type (Table 1).
To test mating ability, unilateral and bilateral mutant crosses (together with wild-type bilateral control crosses) were performed on MS mating media (see Materials and Methods). In wild-type crosses, dikaryotic mating hyphae formed and eventually produced aerial hyphae, which in response to an unknown signal form bulb-like basidia at their apices. Four chains of basidiospores then emerged from the surface of each basidium. In unilateral crosses, α wild-type × a kar7 or α kar7 × a wild-type, the overall level of mating hyphae was reduced compared to those observed in bilateral wild-type crosses (Fig. 2A). However, we were still able to observe mating hyphae as well as basidia decorated with four spore chains, indicating that one wild-type allele of KAR7 from the wild-type mating partner is sufficient for sexual development to progress at a wild-type or near-wild-type level. In contrast, in bilateral α kar7 × a kar7 mutant crosses, a significantly reduced level of mating hyphae was observed (Fig. 2A). Although aerial hyphae were formed followed by basidium formation, no obvious basidiospore chains were observed (bald basidia). In some exceptional cases, a single basidiospore from one or each chain formation center was attached to a basidium without progressing to form mature spore progeny (Fig. 2B). These results indicate that Kar7 is required for the formation of spore chains and also plays a role in hypha formation at the early stages of sexual development during opposite-sex mating.
Fig 2.
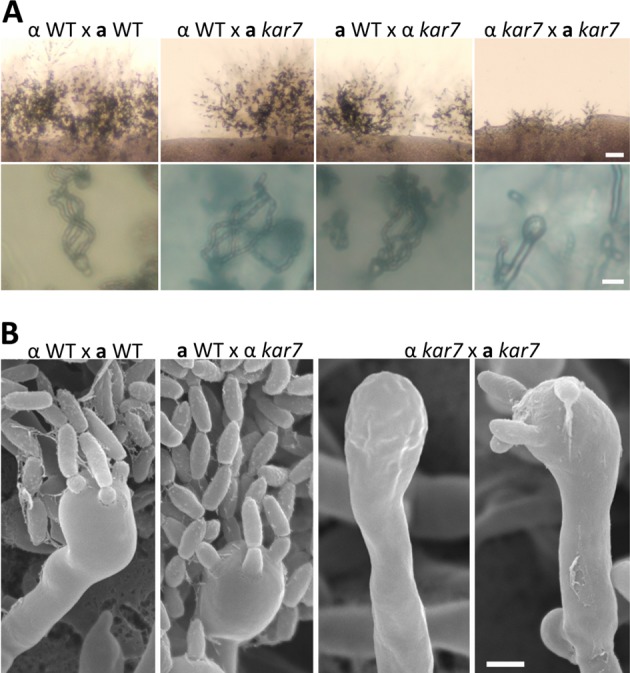
KAR7 is required for opposite-sex mating in C. neoformans. (A) Wild-type (WT) mating produces abundant dikaryotic hyphae decorated with fused clamp connections, basidia, and chains of spores. A unilateral mating between wild-type and kar7 mutant strains also produces mating hyphae that are less abundant than those in wild-type crosses. In the bilateral kar7 × kar7 mutant crosses, few hyphae were observed. (B) For both wild-type × wild-type and wild-type × kar7 crosses, four complete chains of basidiospores were observed; however, in the bilateral kar7 × kar7 mutant crosses, bald basidia lacking any spores or occasionally basidia with only a single immature spore attached were observed. Scale bars = 100 μm for the upper row and 10 μm for the bottom row in panel A and 5 μm for panel B.
Nuclear localization during wild-type and kar7 mutant opposite-sex matings.
Monitoring nuclear positioning provides a way to elucidate the role of karyogamy genes during mating. To assess this in live cells, we fused the Nop1 nucleolar protein with mCherry (C-terminus tagged) or GFP (N-terminus tagged). In KN99α cells, the NOP1-mCherry gene was introduced into the native NOP1 gene and homologous recombination was confirmed by PCR (data not shown). In KN99a cells, the GFP-NOP1 gene was ectopically integrated (Table 1). Wild-type a and α strains expressing either Nop1-mCherry or GFP-Nop1, respectively, were crossed and their nuclear positions were monitored during the mating process. In wild-type crosses, we observed that the mCherry and GFP signals are largely superimposed, indicating that cell-cell cytosolic fusion occurs during opposite-sex mating, allowing mixing of the nuclear signals in the dikaryon, and thus the nucleus positioning to be observed is only for opposite-sex mating (Fig. 3). Microscopic observations with live cell matings were conducted, and representative nuclear positions during wild-type opposite-sex mating are presented in Fig. 3A as follows. (i) In dikaryotic (1N + 1N) hyphae, a nucleus can be observed moving through the clamp connection from the apical hyphal compartment to the subapical hyphal cell, (ii) at the apical hyphal tip, a basidium formed, wherein the two nuclei congress, and (iii) fusion of two nuclei occurs, followed by (iv) meiosis.
Fig 3.
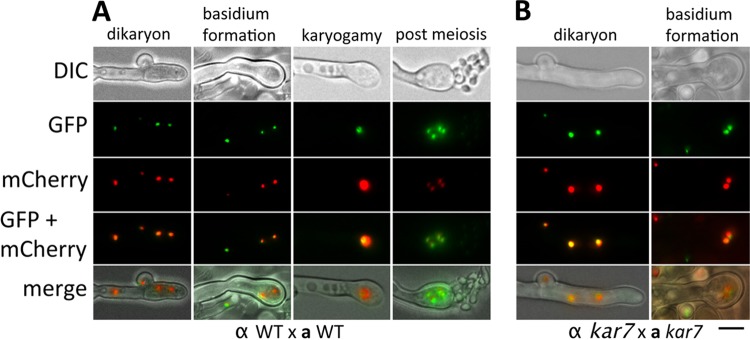
Nuclear positioning during mating in the wild type (A) and kar7 mutant (B). DIC, differential interference contrast. a mating type cells producing Nop1-GFP and α mating type cells producing Nop1-mCherry were cocultured on MS mating medium. The two fluorescent signals were observed to merge, indicating that cytosolic fusion occurs during the process of a-α opposite-sex mating. The two nuclei remain separated in the dikaryotic hyphae. In the basidium, the two nuclei migrate toward each other and karyogamy occurs, resulting in diploidization. Meiosis follows, and chains of basidiospores are produced (A). In the crosses between two kar7 mutants, the dikaryotic hyphae produced are similar to those in the wild type; however, in the mutant basidia, the two congressed nuclei do not undergo fusion, resulting in a failure to proceed into meiosis or produce spore progeny (B). Scale bar = 5 μm.
C. neoformans forms holobasidia (single-celled basidia), unlike the closely aligned species Cryptococcus heveanensis, where phragmobasidia (basidia divided into four cells by septa) are formed (41). We also found that karyogamy could occur in the subapical area of hyphae of the basidium as well as more apically within the basidium proper, as seen in Pisolithus microcarpus (9) (Fig. 3; see Fig. S1 in the supplemental material). In bilateral α kar7 × a kar7 mutant crosses, although we were able to observe early stages of nuclear dynamics, including nuclear migration through a clamp connection and nuclear congression in the basidia, no apparent nuclear fusion was observed, which likely results in the observed failure to enter into meiosis or form basidiospore chains (Fig. 3B). These results indicate that the KAR7 gene is required for karyogamy in the basidia to complete later steps in the mating process, including nuclear fusion to form the diploid, entry into meiosis, and production of sexual spore progeny in C. neoformans.
KAR7 is necessary for unisexual reproduction.
It is of interest to establish whether KAR7 is also involved in unisexual mating in C. neoformans. Same-sex mating under laboratory conditions is not yet known in serotype A; therefore, we employed the serotype D α strain, XL280, which displays robust unisexual reproduction/same-sex mating (30–32). The KAR7 locus ORF was replaced with a NEO resistance cassette by biolistic transformation and homologous recombination, as described above for the serotype A strains. The wild type and the independently derived kar7 mutants were obtained and cultured individually to undergo unisexual mating on V8 medium (pH 7) in the dark. In solo culture, the wild-type strain XL280 produced hyphae, basidia, and basidiospores at the edge of the colonies (unisexual reproduction), whereas the independent kar7 mutants all exhibited a severe defect in producing unisexual hyphae (Fig. 4A) compared to the wild type. These findings indicate a role for Kar7 in the early stages of hypha formation during same-sex mating (Fig. 1). Further analyses found that the kar7 mutants can produce hyphae and basidia but are defective in basidiospore chain formation, indicating that Kar7 is also required during same-sex mating to complete karyogamy, meiosis, and spore progeny production, similar to its role in a-α opposite sex mating (Fig. 4BB and C). Scanning electon microscopy analysis further verified that bald basidia, or in some cases basidia decorated with only one or two immature basidiospores, were formed without completing sexual reproduction. These results indicate that Kar7 function is necessary for same-sex as well as opposite-sex mating.
Fig 4.
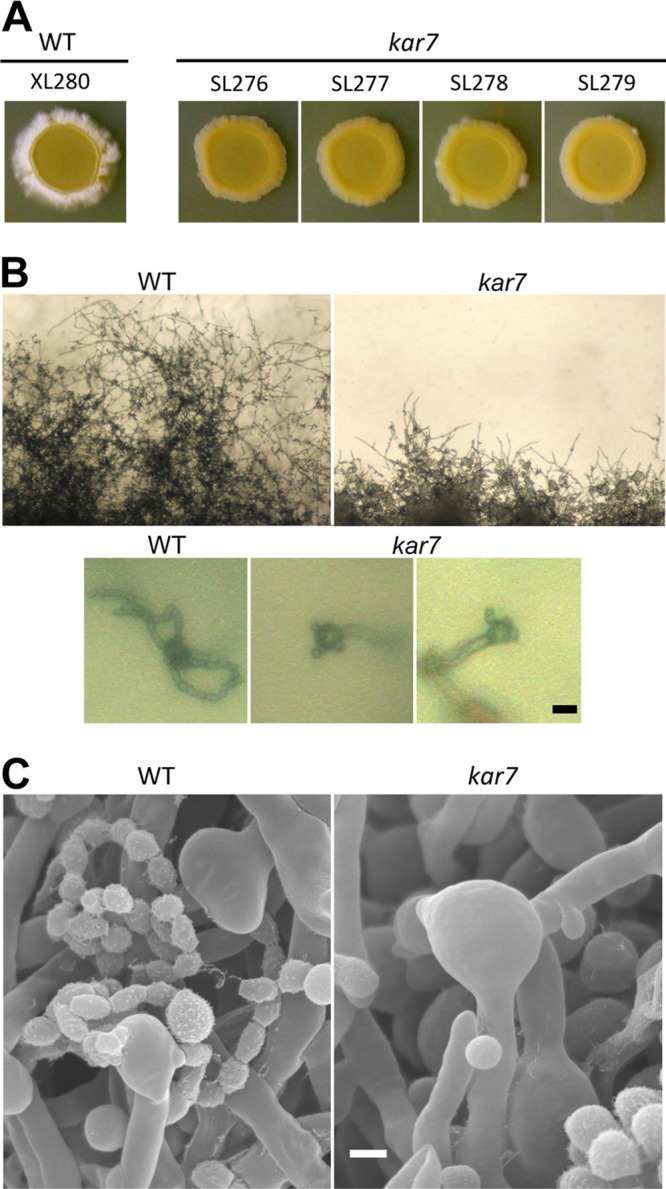
KAR7 is necessary to complete unisexual reproduction in C. neoformans. The edges of colonies of wild-type (WT) strain XL280 exhibit abundant monokaryotic hyphae; however, two independent sets of kar7 mutants (the first and second ones are independent from the third and fourth from left to right) produced considerably less hyphae (A). The wild type produces four intact chains of basidiospores, whereas in the kar7 mutant, unisexual reproduction was arrested (possibly before meiosis), resulting in no spore progeny production. Images in panels B and C show kar7 mutant basidia with a single attached immature spore. Scale bars = 10 μm for panel B and 5 μm for panel C.
Nuclear positioning during same-sex mating.
To observe nuclear positions during unisexual mating, we integrated the GFP-Nop1-encoding gene ectopically into the genomes of the wild-type XL280 and kar7 mutant strains. GFP signal-positive strains were chosen and grown on V8 medium (pH 7) to stimulate same-sex mating. Nuclear positions during mating were monitored (Fig. 5). Based on the GFP-Nop1 signal, the same-sex mating hyphae are monokaryotic (1N) and the clamp connections are unfused (see the monokaryon panel in Fig. 5A). This is in contrast to opposite-sex mating (Fig. 3A), during which dikaryotic hyphae are produced with two separate fluorescent Nop1 signals that were observed in a single hyphal compartment and one labeled nucleus moves through clamp cells that then fuse. At the terminal same-sex mating hyphal compartment, a nucleus underwent mitosis to generate a transient dikaryotic stage (1N + 1N), and karyogamy then occurred inside the basidia or subapical hyphae (Fig. 5; see Fig. S1 in the supplemental material). Meiosis and complete basidiospore chain formation then followed (Fig. 5A). However, in the kar7 mutant same-sex mating assays, two separate nuclei were observed that persisted in the basidia and did not fuse. As a result, diploidization did not occur, causing a failure to enter into meiosis or form basidiospore chains (Fig. 5B). This observation leads us to conclude that diploidization during XL280 same-sex mating can occur in the basidia as a late step in the pathway and that these steps (karyogamy and meiosis) may be necessary for nuclei to be packaged into spores. However, we also observed that in the serotype D strain JEC21, the kar7 mutant was completely unable to initiate filamentation, indicating that karyogamy and diploidization can also occur as an early stage required for unisexual mating hyphal development (see Fig. S2 in the supplemental material).
Fig 5.
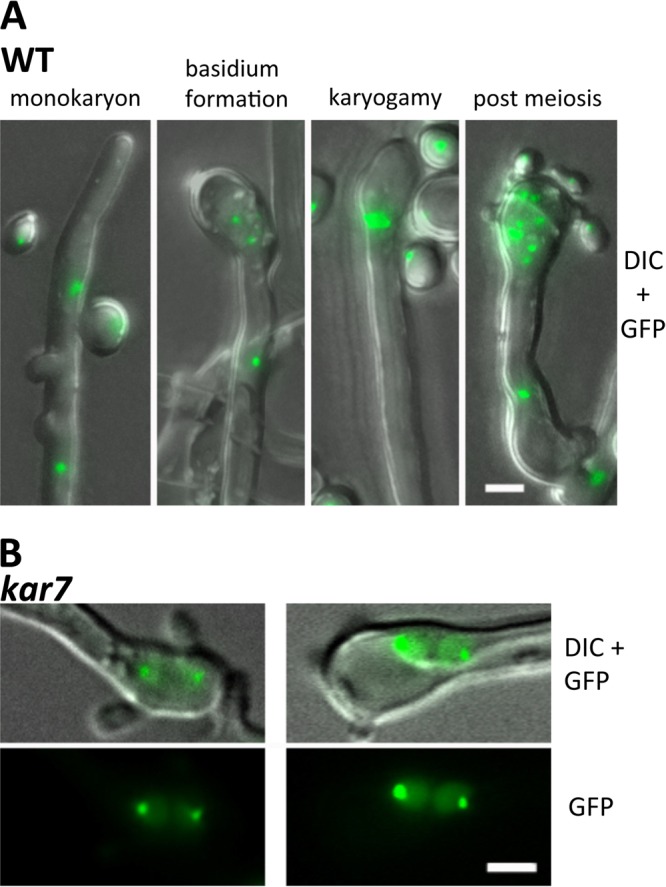
Nuclear positions during unisexual reproduction in the wild type and kar7 mutant. Nuclei were visualized with the GFP-Nop1 protein. (A) In the wild-type XL280 strain, sexual hyphae are monokaryotic with unfused clamp connections. Two nuclei congress in the basidia and undergo karyogamy, meiosis occurs, and basidiospores are produced. DIC, differential interference contrast. (B) In contrast, in the kar7 mutants, the two nuclei congress but do not undergo karyogamy, resulting in a failure to produce the diploid nucleus or undergo meiosis, and consequently, no spore chains are produced. Scale bars = 5 μm.
Diploid kar7/kar7 mutants are defective in unisexual reproduction.
Our observation that kar7 mutants failed to complete unisexual reproduction prompted us to test the impact of kar7 in an α/α diploid strain. If diploidization in the basidia is essential to complete mating, and the sole role of Kar7 is to promote karyogamy and diploidization, then kar7/kar7 diploid mutants might bypass the normal requirement for Kar7 for diploidization and complete unisexual reproduction. On the other hand, if the kar7/kar7 diploid mutants are still defective in unisexual mating, it would suggest that Kar7 plays an additional role beyond karyogamy/diploidization during same-sex mating.
To test these hypotheses, we constructed an α/α diploid derivative of strain XL280. First, the URA5 locus was replaced with either the NEO or NAT resistance cassette. The resulting haploid strains, XL280 ura5::NEO and XL280 ura5::NAT, were then fused to produce the ura5::NEO/ura5::NAT strain MN142.3 (Table 1). The ploidy of this isolate was confirmed to be diploid by fluorescence-activated cell sorter (FACS) and nucleus staining with DAPI (4′,6-diamidino-2-phenylindole) (data not shown). The MN142.3 α/α diploid strain exhibits robust same-sex mating (Fig. 6), as expected based on previous studies (31). We disrupted both alleles of the KAR7 gene in strain MN142.3 by gene replacement with the hygromycin resistance gene cassette and the wild-type URA5 gene (Table 1) as markers. The mutants were confirmed by Southern blotting (see Fig. S3 in the supplemental material). The heterozygous kar7::HYG/KAR7 mutants displayed wild-type unisexual hyphal growth and basidiospore chain formation (Fig. 6). However, interestingly, independently derived homozygous kar7/kar7 diploid mutants displayed reduced filamentation and defects in basidiospore chain formation (Fig. 6). This result suggests that Kar7 may play an additional role(s) during mating beyond its role in karyogamy and diploidization. As a note, it is also formally possible that Kar7 functions early in some cells/hyphae and late in others such that there is a mixed culture of haploid and diploid monokaryotic hyphae produced. Nuclear position monitoring will provide further insight into the distinct roles of Kar7 during these developmental processes.
Fig 6.
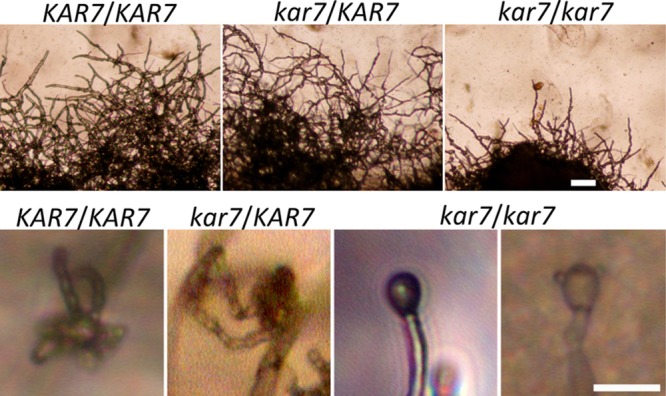
Unisexual reproduction of a diploid kar7/kar7 strain. In the diploid strain, both alleles of the KAR7 gene were disrupted. The wild type and the heterozygous KAR7/kar7::HYG mutant undergo complete same-sex mating, including abundant sexual hypha formation and basidium formation followed by spore chain production. However, kar7::HYG/kar7::URA5 mutants display mating defects, as seen in the haploid kar7 mutants: for example, less abundant sexual hyphae formation and bald or single spore-attached basidium formation as a consequence of a failure to enter into meiosis or produce spore chains. Scale bars = 100 μm for the upper row and 10 μm for the bottom row.
KAR2 is essential for viability, but its role in C. neoformans mating is unknown.
Exhaustive trials to disrupt the KAR2 gene in C. neoformans were unsuccessful. Thus, we tested if the KAR2 gene is essential for viability. One KAR2 allele from the diploid strain MN142.3 was replaced with the HYG drug resistance cassette. We obtained a heterozygous kar2::HYG/KAR2 diploid isolate that underwent unisexul reproduction, from which spore-derived progeny were analyzed based on hygromycin resistance. Of 12 haploid progeny, none were kar2 mutants (n = 12; P = 0.0005), indicating KAR2 is essential. We did not observe any hyphal morphogenesis defect of the heterozygous kar2/KAR2 mutant strains.
KAR3, KAR4, and KAR8 are dispensable during mating.
The functions of three other KAR genes (KAR3, KAR4, and KAR8) during mating were assessed. The KAR3 gene was disrupted in the C. neoformans serotype A strain pair KN99α and KN99a. kar3::NAT and kar3::NEO mutants crossed unilaterally (mutant × wild type) or bilaterally (mutant × mutant) underwent opposite-sex mating successfully (Fig. 7A). Because Kar3 is a microtubule motor protein involved in nuclear congression in S. cerevisiae (Fig. 1 and Table 2), we tested if there is any nuclear distance difference in mating hyphae between the wild type and α kar3 × a kar3 crosses. To monitor nuclei, α mating type kar3 mutants (SL295 and SL296) were transformed with the GFP-Nop1 plasmid (pSL04-1), and the resulting α kar3 strains expressing GFP-Nop1 (Table 1) were crossed with a kar3 mutants. The distance between two nuclei in one dikaryotic hyphal compartment was not significantly different between wild-type and α kar3 × a kar3 matings (5.57 ± 0.47 μm versus 5.53 ± 0.76 μm, respectively) (see Fig. S4 in the supplemental material). These results indicate that Kar3 is not necessary to complete the mating process or govern nuclear dynamics in the dikaryotic hyphae.
Fig 7.
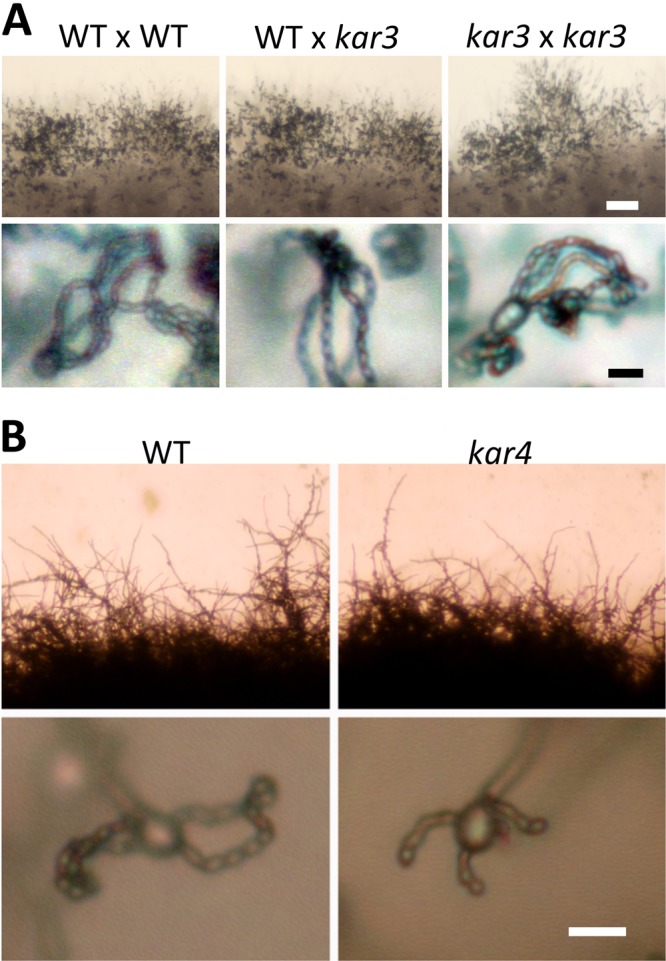
KAR3 and KAR4 are dispensable for mating in C. neoformans. (A) Serotype A kar3 mutants complete opposite-sex mating without any marked defects observed in either unilateral or bilateral mutant crosses. (B) Unisexual reproduction of kar4 mutants produces four chains of basidiospores. V8 (pH 7) and MS media for serotypes D and A, respectively, were used for mating and incubated at room temperature in the dark for 3 weeks before observation. Scale bars = 50 μm for the upper row and 10 μm for the bottom row in panel A and 10 μm for panel B.
We initially speculated that the C. neoformans KAR4 might be essential because gene disruption attempts were not successful, even though the kar4 mutant is viable in S. cerevisiae. However, we did obtain viable kar4 mutants through a diploid unisexual reproduction approach (see above and Materials and Methods). One KAR4 allele in the diploid strain MN142.3 was replaced with the hygromycin resistance gene, and the resulting heterozygous kar4/KAR4 strains underwent hyphal morphogenesis and sporulation on V8 medium (pH 7) in the dark. Spores were dissected and we recovered 7 kar4::HYG haploid progeny out of 13 progeny analyzed. Based on PCR, these kar4 mutant progeny lack the ORF of the wild-type gene (data not shown). The ploidy of the progeny was confirmed to be haploid, based on FACS analysis of cells stained with propidium iodide (see Fig. S5 in the supplemental material), indicating that the KAR4 gene is not essential. However, kar4 mutants did not exhibit any unisexual mating defects (Fig. 7B) compared to the wild type, indicating that KAR4 is also dispensable for unisexual mating.
The KAR8 gene encoding a putative protein chaperone in the ER membrane was replaced with the NEO or NAT cassette in the congenic serotype D strains JEC21α and JEC20a (Table 1). In the unilateral kar8 × wild-type and bilateral kar8 × kar8 mutant crosses, we found that disruption of KAR8 did not result in mating defects, wherein the mutants were able to complete mating and produce spore progeny (Fig. 8A). We also tested the role of Kar8 in same-sex mating. The XL280 strain was used, and the KAR8 locus was replaced with the NAT marker gene. The kar8::NAT mutant exhibited morphologically complete same-sex mating similar to that of the wild-type parental strain (Fig. 8B). These results indicate that Kar8 is dispensable for both opposite-sex and unisexual mating.
Fig 8.
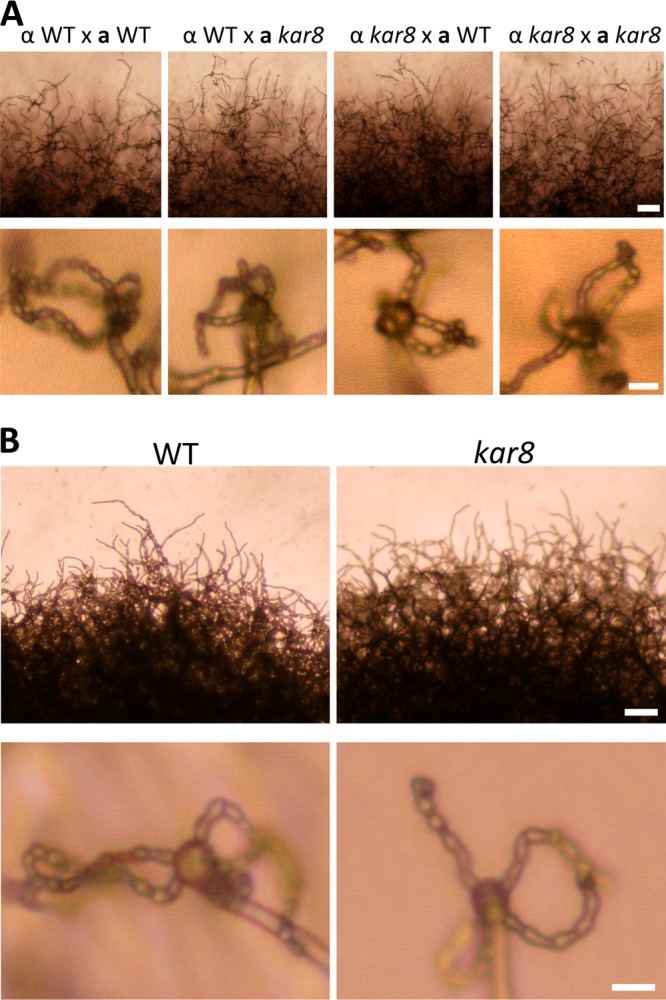
KAR8 is not required for opposite- or same-sex mating. (A) Serotype D kar8 mutants exhibit no apparent defects in sexual development during a-α opposite-sex mating. (B) An XL280-derived kar8 mutant also displays no apparent mating defects. V8 medium (pH 7) was used for mating and incubated at room temperature in the dark for 3 weeks before observation. Scale bars = 100 μm for the upper row and 5 μm for the bottom row in both panels A and B.
DISCUSSION
C. neoformans is thought to be predominantly a haploid organism. However, both clinical and environmental diploid isolates also occur and represent ∼10% of isolates analyzed (34). One central question is how are diploids in general formed? There are two possibilities: (i) endoreplication and (ii) pathways involving karyogamy. Endoreplication can produce diploids with homozygous alleles, and some known αAAα, aAAa, αDDα, aDDa, isolates might have arisen via this process (12, 29, 33). However, other diploids are clearly heterozygous, including αAAa, αDDa, and also αADa, aADα, and αADα hybrid isolates (12, 34, 35). In these cases, endoreplication is not sufficient to explain their occurrence, and cell-cell and nuclear fusion (karyogamy) are an alternative route to explain the existence of these heterozygous diploids. Karyogamy in basidiomycetes is associated with mating. However, a question remaining unanswered is when diploidization occurs during opposite- or same-sex mating—i.e., early karyogamy before hyphal formation or late karyogamy just prior to meiosis. While karyogamy during opposite-sex mating is typically modeled as occurring late, under laboratory conditions, two haploid cells can undergo cell-cell and nuclear fusion at 37°C, which is how diploids are constructed and isolated in the laboratory (19, 20). Interestingly, a/α diploid strains exhibit thermal dimorphism, in which the diploid cell grows as yeast at 37°C, whereas at 24°C, the diploid cell undergoes filamentation to produce monokaryotic hyphae and complete sexual reproduction (56). This is an example of an “early” karyogamy event occurring during opposite-sex mating (Fig. 1). By analogy, karyogamy could also occur early, late, or both early and late, during unisexual reproduction. Examination of karyogamy genes during opposite-sex and unisexual mating can further provide insight to understand when and how diploidization occurs in C. neoformans.
Although the mechanics of karyogamy are well established in S. cerevisiae (52), the roles of karyogamy genes are less well understood in basidiomycetes, including C. neoformans. In this study, we found that Kar7/Sec66 plays a key role in completion of both opposite-sex and unisexual mating. Mutants lacking this protein exhibit a failure of karyogamy in C. neoformans during both types of mating. Therefore, the KAR7 gene is essential for diploidization. In the analysis of unisexual mating of the kar7 mutants, the mating process was arrested at the basidium stage without karyogamy (Fig. 2 to 5). Thus, in the XL280 strain, diploidization can occur late in the basidium just prior to meiosis. However, we cannot exclude the possibility that the KAR7 gene functions through processes in addition to karyogamy during mating. Our study with diploids homozygous for the kar7 mutation revealed that diploidy itself is not sufficient to overcome the requirement for KAR7 for meiotic entry and basidiospore chain formation (Fig. 6). The underlying role in this case remains to be elucidated. In addition, in the other serotype D strain analyzed (JEC21), a kar7 mutant exhibited a complete defect in unisexual filamentation (see Fig. S2 in the supplemental material). In this case, early karyogamy and diploidization could be a prerequisite step for early hyphal growth. This hyphothesis is supported by the findings that (i) diploid α/α strains are more filamentous than haploid α strains, and (ii) blastospores emerging from the hyphae of the haploid strain JEC21 can be diploid (31).
Kar7/Sec66 is a component of the Sec63 complex that functions in protein translocation into the ER lumen (23, 65). The exact mechanistic role of the Kar7 protein in karyogamy is unknown: the target proteins for Kar7 and the Sec63 complex that are translocated into the ER and those in the ER transported to cytosol by the Kar7 and Sec63 complex remain to be identified. One possible function of Kar7 is that it is involved in transport of proteins to initiate and complete nuclear fusion and, potentially, the transport of proteins associated with triggering meiosis. We also observed that the kar7 mutants exhibit some detrimental phenotypes during vegetative yeast growth (see Fig. S6 in the supplemental material), which indicates that the protein trafficking function may affect growth. It is also possible that Kar7 has a karyogamy-specific function other than protein trafficking in the ER membrane (45).
Among the KAR genes involved in nuclear congression, the C. neoformans genome lacks KAR1 and KAR9 genes, and, more interestingly, our study found that the KAR3 and KAR4 genes are not essential for sexual reproduction (Fig. 7), although in S. cerevisiae, both KAR3 and KAR4 are essential for karyogamy and mating (13, 23, 24, 40), and in C. albicans, KAR3 is required to complete sexual development (4). It is of interest that the C. neoformans Kar3 and Kar4 proteins appear to play no role in mating and therefore are dispensable for karyogamy. C. neoformans is a basidiomycete, and its mating is characterized by an extended dikaryotic hyphal stage, wherein two parental nuclei are paired but unfused in a single hyphal compartment. Nuclear fusion (karyogamy) occurs inside the basidia at the apex of the aerial hypha (2, 11). It is possible that nuclear congression is a less relevant step in C. neoformans mating during which the dikaryotic stage is prolonged. In ascomycetes, including S. cerevisiae, the dikaryotic stage is transient and fleeting during mating, and thus karyogamy genes that function in nuclear congression are of substantial consequence during mating to ensure the rapidity of the process. This phenotypic difference in nuclear positions during mating and the timing of nuclear fusion may impact the different evolutionary trajectories of the KAR genes involved in nuclear migration in the two phyla.
Among the two KAR genes involved in nuclear membrane fusion, C. neoformans KAR8 was found to be dispensable for sexual reproduction (Fig. 8). Kar8 is a DnaJ-like protein anchored to the ER membrane, where it functions together with Hsp70. In S. cerevisiae, a kar8 deletion mutant displays defects in karyogamy (46). However, in C. neoformans, only KAR7 was found to be essential for C. neoformans mating, indicating a differential adaptation of the KAR genes in this fungus. For example, S. cerevisiae karyogamy genes, including KAR4 and KAR5, are regulated by pheromone (Table 2); however, in C. neoformans, the pheromone signal is involved in the initial cell-cell fusion step (and subsequent clamp cell fusion events), which is separated from karyogamy by the extended dikaryotic hyphal stage (see Fig. S7 in the supplemental material).
Our finding that some KAR genes are dispensable for sexual reproduction in C. neoformans indicates that other novel proteins may be involved in C. neoformans karyogamy. Future work that analyzes mating defects, especially in mutants arrested at the basidium formation step without further progression into meiosis and sporulation, has the potential to identify novel genes involved in karyogamy in C. neoformans. Kar2 is an ATPase involved in protein trafficking in the ER, and in C. neoformans, KAR2 is essential as in S. cerevisiae. The possible roles of KAR2 in C. neoformans nuclear dynamics remain to be elucidated.
It is curious that some of the genes one would expect to be involved in karyogamy and sexual reproduction don't seem to play such a role in C. neoformans. This may indicate that karyogamy is more plastic than one might have anticipated. Analogies are found in meiosis in other fungi and insects. The Candida lusitaniae genome is missing many key meiotic genes, but it still undergoes meiosis during sexual reproduction (8, 50). Caenorhabditis elegans, Drosophila melanogaster, Neurospora crassa, Ustilago maydis, and C. lusitaniae all lack the meiosis-specific Dmc1 protein that functions in DNA double-strand-break repair during meiosis in other species, and yet all still successfully complete meiosis (5, 15, 61). In addition, in Schizosaccharomyces pombe synaptonemal-complex component protein genes are missing, and instead LinE elements are involved in chromosomal pairing and migration (37).
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Mark Rose for valuable discussions, Xuying Wang for technical support for spore dissection, Min Ni for constructing the MN142.3 diploid strain, Sujal Phadke for critical reading, and Anna Averette for technical support. We are also grateful to Valerie Knowlton and the NC State EM core laboratory for SEM analysis and to Yasheng Gao for assistance with light microscopy.
S.C.L is supported by the NIH Molecular Mycology and Pathogenesis Training Program (AI52080). This work was supported by NIH/NIAID R37 grant AI39115-14 to J.H.
Footnotes
Published ahead of print 27 April 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Anderson JB, Kohn LM. 2007. Dikaryons, diploids, and evolution, p 317–331. In Heitman J, Kronstad JW, Taylor JW, Casselton LA. (ed), Sex in fungi. ASM Press, Washington, DC [Google Scholar]
- 3. Beh CT, Brizzio V, Rose MD. 1997. KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J. Cell Biol. 139:1063–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bennett RJ, Miller MG, Chua PR, Maxon ME, Johnson AD. 2005. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol. Microbiol. 55:1046–1059 [DOI] [PubMed] [Google Scholar]
- 5. Borkovich KA, et al. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Botts MR, Hull CM. 2010. Dueling in the lung: how Cryptococcus spores race the host for survival. Curr. Opin. Microbiol. 13:437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brizzio V, et al. 1999. Genetic interactions between KAR7/SEC71, KAR8/JEM1, KAR5, and KAR2 during nuclear fusion in Saccharomyces cerevisiae. Mol. Biol. Cell 10:609–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler G, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campos AN, Costa MD. 2010. Basidiosporogenesis, meiosis, and post-meiotic mitosis in the ectomycorrhizal fungus Pisolithus microcarpus. Fungal Genet. Biol. 47:477–483 [DOI] [PubMed] [Google Scholar]
- 10. Casadevall A, Perfect JR. 1998. Cryptococcus neoformans. ASM Press, Washington, DC [Google Scholar]
- 11. Clark TA, Anderson JB. 2004. Dikaryons of the basidiomycete fungus Schizophyllum commune: evolution in long-term culture. Genetics 167:1663–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cogliati M, Esposto MC, Clarke DL, Wickes BL, Viviani MA. 2001. Origin of Cryptococcus neoformans var. neoformans diploid strains. J. Clin. Microbiol. 39:3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Conde J, Fink GR. 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. U. S. A. 73:3651–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davidson RC, et al. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607–2615 [DOI] [PubMed] [Google Scholar]
- 15. Donaldson ME, Saville BJ. 2008. Bioinformatic identification of Ustilago maydis meiosis genes. Fungal Genet. Biol. 45(Suppl 1):S47–S53 doi:10.1016/j.fgb.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 16. D'Souza CA, et al. 2011. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. mBio 2:e00342–10 doi:10.1128/mBio.00342–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fraser JA, Subaran RL, Nichols CB, Heitman J. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2:1036–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giles SS, Dagenais TRT, Botts MR, Keller NP, Hull CM. 2009. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect. Immun. 77:3491–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hull CM, Davidson RC, Heitman J. 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev. 16:3046–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hull CM, Heitman J. 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36:557–615 [DOI] [PubMed] [Google Scholar]
- 21. Kavanaugh LA, Fraser JA, Dietrich FS. 2006. Recent evolution of the human pathogen Cryptococcus neoformans by intervarietal transfer of a 14-gene fragment. Mol. Biol. Evol. 23:1879–1890 [DOI] [PubMed] [Google Scholar]
- 22. Kozubowski L, Heitman J. 2010. Septins enforce morphogenetic events during sexual reproduction and contribute to virulence of Cryptococcus neoformans. Mol. Microbiol. 75:658–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD. 1994. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J. Cell Biol. 126:911–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kurihara LJ, Stewart BG, Gammie AE, Rose MD. 1996. Kar4p, a karyogamy-specific component of the yeast pheromone response pathway. Mol. Cell. Biol. 16:3990–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kwon-Chung KJ. 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68:821–833 [PubMed] [Google Scholar]
- 26. Kwon-Chung KJ. 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67:1197–1200 [PubMed] [Google Scholar]
- 27. Kwon-Chung KJ. 1976. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68:943–946 [PubMed] [Google Scholar]
- 28. Lee SC, Ni M, Li W, Shertz C, Heitman J. 2010. The evolution of sex: a perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. 74:298–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li W, et al. 2012. Genetic diversity and genomic plasticity of Cryptococcus neoformans AD hybrid strains. G3 (Bethesda) 2:83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin X, Huang JC, Mitchell TG, Heitman J. 2006. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATalpha allele enhances filamentation. PLoS Genet. 2:e187 doi:10.1371/journal.pgen.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin X, Hull CM, Heitman J. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017–1021 [DOI] [PubMed] [Google Scholar]
- 32. Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. 2010. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet. 6:e1000953 doi:10.1371/journal.pgen.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin X, et al. 2007. αADα hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 3:e186 doi:10.1371/journal.pgen.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin X, et al. 2009. Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Pathog. 5:e1000283 doi:10.1371/journal.ppat.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Litvintseva AP, Lin X, Templeton I, Heitman J, Mitchell TG. 2007. Many globally isolated AD hybrid strains of Cryptococcus neoformans originated in Africa. PLoS Pathog. 3:e114 doi:10.1371/journal.ppat.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loftus BJ, et al. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lorenz A, et al. 2004. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J. Cell Sci. 117:3343–3351 [DOI] [PubMed] [Google Scholar]
- 38. Makio T, Nishikawa S, Nakayama T, Nagai H, Endo T. 2008. Identification and characterization of a Jem1p ortholog of Candida albicans: dissection of Jem1p functions in karyogamy and protein quality control in Saccharomyces cerevisiae. Genes Cells 13:1015–1026 [DOI] [PubMed] [Google Scholar]
- 39. Martin-Castellanos C, et al. 2005. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr. Biol. 15:2056–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meluh PB, Rose MD. 1990. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell 60:1029–1041 [DOI] [PubMed] [Google Scholar]
- 41. Metin B, Findley K, Heitman J. 2010. The mating type locus (MAT) and sexual reproduction of Cryptococcus heveanensis: insights into the evolution of sex and sex-determining chromosomal regions in fungi. PLoS Genet. 6:e1000961 doi:10.1371/journal.pgen.1000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Middleton K, Carbon J. 1994. KAR3-encoded kinesin is a minus-end-directed motor that functions with centromere binding proteins (CBF3) on an in vitro yeast kinetochore. Proc. Natl. Acad. Sci. U. S. A. 91:7212–7216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller RK, Matheos D, Rose MD. 1999. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol. 144:963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller RK, Rose MD. 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140:377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ng DT, Walter P. 1996. ER membrane protein complex required for nuclear fusion. J. Cell Biol. 132:499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nishikawa S, Endo T. 1997. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J. Biol. Chem. 272:12889–12892 [DOI] [PubMed] [Google Scholar]
- 47. Park BJ, et al. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530 [DOI] [PubMed] [Google Scholar]
- 48. Price MS, Nichols CB, Alspaugh JA. 2008. The Cryptococcus neoformans Rho-GDP dissociation inhibitor mediates intracellular survival and virulence. Infect. Immun. 76:5729–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raju NB. 1992. Genetic control of the sexual cycle in Neurospora. Mycol. Res. 96:241–262 [Google Scholar]
- 50. Reedy JL, Floyd AM, Heitman J. 2009. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr. Biol. 19:891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Romisch K. 1999. Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J. Cell Sci. 112:4185–4191 [DOI] [PubMed] [Google Scholar]
- 52. Rose MD. 1996. Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu. Rev. Cell Dev. Biol. 12:663–695 [DOI] [PubMed] [Google Scholar]
- 53. Rose MD, Fink GR. 1987. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell 48:1047–1060 [DOI] [PubMed] [Google Scholar]
- 54. Rose MD, Misra LM, Vogel JP. 1989. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57:1211–1221 [DOI] [PubMed] [Google Scholar]
- 55. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 56. Sia RA, Lengeler KB, Heitman J. 2000. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet. Biol. 29:153–163 [DOI] [PubMed] [Google Scholar]
- 57. Spang A, Courtney I, Grein K, Matzner M, Schiebel E. 1995. The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J. Cell Biol. 128:863–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tange Y, et al. 1998. A novel fission yeast gene, tht1+, is required for the fusion of nuclear envelopes during karyogamy. J. Cell Biol. 140:247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tokunaga M, Kawamura A, Kohno K. 1992. Purification and characterization of BiP/Kar2 protein from Saccharomyces cerevisiae. J. Biol. Chem. 267:17553–17559 [PubMed] [Google Scholar]
- 60. Velagapudi R, Hsueh Y-P, Geunes-Boyer S, Wright JR, Heitman J. 2009. Spores as infectious propagules of Cryptococcus neoformans. Infect. Immun. 77:4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Villeneuve AM, Hillers KJ. 2001. Whence meiosis? Cell 106:647–650 [DOI] [PubMed] [Google Scholar]
- 62. Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. 2004. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5:567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wickes BL, Mayorga ME, Edman U, Edman JC. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc. Natl. Acad. Sci. U. S. A. 93:7327–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xue C, Tada Y, Dong X, Heitman J. 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1:263–273 [DOI] [PubMed] [Google Scholar]
- 65. Young BP, Craven RA, Reid PJ, Willer M, Stirling CJ. 2001. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 20:262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


