Abstract
Synthesis of functional mRNA in eukaryotes involves processing of precursor transcripts, including the addition of a poly(A) tail at the 3′ end. A multiprotein complex recognizes a polyadenylation signal, generally the hexanucleotide AAUAAA in metazoans, to direct processing of the pre-mRNA. Based on sequence analysis of several cDNAs, we have previously suggested that the UAAA tetranucleotide (which may include the UAA translation stop codon) could be the polyadenylation signal in Trichomonas vaginalis, a parasitic protozoon that causes human trichomoniasis. This proposal is analyzed here with the aid of a transient-expression system of a reporter gene (cat flanked by T. vaginalis actin noncoding sequences). When cells were transfected with a plasmid bearing the original 3′ untranslated region (UTR) sequence containing the UAAA motif, the resulting cat mRNA was polyadenylated similarly to the endogenous actin mRNA. Base changes in the UAAA sequence produced alterations to the polyadenylation site of the reporter mRNAs, while nucleotide substitutions at either side of UAAA did not. Furthermore, relocation of the UAAA motif redirected the processing and polyadenylation of the reporter mRNA. In addition, a pre-mRNA cleavage site for polyadenylation was defined. Interaction of T. vaginalis proteins with the UAAA motif was shown by electrophoretic mobility shift assays. Based on our findings, we provide evidence that in T. vaginalis the UAAA tetranucleotide has a role equivalent to that of the metazoan consensus AAUAAA polyadenylation signal.
INTRODUCTION
Trichomonas vaginalis is a parasitic protozoon that causes trichomoniasis, one of the most common sexually transmitted infections in humans (34). Phylogenetic analyses based on small-subunit (SSU) rRNA have placed this organism among the earliest-diverging eukaryotic branches (27) and, more recently, in the Excavata supergroup according to the phylogenetic egalitarian model (7). Synthesis of functional mRNA in eukaryotes involves polyadenylation of the precursor transcripts. T. vaginalis mRNAs are polyadenylated, but this process has not been experimentally addressed. This situation contrasts with the considerable effort directed toward the analysis of transcription by RNA polymerase II in this organism: core promoters have been found to consist solely of a highly conserved initiator element (Inr), IBP39 is a protein involved in promoter recognition, and no homologues have been identified in other species (14). Regarding RNA splicing, relatively few genes (about 65) in the T. vaginalis genome appear to contain an intron (4), and the ones recognized are short and characterized by a conserved 12-nucleotide (nt) sequence (30); spliceosomal components also have been identified (26).
The 3′ poly(A) tail of mRNA is involved in regulatory mechanisms such as mRNA stability, export of mRNA from the nucleus to the cytoplasm, and initiation of translation (22, 33). Recognition of the polyadenylation site is coordinated by interactions between processing factors and RNA sequence elements (cis elements) located both upstream and downstream from the poly(A) addition site. The consensus polyadenylation signal in metazoa is the hexanucleotide AAUAAA, which is located within the transcribed 3′ untranslated region (3′ UTR) of the mRNA, 10 to 30 nucleotides upstream of the actual cleavage site for polyadenylation. This motif is recognized by the cleavage/polyadenylation specificity factor 160 subunit (CPSF-160). Additional sequence motifs have been described as being relevant for the polyadenylation process; the dinucleotide CA has been identified as the cleavage site, recognized by the CPSF-73 endonuclease. A U-rich or GU-rich region located up to 30 bases downstream from the cleavage site is also relevant in metazoans. This U-rich element is recognized by cleavage stimulation factor (CstF), which interacts with CPSF to cleave the mRNA and start the addition of the poly(A) tail using the poly(A) polymerase in a template-independent manner (5, 17, 33). In mammals, UGUA elements are generally present within 40 to 100 nucleotides upstream of the poly(A) site (12, 31). In the yeast Saccharomyces cerevisiae, sequences directing this process are less conserved than in metazoans. Nonetheless, four sequence elements have been identified: the AU-rich element, the A-rich positioning element, the cleavage site, and the U-rich elements flanking the cleavage site (11, 20). The motif UA(A/U)UU in Entamoeba histolytica and the sequence AGURAAY in Giardia spp. have been proposed as the equivalent polyadenylation sequences (1, 15), although no experimental evidence that demonstrates their function has been provided so far.
Based on a comparative sequence analysis of the 3′ UTRs of several T. vaginalis cDNAs, we have previously proposed the UAAA tetranucleotide to be the polyadenylation signal in T. vaginalis (9). It was shown there that the mRNA in this organism posses a very short 3′ UTR and that the consensus eukaryotic polyadenylation signal (AAUAAA) is rarely found. The single identified consensus sequence related to this motif was the UAAA tetranucleotide in the 3′ UTR, which may overlap the UAA translation stop codon. Additional cis elements were also proposed: Y↓(A)2-5UU as the site for cleavage and a tract of Us downstream from the cleavage site (9). Here, the recognition of the UAAA motif as the polyadenylation signal is experimentally addressed. Based on the evidence presented, we propose that the UAAA tetranucleotide in T. vaginalis is functionally equivalent to the canonical metazoan AAUAAA polyadenylation signal.
MATERIALS AND METHODS
In silico analysis.
Sixty T. vaginalis cDNA sequences were identified and retrieved from the NCBI database. Additionally, we searched for the presence of the UAAA motif in the 3′ noncoding regions of genes that have evidence of expression. These genes were obtained from the expressed sequence tag (EST) database available at www.trichdb.org. Only genes with a 100% match to an EST were included in the analysis (780 genes). The region downstream from the translation stop codon was obtained using the tool retrieve sequence by gene IDs available at www.trichdb.org and analyzed using Excel (Microsoft Inc.). Sequence logo representations of multiple-sequence alignments were prepared with the WebLogo tool, version 2.8.2 (http://weblogo.berkeley.edu/) (6, 24).
Parasites and culture conditions.
The T. vaginalis isolate CNCD147 was used throughout the experimental work. Parasites were grown in Trypticase-yeast extract-maltose (TYM) medium supplemented with 10% (vol/vol) heat-inactivated horse serum (GIBCO) as previously described (8).
Construction of a transient-expression vector for T. vaginalis.
An SpeI-EcoRI-digested fragment (2,827 bp) from a T. vaginalis genomic actin clone (GenBank access number AF237734) was cloned in pBluescript II KS(−) (Stratagene) (see Fig. 2A). The actin-coding sequence was substituted for the chloramphenicol acetyltransferase (CAT) gene (cat) coding sequence by PCR amplification, which also included the addition of RcaI and SacI restriction sites. In this construct, the UAG stop codon was used for the cat gene, while the original actin 3′ UTR (which included the UAA stop codon) was moved 9 bp downstream (see Fig. 2B). The purpose of this strategy was to separate the stop codon for the cat gene from the analyzed UAAA motif because this tetranucleotide overlapped the actin UAA stop codon in the original genomic clone. The resulting pcat-UAAA plasmid was used as the reference plasmid from which all derived mutant plasmids were constructed (Table 1). From the mutant pcat-UCAA plasmid, three plasmids were derived: the revertant pcat-revUAAA, in which the UCAA motif was reverted to UAAA by site-directed mutagenesis, and two plasmids containing the relocated UAAA motif, pcat-UAAA74 (proximal) and pcat-UAAA118 (distal); in the pcat-UAAA118 plasmid a putative cleavage site was added 20 nt downstream from the relocated UAAA motif, thus rendering the pcat-UAAA118CS plasmid (Table 1; see Fig. 3 and Fig. 5). The QuikChange site-directed mutagenesis kit (Stratagene) was used for these constructs following the manufacturer's directions, and the specific oligonucleotides that were used for mutagenesis are listed in Table 2. All plasmids were verified by sequencing.
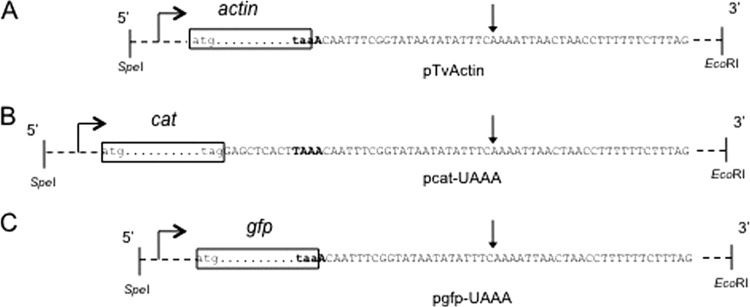
Fig 2.
The reporter system to study RNA motifs involved in polyadenylation. The actin gene coding region from a genomic T. vaginalis clone (pTvActin) (A) was replaced with the cat coding region (pcat-UAAA) (B) or with the GFP gene coding region (pgfp-UAAA) (C). The coding regions are represented as dots between the initiation (atg) and the stop (taa or tag) codons inside boxes. Dashed lines represent upstream (934-bp) and downstream (762-bp) noncoding regions of the T. vaginalis actin gene. The DNA sequence is shown. The TAAA motif is in bold. In the pcat-UAAA plasmid, the TAAA motif was placed 9 nucleotides downstream from the stop codon, and TAG was introduced as the stop codon. Horizontal arrows represent the start of transcription. The vertical arrow marks the site where mRNAs are polyadenylated.
Table 1.
Plasmids used in this study
| Plasmid name | Sequence surrounding UAAA motifa | Description |
|---|---|---|
| pcat-UAAA | ...CACUUAAACAAU... | Parental plasmid described in Fig. 1 |
| pcat-GCCC | ...CACUGCCCCAAU... | Change of the four bases of the UAAA motif |
| pcat-GAAA | ...CACUGAAACAAU... | Change of the first base of the UAAA motif |
| pcat-UCAA | ...CACUUCAACAAU... | Change of the second base of the UAAA motif |
| pcat-UACA | ...CACUUACACAAU... | Change of the third base of the UAAA motif |
| pcat-UAAC | ...CACUUAACCAAU... | Change of the fourth base of the UAAA motif |
| pcat-GUAAA | ...CACGUAAACAAU... | Change of the upstream base adjacent to the UAAA motif |
| pcat-UAAAG | ...CACUUAAAGAAU... | Change of the downstream nucleotide adjacent to the UAAA motif |
| pcat-AAUAAA | .CUUCAAUAAACAAU... | Introduction of two A nucleotides to generate the eukaryotic polyadenylation signal AAUAAA |
| pcat-revUAAA | ...CACUUAAACAAU... | Revertant construct from pcat-UCAA |
| pcat-UAAA74 | ...UGUAAAUCACUUCAUUAAUAUUGUGUUUUAAUG... | Relocation of the UAAA motif 74 nt downstream from the stop codon in pcat-UCAA |
| pcat-UAAA118 | ...ACUAAAACGAUGUACCCUGAACAUUUCUUUUCCA... | Relocation of the UAAA motif 118 nt downstream from the stop codon in pcat-UCAA |
| pcat-UAAA118CS | ...ACUAAAACGAUGUACCCUGAACAUUUCAAUUCCA... | Addition of a cleavage site in the pcat-UAAA118 plasmid |
The UAAA motif is in bold. Underlining denotes a nucleotide changed from that in the parental plasmid UAAA.
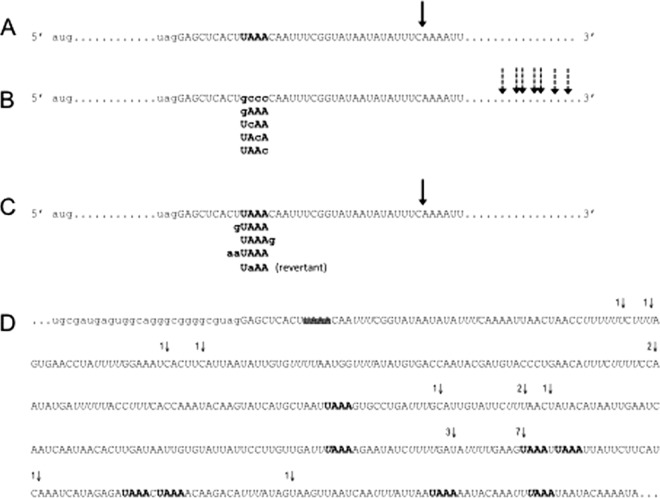
Fig 3.
Schematic representation of the polyadenylation site in mRNAs derived from parental and mutant pcat plasmids. (A) Partial sequence of the 3′ UTR of cat transcripts from T. vaginalis cells that were transfected with the pcat-UAAA plasmid. The UAAA motif is in bold, and the arrow denotes the site at which all mRNAs were found to be polyadenylated. (B) Schematic representation of the 3′ UTR of cat transcripts from cells transfected with the different pcat mutant plasmids. In these plasmids, the UAAA motif was changed to the indicated sequences. The arrows represent the sites at which mRNAs were found to be polyadenylated (fully described in panel D). The mutated nucleotides are shown in bold lowercase letters. (C) Sequence of the 3′ UTR of cat transcripts from cells transfected with pcat mutant plasmids in which the UAAA motif remained unaltered but the adjacent bases were changed as indicated. The UaAA (revertant) is a revertant construct derived from pcat-UCAA by site-directed mutagenesis. The arrow denotes the site at which all the mRNAs analyzed were found to be polyadenylated and is the same position as in the wild type (A). The mutated nucleotides are shown in bold lowercase letters. (D) RNA sequence from the pcat mutant plasmids in panel B, showing the actual positions where mRNAs were found to be polyadenylated. Part of the cat coding sequence is in lowercase letters. The 3′ region is in uppercase. The parental UAAA motif is not present in the pcat mutant plasmids and is therefore crossed out; this tetranucleotide was replaced by the bold motifs depicted in panel B. The last nucleotides identified before the poly(A) tail in cat cDNA clones are identified by arrows. The number of clones found for each position is shown next to the vertical arrows. Additional UAAA motifs in the 3′ noncoding region are shown in bold, and U-rich elements are in italic. In all cases, the sites where the mRNAs were polyadenylated were deduced from sequencing oligo(dT)-derived cDNA clones.
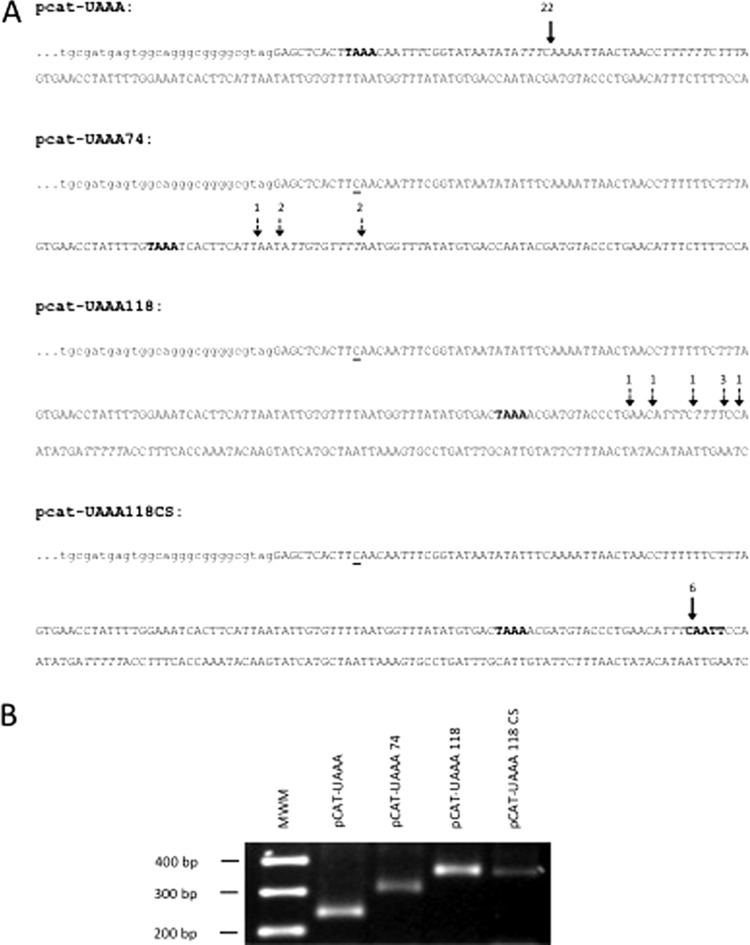
Fig 5.
Relocation of the UAAA motif redirects the polyadenylation site. (A) DNA sequences of the plasmid constructs derived from the pcat-UCAA mutant, in which the UAAA motif was relocated downstream from the parental site. pcat-UAAA, parental sequence. The arrow denotes the site where all mRNAs were found to be polyadenylated. pcat-UAAA74, the UAAA motif (bold) was placed 74 nt downstream from the stop codon, and the parental UAAA motif was mutated (underlined C). The arrows represent the close-by sites where mRNAs were found to be polyadenylated. pcat-UAAA118, the UAAA motif (bold) was placed 118 nt downstream from the stop codon, and the parental UAAA motif was mutated (underlined C). The arrows represent close-by sites where mRNAs were found to be polyadenylated. pcat-UAAA118CS, the CAATT motif (bold), proposed as one of the cleavage site consensus motifs, was placed 20 nt downstream from the UAAA sequence in the pcat-UAAA118 plasmid. The arrow represents the site where all mRNAs were polyadenylated. The numbers above the arrows represent the number of clones found polyadenylated at each position. All polyadenylation sites were inferred from cDNA cloning and sequencing. (B) Evaluation of the size and homogeneity of the cat mRNAs by RT-PCR. Bands of the expected size were obtained, as predicted from the position where the UAAA motifs were introduced. Amplification products were analyzed on a 2.2% agarose gel and stained with ethidium bromide.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| CATSTOP 5 | 5′GCGTAGGAGCTCACTGAAACAATTTCGGTA3′ |
| CATSTOP 6 | 5′TACCGAAATTGTTTCAGTGAGCTCCTACGC3′ |
| CATSTOP 7 | 5′CGTAGGAGCTCACTTCAACAATTTCGGTAT3′ |
| CATSTOP 8 | 5′ATACCGAAATTGTTGAAGTGAGCTCCTACG3′ |
| CATSTOP 9 | 5′GTAGGAGCTCACTTACACAATTTCGGTATA3′ |
| CATSTOP 10 | 5′TATACCGAAATTGTGTAAGTGAGCTCCTAC3′ |
| CATSTOP 11 | 5′TAGGAGCTCACTTAACCAATTTCGGTATAA3′ |
| CATSTOP 12 | 5′TTATACCGAAATTGGTTAAGTGAGCTCCTA3′ |
| CATSTOP 13 | 5′TAGGAGCTCACTGCCCCAATTTCGGTATAA3′ |
| CATSTOP 14 | 5′TTATACCGAAATTGGGGCAGTGAGCTCCTA3′ |
| CATSTOP15 | 5′AGGAGCTCACTTAAAGAATTTCGGTATAAT3′ |
| CATSTOP16 | 5′ATTATACCGAAATTCTTTAAGTGAGCTCCT3′ |
| GTAAA | 5′GCGTAGGAGCTCACGTAAACAATTTCGGTATAATA3′ |
| RC GTAAA | 5′TATTATACCGAAATTGTTTACGTGAGCTCCTACGC3′ |
| INS TAAA | 5′GCGTAGGAGCTCACTTCAATAAACAATTTCGGTATAATA3′ |
| RC INS | 5′TATTATACCGAAATTGTTTATTGAAGTGAGCTCCTACGC3′ |
| F-REGSP | 5′GCGTAGGAGCTCACTTAAACAATTTCGGTATAATA3′ |
| RC-REGSP | 5′TATTATACCGAAATTGTTTAAGTGAGCTCCTACGC3′ |
| PAS | 5′TGGTTTATATGTGACTAAAACGATGTACCCTGAAC3′ |
| RC-PASN | 5′GTTCAGGGTACATCGTTTTAGTCACATATAAACCA3′ |
| CS | 5′TACCCTGAACATTTCAATTCCAATATGATTTTTAC3′ |
| RC-CS | 5′GTAAAAATCATATTGGAATTGAAATGTTCAGGGTA3′ |
| F-NSP | 5′GTGAACCTATTTTGTAAATCACTTCATTAATATTG3′ |
| RC-NSP | 5′CAATATTAATGAAGTGATTTACAAAATAGGTTCAC3′ |
| AP | 5′GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT3′ |
| AUAP-B | 5′GGATCCGGCCACGCGTCGACTAGTAC3′ |
| CAT10 | 5′CACATTCTTGCCCGCCTG3′ |
| CAT11 | 5′GAGCTGGTGATATGGGAT3′ |
| CAT 20 | 5′AAGTTGTCCATATTGGCC3′ |
| For-invit | 5′TAATACGACTCACTATAGGGAATTACAACAGTACTGCGAT3′ |
| Rev-invit | 5′ATTTCCAAAATAGGTTCACT3′ |
| For-CATqPCR | 5′CGCCCCCGTTTTCACCATGG3′ |
| For-GFPqPCR | 5′CCGTGCTGCTGCCCGACAA3′ |
Transfection of T. vaginalis cells.
T. vaginalis cultured cells were transfected by electroporation (28). Briefly, T. vaginalis cells were grown to a density of 1.5 ×106 cells/ml in TYM medium. The cells were collected and washed in cold Zimmerman solution [1.5 mM KH2PO4, 132 mM NaCl, 8 mM KCl, 8 mM Na2HPO4, 0.5 mM (CH3COO)2Mg, 90 mM (CH3COO)2Ca] and brought to 6 × 108 cells/ml. From this suspension, 400 μl of cells was mixed with 50 μl of plasmid DNA (1 μg/μl in Tris-EDTA [TE] buffer [10 mM Tris, 0.2 mM EDTA, pH 7.4]) and incubated for 5 min on ice. Mock-transfected cells with 50 μl of TE buffer were used as controls. Cells were electroporated at 360 V, 1,500 μF, and 13 Ω using a BTX600 Electro Cell Manipulator (BTX). Following electroporation, transfected cells were incubated in 30 ml of serum-enriched TYM medium at 37°C for 22 h. Extracts from transfected cells were prepared as described below and were used to measure the biochemical activity of CAT and to obtain the RNA poly(A)+ fraction for reverse transcription-PCR (RT-PCR) assays. Transfection experiments, isolation of RNA poly(A)+ fractions for RT-PCR analyses, and measurements of CAT activity were repeated at least three times for each plasmid construct. For all transfection experiments, both a positive control (the pcat-UAAA plasmid) and a negative control (cells transfected with TE buffer) were included. For the cotransfection experiments, 25 μg of each plasmid (pgfp-UAAA plasmid as the internal control and the pcat-derived tested plasmid) were used under the same conditions as described above. The highest CAT activity was consistently obtained in cell extracts at 22 h after transfection.
Analysis of cat transcripts from transfected T. vaginalis cells.
Trichomonads from 22 h after transfection were collected by centrifugation and washed with phosphate-buffered saline (PBS). Total RNA was prepared using TRIzol (Invitrogen). The RNA poly(A)+ fraction was isolated from total RNA using the QuickPrep Micro mRNA purification kit (Amersham Biosciences) according to the manufacturer's instructions. The 3′ rapid amplification of cDNA ends (3′ RACE) method was used to identify the 3′ end of the cat mRNA and, consequently, the polyadenylation site (10). Briefly, for the synthesis of cDNA, 500 ng of poly(A)+ RNA and 18 pmol of AP adapter primer (Table 2) were heat denatured and used for the reverse transcriptase reaction (1× First Strand buffer, 0.5 mM each deoxynucleoside triphosphate [dNTP], 5 mM dithiothreitol [DTT], 100 units of SuperScript III reverse transcriptase [Invitrogen]). The reaction was carried out for 50 min at 42°C, followed by heat inactivation of the enzyme and degradation of the RNA with RNase H. The amplification of cat cDNA 3′ ends from the total cDNA pool was performed using the CAT10 primer and the AUAP-B adapter primer (Table 2) in a 100-μl PCR mixture (40 pmol of each primer, 1× PCR buffer, 0.6 mM each dNTP, 3 mM MgCl2, 5 U of Taq DNA polymerase [Invitrogen]). The amplification reaction consisted of 30 cycles of denaturing at 94°C for 1 min, annealing at 45°C for 2 min, and extension at 72°C for 3 min, followed by a last extension cycle of 5 min at 72°C. To increase the specificity of the amplicons, a second (seminested) PCR amplification was performed on the initial PCR product using the CAT11 primer and the AUAP-B adapter primer (Table 2) under similar conditions. PCR products were cloned in the Zero Blunt TOPO PCR cloning system (Invitrogen) and sequenced. Plasmid preparations and other common molecular biology techniques were performed essentially as previously described (23).
The relative homogeneity of cat mRNA was estimated by qualitative RT-PCR from cotransfected cells. The cDNA was synthesized from total RNA (5 μg) and 18 pmol of AP adapter primer, heat denatured, and used for the reverse transcriptase reaction (1× First Strand buffer, 0.5 mM each dNTP, 5 mM DTT, 100 units of SuperScript III reverse transcriptase [Invitrogen]). The reaction was carried out for 50 min at 42°C, followed by heat inactivation of the enzyme and degradation of the RNA with RNase H. The cDNA was digested overnight with appropriate restriction endonucleases to remove any possible contaminating DNA. The cDNA was extracted with phenol-chloroform (1:1) and precipitated overnight at −20°C with 2-propanol, 10 μg of tRNA, and 0.3 M sodium acetate. The cDNA was suspended in water. A multiplex PCR was used to simultaneously amplify cat and green fluorescent protein (GFP) cDNA 3′ ends from the total cDNA pool using the For-CATqPCR primer, For-GFPqPCR, and the AUAP-B adapter primer (Table 2). PCRs were performed in 25 μl (20 pmol of each primer, 1× PCR Buffer, 0.6 mM each dNTP, 3 mM MgCl2, 1.25 U of Taq DNA polymerase [Invitrogen]) and consisted of 16, 20, 24, and 30 cycles of denaturing at 92°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 2 min, followed by a last extension cycle of 10 min at 72°C. Five microliters of the PCR products was electrophoresed in 5% polyacrylamide gels stained with ethidium bromide and visualized by UV light. The size and homogeneity of the amplified products were evaluated with this approach. As an additional way to determine the sites where the analyzed mRNAs were polyadenylated, these PCR products were cloned in the pGEM-T Easy vector system (Promega) and some of the clones were sequenced.
Preparation of RNA probes by in vitro transcription.
Single-stranded RNA probes (121 bases long) were used for RNA gel mobility shift assays. These RNA probes were synthesized by in vitro transcription and contained 39 nt upstream and 79 nt downstream of the cat stop codon. The DNA templates for the transcription were PCR products that were amplified from the pcat-UAAA and the pcat-GCCC plasmids with the For-invit and Rev-invit primers (Table 2). The forward primer includes the T7 RNA polymerase promoter sequence (TAATACGACTCACTATAGG) for in vitro transcription. The PCR products were cloned in the Zero Blunt TOPO cloning system (Invitrogen), and plasmids from single colonies were sequenced. The inserts were purified from the TOPO clones and were used as templates for in vitro RNA synthesis using T7 RNA polymerase with the MAXIscript kit (Ambion), following the manufacturer's instructions. The transcription reaction mixture was incubated at 37°C for 10 min and included 20 μCi [α-32P]UTP (3,000 Ci/mmol; Perkin-Elmer). The RNA was precipitated with 2-propanol and dissolved in nuclease-free water. The size and integrity of the transcripts were verified on a 4% polyacrylamide–1× Tris-borate-EDTA (TBE)–8 M urea gel.
RNA mobility shift assays.
T. vaginalis proteins were prepared essentially as described for the binding assays (16, 18, 19) with minor modifications as follows. T. vaginalis cells were grown to a density of 1.5 ×106 cells/ml in TYM medium. The cells (4 × 109 cells/ml) were collected by centrifugation, washed two times with PBS, suspended in 4 packed cell volumes (PCV) of hypotonic buffer (10 mM HEPES-KOH [pH 7.9], 1 mM EDTA, 5 mM DTT, 0.02 mM leupeptin, 0.2 mM Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK], 1 mM EGTA, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and incubated for 20 min on ice. Cells were gently lysed with a Dounce homogenizer, 4 PCV of lysis buffer (50 mM HEPES-KOH [pH 7.9], 10 mM MgCl2, 2 mM DTT, 25% sucrose, 50% glycerol, 0.02 mM leupeptin, 0.2 mM TLCK, 1 mM PMSF) was added, 1 PCV of 4 M ammonium sulfate was slowly added with gentle stirring, and the solution was stirred for another 30 min. The cell lysate was centrifuged for 3 h at 190,000 × g. Proteins from the supernatant were precipitated with 60% ammonium sulfate and were collected by centrifugation for 20 min at 10,000 × g. Proteins were dissolved in 5% of the original volume of F buffer (25 mM HEPES-KOH [pH 7.9], 75 mM KCl, 12 mM MgCl2, 0.25 mM EDTA, 2.5 mM DTT, 10% glycerol, 10 μg/ml leupeptin, 50 μg/ml TLCK, 0.2 mM EGTA, 1 mM PMSF) and were dialyzed overnight with 100 volumes of F buffer. A sample from the protein extracts was analyzed by electrophoresis and quantified by the Bradford method (3). Aliquots were frozen with dry ice-ethanol and stored at −70°C. Binding reaction mixtures contained 5 μg of T. vaginalis protein extract, 5,000 cpm of RNA labeled probe, 1.6 mM ATP, 20 mM phosphocreatine, 0.5 mM MgCl2, 10 mM HEPES-KOH (pH 7.9), 50 mM KCl, 0.05 mM EDTA, 2 mM DTT, 0.1 mg/ml tRNA, 40 U RNase inhibitor (RNase Out; Invitrogen), and 10% glycerol in a final volume of 30 μl. The reaction mixtures were incubated for 10 min at 4°C followed by 10 min at 30°C. Samples were heated prior to loading on a 4% native polyacrylamide 0.5× TBE gel. Gels were dried, and the bands were detected by autoradiography. The assays were independently repeated at least three times with similar results.
Evaluation of CAT activity.
Protein extracts were prepared from 15 ml of the transfected T. vaginalis cultures described above. Cells were collected by centrifugation, washed twice in 1 ml TEN buffer (40 mM Tris-HCl [pH 8], 1 mM EDTA [pH 8], 15 mM NaCl) and resuspended in 75 μl of 25 mM Tris-HCl (pH 8). Cells were lysed by freezing in liquid nitrogen followed by thawing at 37°C in a water bath three times, followed by 10 min of incubation at 65°C. The soluble material was recovered from the supernatant of a 10-min centrifugation at 14,000 rpm and 4°C in a microcentrifuge. The supernatant was used to test for the enzymatic activity of the reporter protein (chloramphenicol acetyltransferase [CAT]). The protein concentration in the extracts was determined colorimetrically by the Bradford method (3) with the aid of a Bio-Rad protein assay solution. The reactions to measure the CAT activity were carried out with 5 μl of the extracted cellular protein fraction, which was incubated for 2.5 h at 37°C in the presence of 0.2 μCi [3H]chloramphenicol (New England Nuclear) and 250 μM N-butyryl coenzyme A in a final volume of 100 μl, as previously described (25). Thereafter, the samples were extracted with xylene, and the radioactivity in the xylene phase was measured with a liquid scintillation counter. The radioactivity values were adjusted for the protein concentration. For comparison among experiments, the CAT activity is reported as a percentage of activity compared to that of the positive control. Transfection assays, followed by the determination of CAT activity, were independently repeated at least four times for each plasmid construct, with consistent results.
RESULTS AND DISCUSSION
Presence of the UAAA motif in the 3′ UTRs of T. vaginalis mRNAs and expressed genes.
Based on the sequence comparison of the 3′ UTRs from 13 T. vaginalis cDNA clones, the UAAA tetranucleotide was previously proposed as the potential polyadenylation signal in this organism (9). Since then, many more mRNA sequences have been made available, as is the case for EST database and the complete T. vaginalis genome (4), which allowed for an extended bioinformatics search of this sequence element. For this analysis we considered that if UAAA is the polyadenylation signal in T. vaginalis, then this motif should be present in all the mRNA 3′ UTRs.
In a first approach, we searched for the UAAA motif in 67 T. vaginalis cDNA sequences accessed in the NCBI database. These sequences show the site where the poly(A) tail is added to the mRNA. It was found that all the cDNAs analyzed have a UAAA tetranucleotide in the 3′ UTR within 11 to 30 nucleotides upstream from the poly(A) tail. Interestingly, in about one-third of the cDNAs studied, the UAAA overlapped the UAA stop codon. A particular case is that of the T. vaginalis syntaxin 16/TLG2-like protein cDNA (accession number AY344235), where the UAAA was not found in the short (21-nt) 3′ UTR sequence; nevertheless, this motif is comprised within the last two codons preceding the stop codon (UCU AAA UAA UUU). A sequence logo diagram (http://weblogo.berkeley.edu/logo.cgi) was generated for the UAAA motifs and the adjacent bases in the 67 cDNA sequences; it shows that no additional bases are conserved apart from this tetranucleotide, and therefore there is no preference for penta- or hexanucleotide variants. This alignment also revealed a preference for Y↓(A)2-5UU as the cleavage site and the presence of a downstream U-rich region (Fig. 1). These findings are in good agreement with our previous proposal (9).
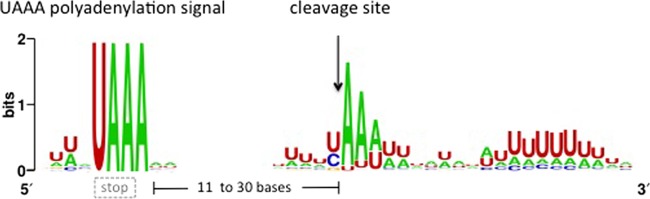
Fig 1.
Sequence logo obtained by alignment of the 3′ UTRs from 67 cDNAs and their corresponding genes. The overall height of the stack indicates the sequence conservation at that position, while the height of symbols within the stack indicates the relative frequency of each nucleotide at that position. The dashed gray box indicates that the UAAA motif may overlap the UAA translation stop codon.
As a second genomic approach, a similar presumption was made: the UAAA element should be present in essentially all T. vaginalis genes that are expressed, as inferred from the trichdb EST database (http://www.trichdb.org/trichdb/). However, for most of the genes that are annotated here, the polyadenylation site has not been experimentally determined. We therefore considered that in these genes the UAAA motif should be present in the 3′ intergenic region (including the stop codon). A total of 780 T. vaginalis genes with a 100% match with an EST were selected for analysis, and it was found that 99.5% of them do possess a UAAA motif either overlapping or downstream from the translation stop codon; Table 3 shows the position of the first tetranucleotide found in the 3′ intergenic region. UAA is by far the most common stop codon found in T. vaginalis; it is used in 695 (89%) of the analyzed genes, and among these, 455 are followed by an A, thus forming a UAAA motif that might be used as the polyadenylation signal. When this is not the case or when UAG or UGA is used as a stop codon, one or more UAAA motifs can be found nearby. Again, no preference for pentanucleotides or hexanucleotides was found. This finding agrees with the known observation that T. vaginalis mRNAs have short 3′ UTR sequences (29). It is important to point out that in this organism the single occurrence of a UAAA tetranucleotide is not sufficient to direct polyadenylation in a precise manner and that additional downstream sequences appear to be important for this process (see below). Relative to these findings, a short polyadenylation signal has been identified in Spironucleous salmonicida, a diplomonad that infects salmon. The position of the poly(A) tail in relation to the termination codon was mapped in 134 EST clones, revealing very short 3′ UTRs. The bioinformatics analyses did not identify any strong consensus sequence outside the conserved unique termination codon UGA, which is also the only motif conserved as a polyadenylation signal. Furthermore, a preference for a U-rich region close to the poly(A) tail is also apparent (2). Similar very short 3′ UTRs have also been observed in Giardia lamblia, where AGURAAY has been identified as a consensus polyadenylation signal. Interestingly, the three most conserved bases are the central URA nucleotides, which may overlap the translation stop codons of Giardia mRNAs (1, 21).
Table 3.
Position of the first UAAA motif in the 3′ intergenic regions of 780 T. vaginalis expressed genes
| Position of UAAA motif downstream from stop codon | % of genes |
|---|---|
| Overlapping UAA stop codon | 58.3 |
| 1–25 bases | 18.5 |
| 26–50 bases | 14.8 |
| 51–100 bases | 5.5 |
| 101–400 bases | 2.4 |
Experimental system designed to analyze polyadenylation sequences.
As an approach to dissect the components of the proposed UAAA motif, a transient-expression vector was designed based on a bacterial coding sequence (cat) flanked by the 5′ and 3′ noncoding regions of a T. vaginalis actin gene known to be expressed. The 5′ region contains the consensus sequence for the initiation of mRNA transcription (13), and the 3′ region contains the UAA stop codon and 762 bp downstream (Fig. 2A). For the construction of the parental pcat-UAAA expression vector, the UAG stop codon was used for the cat reporter gene, followed by the 3′ region of the actin gene (Fig. 2B). This strategy was used to separate the cat stop codon from the UAAA motif, which originally overlapped the actin UAA stop codon. From the pcat-UAAA parental plasmid, several derivatives were constructed by site-directed mutagenesis and are referred to here as pcat vectors. The purpose of these constructs was to define and delimit the role of the UAAA motif in the polyadenylation process by evaluating the effects of (i) the change of the four nucleotides, (ii) the single mutation of each of the four bases, (iii) the specific mutation of either the upstream or the downstream nucleotide adjacent to the UAAA motif, (iv) the insertion of two adenosines to introduce the consensus metazoan AAUAAA polyadenylation signal, and (v) a revertant construct (UCAA to UAAA) to recover the original UAAA motif. These plasmids are listed and described in Table 1. The ability of the UAAA motif to direct polyadenylation was evaluated by identifying the polyadenylation sites of the cat transcripts derived from the pcat vectors.
Four phenotypes associated with changes to the UAAA motif were additionally analyzed by (i) evaluating the cat mRNA abundance and size homogeneity relative to a second transcriptional reporter, (ii) repositioning the UAAA motif further downstream from the intergenic region and evaluating the effect of its relocation upon mRNA polyadenylation, (iii) assessing the ability of an RNA fragment (containing the UAAA motif) to interact with a protein extract from T. vaginalis, and (iv) estimating the levels of CAT activity in cells transfected with the different pcat vectors.
Identification of the polyadenylation site in the reporter cat mRNA.
The correlation between the UAAA motif and the selection of the polyadenylation site of the reporter cat mRNA was evaluated by cloning and sequencing 22 cat cDNA clones derived from the parental construct pcat-UAAA. These clones were readily obtained, and in all cases polyadenylation occurred 22 nt downstream the UAAA motif, at the same position as the endogenous actin mRNA from which the reporter vector was derived (Fig. 3A). The processing site occurred after the C in the CAAAAUU sequence, which is one of the proposed cleavage sites in T. vaginalis [Y↓(A)2-5UU].
In contrast, any change within the UAAA motif altered the polyadenylation site of the cat mRNA, as shown in the schematic representation in Fig. 3B. The identified processing sites are indicated in the genomic sequence in Fig. 3D. Transcripts were found to be polyadenylated at several positions downstream from the parental site, giving rise to mRNAs with longer and heterogeneous 3′ UTRs. These cDNA clones were difficult to obtain, likely because of the scarcity of poly(A) transcripts derived from these mutant constructs. The occurrence of potential nonpolyadenylated transcripts cannot be ruled out, but these would escape our detection system based on oligo(dT) initial priming. The effect of changing any of the four nucleotides is similar, with no apparent correlation between the mutated position and the site of polyadenylation. It is important to point out that several UAAA motifs are found in the 3′ noncoding region, downstream from the mutated one, but none is able to unambiguously direct polyadenylation to a single site in the mutant mRNAs. However, more cDNA clones (16 out of 22) were found to be polyadenylated close to and downstream from UAAA motifs than otherwise. Particularly, one cleavage site seemed to be preferred, with seven cDNA clones identified. This site is in good agreement with the proposed cleavage site Y↓(A)2-5UU for T. vaginalis (9). Nonetheless, the proposed U-rich region downstream from the processing site is not present (Fig. 3D).
The function of the UAAA motif is restricted to the tetranucleotide, since changing the contiguous bases showed no difference in the polyadenylation site selection compared to that of the parental control. This was also the case for the pcat-AAUAAA plasmid, which contained the consensus metazoan polyadenylation signal (AAUAAA) (Fig. 3C). This result was expected since the UAAA motif is contained within.
Finally, to corroborate that changes in the polyadenylation site of the transcripts derived from the mutant plasmids were directly related to the UAAA motif, a single-nucleotide revertant plasmid was constructed from the mutant pcat-UCAA plasmid to reconstruct the original UAAA sequence (pcat-revUAAA). This revertant plasmid completely recovered the original phenotype (Fig. 3C).
The above results suggest that although the UAAA motif plays an essential role in defining the polyadenylation site, additional sequences must be involved that help to identify the site for precise and efficient mRNA processing. These sequences could be the cleavage site and a downstream U-rich region.
Evaluation of cat mRNA size homogeneity.
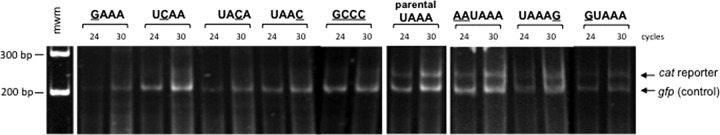
The size homogeneity of cat mRNAs in the transfected cells was evaluated relative to a cotransfected nonrelated gene as a control, transcribed from plasmid pgfp-UAAA. This plasmid is similar to the parental pcat-UAAA, except that the gfp gene was cloned in place of the cat gene (Fig. 2C). Qualitative RT-PCR was used in a multiplex reaction where the 3′ ends of cat and gfp cDNAs were specifically amplified from the total cDNA pool [oligo(dT)-derived]. Since the expected sizes of individual PCR products differ by 35 bp, it was then possible to analyze the size homogeneity and estimate the relative abundance of cat and control gfp mRNAs directly from acrylamide gels. The identity of the amplified products was corroborated by Southern hybridization (data not shown). In all cases the gfp RT-PCR amplification band obtained from the cotransfected control plasmid is well defined (Fig. 4). Also, cat mRNA is expressed as an apparently homogeneous population of transcripts when it is derived from the parental pcat-UAAA vector, as deduced from a well-defined RT-PCR amplification band (Fig. 4, parental UAAA). This is also the case for transcripts derived from all mutant plasmids where the UAAA motif is present (Fig. 4, AAUAAA, UAAAG, and GUAAA). In contrast, cat RT-PCR products derived from plasmids without the UAAA motif appear heterogeneous in size as observed from the blurred amplification products (Fig. 4, GAAA, UCAA, UACA, UAAC, and GCCC). These ill-defined products would reflect differences in the 3′ UTR sizes of mutant mRNAs, which is in good agreement with the variability in the polyadenylation sites of the mutants described earlier (Fig. 2D). In addition, the apparent scarcity of these mRNAs may be due to the presence of nonpolyadenylated transcripts that would not be amplified and would escape our detection system. This result shows that a single change in the UAAA tetranucleotide causes an apparent decrease in the relative abundances of the derived mRNAs, and it reflects the poorly defined site for processing and polyadenylation of these transcripts.
Fig 4.
Size homogeneity of cat mRNAs. T. vaginalis cells were cotransfected with the different pcat plasmids and the pgfp-UAAA plasmid, which was used as an internal control for transfection and transcription. An RT-PCR approach was used, and samples from 24 and 30 cycles of amplification are shown. Amplification products were electrophoresed in a 5% acrylamide gel and stained with ethidium bromide. The lower band of about 200 bp corresponds to the RT-PCR product derived from the gfp mRNA control. This band migrates at the expected size, and it is constant and well defined throughout all the assays. The upper band of about 235 bp corresponds to the RT-PCR product derived from the cat mRNAs. This band is well defined for the cases where the transfected plasmids contain the UAAA motif (parental UAAA, AAUAAA, UAAAG, and GUAAA). RT-PCR products derived from plasmids without the UAAA motif appear as ill-defined bands compared with the gfp control in each lane, thus suggesting mRNAs of heterogeneous size and low abundance (GAAA, UCAA, UACA, UAAC, and GCCC). mwm, molecular size markers.
Repositioning of the UAAA motif relocates the polyadenylation site of transcripts.
To further support the directing role of the UAAA motif in polyadenylation, derivative plasmids in which the UAAA was relocated either 74 or 118 nucleotides downstream from the cat stop codon in the mutant pcat-UCAA plasmid were constructed (Fig. 5A). These two sites were chosen since runs of several Us were present but the UAAA motif was not found close by. The plasmids (pcat-UAAA74 and pcat-UAAA118) were transfected in T. vaginalis cells, and the derived mRNA was characterized by RT-PCR. The expected sizes of the amplified products were about 280 and 320 bp, respectively, if polyadenylation was directed by the inserted UAAA motifs. Figure 5B shows well-defined RT-PCR bands that correspond to the expected sizes, although very small differences in the amplification products would not be appreciated with this approach. By cDNA cloning and sequencing, we determined that mRNAs were processed downstream from the introduced UAAA motifs, but the new processing site was still not well defined since cleavage took place at several close-by positions (Fig. 5A). When the proposed (9) minimal cleavage motif (CAAUU) was introduced downstream the UAAA motif (plasmid pcat-UAAA118CS), all the analyzed clones were polyadenylated at the predicted cytosine (Fig. 5A and B).
It can therefore be concluded that although the UAAA motif is an essential sequence for polyadenylation, it is not enough to direct this process by itself and needs additional downstream sequences to attain its specificity. One of these signals would be the site for endonuclease cleavage (CAAUU and related sequences). Moreover, since the introduced motifs were located upstream from U-rich regions, these sequences might also aid in positioning the polyadenylation machinery, as has been proposed earlier (9, 17, 32). Experiments to further characterize these downstream elements are currently in progress in our laboratory.
RNA mobility shift assays support protein-RNA complex formation mediated by UAAA.
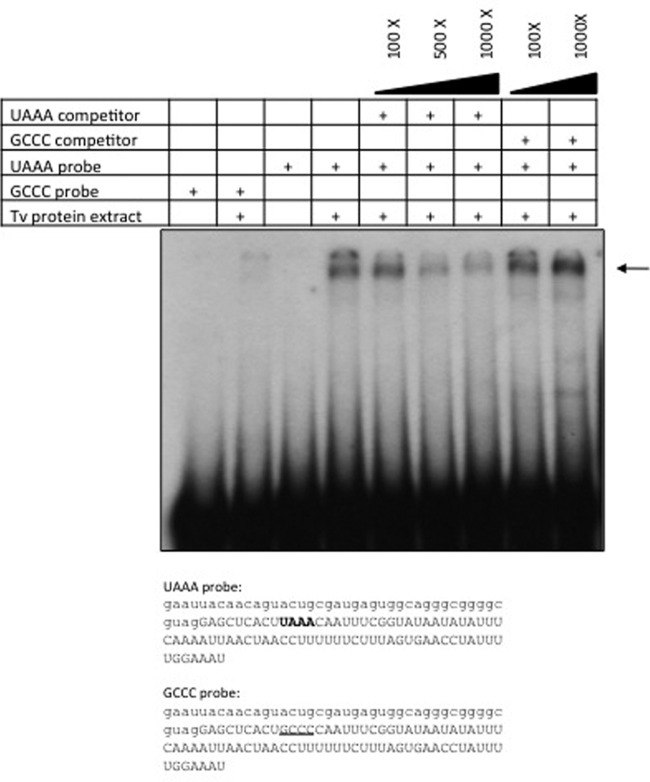
To determine whether the UAAA motif, in the context of the mRNA analyzed here, is recognized by proteins from T. vaginalis, the interaction with an RNA fragment was tested by electrophoretic mobility shift assay (EMSA). For this purpose, 121-nt RNA transcripts were used as probes. These consisted of the last 39 nt of the cat coding region and 79 nt downstream from the cat stop codon; the UAAA probe contained the original UAAA motif, while the GCCC mutant probe was identical except for a four-nucleotide change of UAAA to GCCC (Fig. 6). A high-molecular-weight complex is formed with the probe containing the UAAA motif, while a much less intense shift of the probe is detected with the mutant probe containing the GCCC motif. The finding that the GCCC probe is able to interact with T. vaginalis proteins to a much lesser extent is not surprising given that proposed additional cis elements that could be recognized by protein factors are present in this RNA fragment. The formation of the complex was partially competed with the UAAA unlabeled RNA fragment, whereas no competition was observed with the mutant GCCC competitor (Fig. 6). However, when single point mutations in the UAAA motif were analyzed, an apparent mobility shift profile similar to that for the parental probe was observed (not shown).
Fig 6.
UAAA-specific binding activity is present in T. vaginalis protein extracts. A 121-nt RNA probe (corresponding to positions −51 to +66 relative to the UAAA motif in the pcat-UAAA plasmid) was used in RNA mobility shift assays with 5 μg of T. vaginalis protein extracts. A similar RNA probe in which UAAA was replaced by GCCC was used as a mutant control. The sequences of both probes are shown. The cat coding region is denoted in lowercase letters, the 3′ downstream region is shown in uppercase, the UAAA motif is in bold, and the GCCC motif is underlined. Competition assays were done with 100×, 500×, and 1,000× molar ratios of unlabeled UAAA and GCCC probes. The complexes were analyzed on a 4% acrylamide gel. The arrow shows the migration of the complex.
Regarding the occurrence of potential orthologous proteins for the polyadenylation machinery, a comparative search was done in the T. vaginalis genome database. Several genes with more similarity to the mammalian than to the yeast counterparts were identified; among them were the ones coding for CPSF160, CstF, and symplekin. Of particular relevance is the CPSF160 subunit, since it recognizes the AAUAAA polyadenylation signal in metazoans. Taken together, the EMSAs and the identification of genes coding for putative polyadenylation proteins suggest that T. vaginalis possesses a polyadenylation machinery similar to the metazoan apparatus that can recognize the UAAA motif in a 3′ UTR context. These protein factors are a current topic of study in our laboratory.
Reduced CAT activity from plasmids lacking the UAAA motif.
As an indirect way to estimate the effect of changes in the polyadenylation process of the reporter transcript, the biochemical activity of CAT in the extracts of T. vaginalis cells that were transformed with the pcat vectors was evaluated. In all transfections, both a positive control (the plasmid pcat-UAAA) and a negative control (cells transfected with TE buffer) were included. Table 4 shows that any change to the UAAA motif resulted in a lower (15 to 20%) level of CAT activity than that in the UAAA control. In contrast, cells that were transformed with plasmids bearing changes in the bases adjacent to the UAAA motif or with the revertant plasmid showed CAT activity levels similar to those in cells transformed with the parental pcat-UAAA. The same was observed with the plasmid bearing the consensus metazoan polyadenylation signal (Table 4). Given that all pcat plasmids provide the same promoter and 5′ flanking sequence, we assume that transcription initiation is similar for all cat mRNA. The difference among transcripts would be the polyadenylation site, with aberrantly positioned poly(A) tails or nonpolyadenylated transcripts in the mutants within the UAAA motif. Our proposal is that these mutants would render fewer translatable mRNAs and, in turn, a lower abundance of CAT protein and lower measured activity in the transfected cell extracts.
Table 4.
CAT activities in T. vaginalis cells transfected with the pcat vectors
| Plasmida | Mean (SD) % CAT activity |
|---|---|
| pcat-UAAA | 100 |
| pcat-GCCC | 11.7 (4.05) |
| pcat-GAAA | 10.2 (8.5) |
| pcat-UCAA | 9.9 (3.1) |
| pcat-UACA | 10.3 (5.8) |
| pcat-UAAC | 14.5 (9.3) |
| pcat-GUAAA | 146.9 (7.9) |
| pcat-UAAAG | 101.6 (7.8) |
| pcat-AAUAAA | 127.4 (22.4) |
| pcat-revUAAA | 122.3 (31.5) |
The UAAA motif is in bold.
In conclusion, the above results show that the UAAA tetranucleotide is recognized as the signal that directs the polyadenylation of mRNA in T. vaginalis, with a role equivalent to that of the metazoan AAUAAA polyadenylation signal. Since the UAAA motif is contained within the AAUAAA hexanucleotide, we think that these two signals might be evolutionarily related.
Although UAAA is an apparently strict and essential element for defining the polyadenylation site of mRNA in T. vaginalis, it is not sufficient to direct this process by itself. Our data indicate that at least two additional elements (U-rich regions and a consensus cleavage site) enhance the specificity and direct the polyadenylation process in this early-divergent organism. Further work will address the identification of trans-acting factors of the polyadenylation complex in T. vaginalis.
ACKNOWLEDGMENTS
This work was supported by grant P45037-Q from Consejo Nacional de Ciencia y Tecnología (CONACYT), México, and grant IN220210 from Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT), Universidad Nacional Autónoma de México (UNAM). We acknowledge scholarships to Vanessa Fuentes and Guadalupe Barrera from CONACYT, México.
This work was done by Vanessa Fuentes in partial fulfillment of the requirements for a Ph.D. degree from Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México.
We thank Jessica Guzmán for initial work on the construction of the reporter vector and Patricia de la Torre for help with DNA sequencing.
Footnotes
Published ahead of print 30 March 2012
REFERENCES
- 1. Adam RD. 1991. The biology of Giardia spp. Microbiol. Rev. 55:706–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersson JO, et al. 2007. A genomic survey of the fish parasite Spironucleus salmonicida indicates genomic plasticity among diplomonads and significant lateral gene transfer in eukaryote genome evolution. BMC Genomics 8:51 doi:10.1186/1471-2164-8-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 4. Carlton JM, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colgan DF, Manley JL. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755–2766 [DOI] [PubMed] [Google Scholar]
- 6. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dacks JB, Walker G, Field MC. 2008. Implications of the new eukaryotic systematics for parasitologists. Parasitol. Int. 57:97–104 [DOI] [PubMed] [Google Scholar]
- 8. Diamond LS. 1957. The establishment of various trichomonads of animals and man in axenic cultures. J. Parasitol. 43:488–490 [PubMed] [Google Scholar]
- 9. Espinosa N, Hernandez R, Lopez-Griego L, Lopez-Villasenor I. 2002. Separable putative polyadenylation and cleavage motifs in Trichomonas vaginalis mRNAs. Gene 289:81–86 [DOI] [PubMed] [Google Scholar]
- 10. Frohman MA, Dush MK, Martin GR. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. U. S. A. 85:8998–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo Z, Sherman F. 1995. 3′-end-forming signals of yeast mRNA. Mol. Cell. Biol. 15:5983–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu J, Lutz CS, Wilusz J, Tian B. 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 11:1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liston DR, Lau AO, Ortiz D, Smale ST, Johnson PJ. 2001. Initiator recognition in a primitive eukaryote: IBP39, an initiator-binding protein from Trichomonas vaginalis. Mol. Cell. Biol. 21:7872–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liston DR, Johnson PJ. 1998. Gene transcription in Trichomonas vaginalis. Parasitol. Today 14:261–265 [DOI] [PubMed] [Google Scholar]
- 15. Lopez-Camarillo C, Orozco E, Marchat LA. 2005. Entamoeba histolytica: comparative genomics of the pre-mRNA 3′ end processing machinery. Exp. Parasitol. 110:184–190 [DOI] [PubMed] [Google Scholar]
- 16. Lovrien RE, Matulis D. 2001. Selective precipitation of proteins. Curr. Protoc. Protein Sci. 4:4.5. [DOI] [PubMed] [Google Scholar]
- 17. Mandel CR, Bai Y, Tong L. 2008. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 65:1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manley JL, Fire A, Cano A, Sharp PA, Gefter ML. 1980. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl. Acad. Sci. U. S. A. 77:3855–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manley JL, Fire A, Samuels M, Sharp PA. 1983. In vitro transcription: whole-cell extract. Methods Enzymol. 101:568–582 [DOI] [PubMed] [Google Scholar]
- 20. Millevoi S, Vagner S. 2010. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 38:2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morrison HG, et al. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–1926 [DOI] [PubMed] [Google Scholar]
- 22. Proudfoot NJ, Furger A, Dye MJ. 2002. Integrating mRNA processing with transcription. Cell 108:501–512 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seed B, Sheen JY. 1988. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene 67:271–277 [DOI] [PubMed] [Google Scholar]
- 26. Simoes-Barbosa A, Meloni D, Wohlschlegel JA, Konarska MM, Johnson PJ. 2008. Spliceosomal snRNAs in the unicellular eukaryote Trichomonas vaginalis are structurally conserved but lack a 5′-cap structure. RNA 14:1617–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sogin ML. 1991. Early evolution and the origin of eukaryotes. Curr. Opin. Genet. Dev. 1:457–463 [DOI] [PubMed] [Google Scholar]
- 28. Tsai CD, Liu HW, Tai JH. 2002. Characterization of an iron-responsive promoter in the protozoan pathogen Trichomonas vaginalis. J. Biol. Chem. 277:5153–5162 [DOI] [PubMed] [Google Scholar]
- 29. Vanacova S, Liston DR, Tachezy J, Johnson PJ. 2003. Molecular biology of the amitochondriate parasites, Giardia intestinalis, Entamoeba histolytica and Trichomonas vaginalis. Int. J. Parasitol. 33:235–255 [DOI] [PubMed] [Google Scholar]
- 30. Vanacova S, Yan W, Carlton JM, Johnson PJ. 2005. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. U. S. A. 102:4430–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Venkataraman K, Brown KM, Gilmartin GM. 2005. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 19:1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wahle E, Keller W. 1996. The biochemistry of polyadenylation. Trends Biochem. Sci. 21:247–250 [PubMed] [Google Scholar]
- 33. Wahle E, Ruegsegger U. 1999. 3′-end processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 23:277–295 [DOI] [PubMed] [Google Scholar]
- 34. World Health Organization 2001. Global prevalence and incidence of selected curable sexually transmitted infections. World Health Organization, Geneva, Switzerland [Google Scholar]