Abstract
The regulation of Ace2 and morphogenesis (RAM) network is a protein kinase signaling pathway conserved among eukaryotes from yeasts to humans. Among fungi, the RAM network has been most extensively studied in the model yeast Saccharomyces cerevisiae and has been shown to regulate a range of cellular processes, including daughter cell-specific gene expression, cell cycle regulation, cell separation, mating, polarized growth, maintenance of cell wall integrity, and stress signaling. Increasing numbers of recent studies on the role of the RAM network in pathogenic fungal species have revealed that this network also plays an important role in the biology and pathogenesis of these organisms. In addition to providing a brief overview of the RAM network in S. cerevisiae, we summarize recent developments in the understanding of RAM network function in the human fungal pathogens Candida albicans, Candida glabrata, Cryptococcus neoformans, Aspergillus fumigatus, and Pneumocystis spp.
INTRODUCTION
A wide variety of key physiological processes in eukaryotic cells are regulated by networks of protein kinase-based signaling pathways. Accordingly, the study of these pathways has been the focus of intense research efforts in nearly every area of eukaryotic cell biology. Because a large number of protein kinase signaling pathways and cascades are highly conserved between model organisms and humans, our current understanding of many mechanistic paradigms for protein kinase-based cell signaling has developed from studies of model organisms, such as the budding yeast Saccharomyces cerevisiae (10). These studies not only have contributed to our fundamental understanding of eukaryotic biology but also have had profound biomedical implications (28). For example, protein kinases are frequently dysregulated in cancer cells, and consequently, protein kinase inhibitors such as imatinib have emerged as important components of current anticancer therapies (19).
A second area of eukaryotic biology with important biomedical relevance that has benefited from S. cerevisiae-based studies of protein kinase signaling pathways is the study of pathogenic fungi (26, 45). The ability of modern medicine to support patients with compromised immune function and the development of immunomodulatory therapies have contributed to an increase in the morbidity and mortality attributable to invasive fungal infections (14). As the medical importance of pathogenic fungi has increased, so too has interest in the fundamental biology of these organisms as well as the mechanistic basis of their pathogenesis (68). As a result, the study of protein kinase signaling networks has led to many important insights into fungal virulence, host-pathogen interactions, and, more recently, new approaches to antifungal therapy (1).
An excellent example of a signaling pathway that is highly conserved within all eukaryotes, is well-studied in S. cerevisiae, and is currently being characterized in pathogenic fungi is the regulation of Ace2 and morphogenesis (RAM) network (65). In this minireview, we summarize recent developments in our understanding of the RAM pathway in the human fungal pathogens. As part of this discussion, we also provide a brief overview of S. cerevisiae-based studies; however, a complete review of the S. cerevisiae RAM pathway literature, as well as that of other important model fungi such as S. pombe and N. crassa, is beyond the scope of this minireview; key references to this work are included where appropriate.
OVERVIEW OF RAM PATHWAY
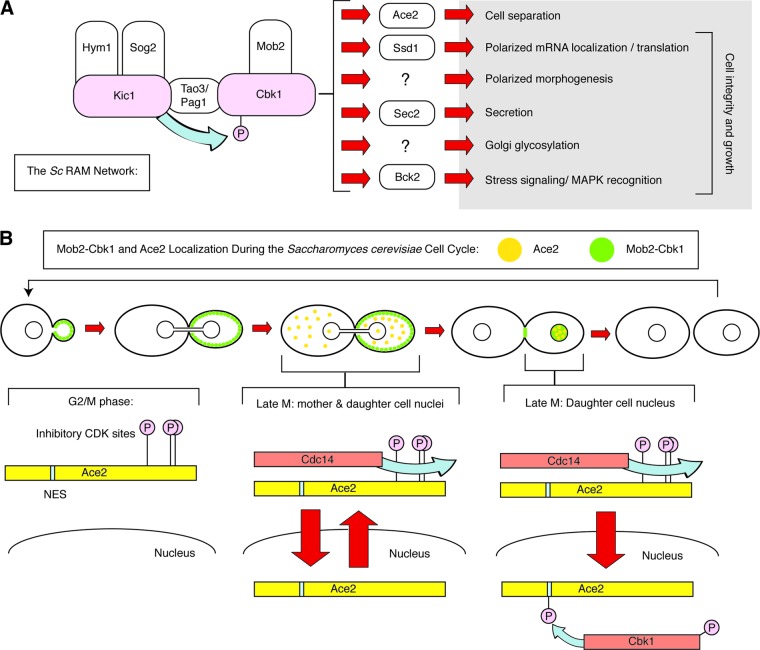
The RAM network has been most extensively studied in S. cerevisiae and has been implicated in the regulation of a number of cellular processes, including mother-daughter cell separation (3, 18, 65, 69), daughter cell-specific gene transcription (12, 81), maintenance of cell wall integrity (18, 50, 77), cell cycle progression (4, 7, 70), mating projection formation and mating efficiency (3, 5, 65, 81), polarized growth (18, 65, 69, 81), and stress signaling (47). As detailed in this review, many of the functions of the RAM pathway characterized in S. cerevisiae are conserved in other model and pathogenic fungi. As an introduction to the components and functions of the RAM pathway, we first summarize the general concepts of RAM function that have emerged from S. cerevisiae-based studies (Fig. 1A).
Fig 1.
Overview of the S. cerevisiae RAM pathway and model of ScAce2 regulation. (A) Schematic of the components of the S. cerevisiae RAM pathway, indicating that ScKic1 may phosphorylate and regulate the key kinase ScCbk1. The functions of the RAM pathway are listed, along with candidate or confirmed substrates involved in each respective function. (B) Model for the mechanism by which the RAM pathway and cyclin-dependent kinases regulate the daughter cell-specific localization and activation of ScAce2.
S. cerevisiae RAM network.
The RAM signaling network is essential for viability (65) and is comprised of two serine/threonine protein kinases, ScCbk1 and ScKic1, and four associated proteins, ScMob2, ScTao3/Pag1, ScHym1, and ScSog2 (Fig. 1A). ScCbk1 is the terminal kinase in the pathway and belongs to the Ndr/LATS subfamily of AGC kinases that function as tumor suppressors in metazoans (29). ScCbk1 is more closely related to Ndr kinases, which play a role in the polarized morphogenesis of a variety of cell types (21, 29, 30, 31). ScMob2 directly binds to, and colocalizes with, ScCbk1 throughout the cell cycle and appears to function as a ScCbk1-regulatory subunit (12, 81). Genetic and biochemical data suggest that ScKic1, a member of the germinal center (GC) kinase family, directly activates ScMob2-Cbk1 by phosphorylating ScCbk1 within the terminal hydrophobic domain (58, 65). Both ScHym1, which is related to mammalian MO25 (63), and the LRR-containing protein ScSog2, a protein apparently unique to yeasts, form a complex with ScKic1 and may facilitate ScKic1-mediated phosphorylation of ScCbk1 (3, 18, 65). ScTao3, also known as ScPag1, is a conserved 270-kDa protein related to Furry (Fry) protein (21) which has been shown to interact with both ScCbk1 and ScKic1 (immunoprecipitation and two-hybrid assays) and may facilitate ScCbk1 activation (18, 65).
Much attention has been devoted to elucidating the function and regulation of ScCbk1, the terminal kinase in the S. cerevisiae RAM network. Genetic and biochemical experiments indicate that ScCbk1 kinase activity and function are dependent on each of the other RAM proteins as well as phosphorylation at two sites that are highly conserved among Ndr/Lats family kinases (5, 38, 65, 67). The details and consequences of ScCbk1 phosphorylation have been reviewed elsewhere and are briefly summarized here (24, 56). Similar to other Ndr kinases, ScCbk1 is phosphorylated in the kinase activation loop (T loop) via an intramolecular, autophosphorylation and in the C-terminal hydrophobic motif (HM) at T743 (7, 38, 62, 75). ScCbk1 T-loop autophosphorylation is important for kinase activity but does not significantly impair ScCbk1 function in vivo (7, 38, 67). In contrast, ScCbk1 HM phosphorylation is required for polarized growth and asymmetric localization of ScCbk1 as well its substrate ScAce2 to daughter cell nuclei (38, 67). ScKic1-related GC kinases have been shown to phosphorylate the HM domain of Ndr kinases in other cell types (24, 31, 56). Consistent with these findings, genetic and in vitro experiments strongly suggest that ScKic1 phosphorylates ScCbk1 on T743 (7, 65, 67).
The S. cerevisiae RAM network is implicated in the regulation of many cellular functions. This function is reflected in the subcellular localization of the RAM network components as well as in the ScCbk1 substrates that have been identified to date. Notably, ScCbk1 and ScMob1 localize to sites of polarized growth as well as to the daughter cell nucleus at the end of mitosis, reflecting roles in both polarized growth and daughter cell-specific gene expression. Like other Ndr/Lats kinases (27, 59), ScCbk1 has been shown to phosphorylate substrates on similar consensus sites (HXRXX[S/T] and RXX[S/T×) (38). However, the physiologic substrates of ScCbk1 are only beginning to be identified. To date, only two ScCbk1 substrates have been well characterized: the zinc finger transcription factor ScAce2 (8, 15) and the mRNA binding protein ScSsd1 (38, 49, 50). There is additional evidence suggesting that ScCbk1 may also regulate several other proteins, including the mRNA-binding protein ScPuf5/Mpt5 (5), the RAB GEF ScSec2 (51), and the cell wall integrity (CWI) pathway MAPKK ScBck2 (47).
In the sections below, we summarize the major cellular functions of the RAM pathway in S. cerevisiae and pathogenic fungi. However, it is important to note that the RAM pathway has also been studied in a number of other model fungi. Indeed, the founding member of the fungal Ndr kinases, COT1, was identified in Neurospora crassa and has been extensively characterized (55, 56, 57, 83). Similarly, the Ndr kinase ortholog in Schizosaccharomyces pombe, Orb6, has also been the subject of intensive study, as have the other genes (Pmo25, Nak1, Mob2, and Mor2) that constitute the morphogenesis Orb6 network (MOR), the S. pombe ortholog of the RAM network (13, 24, 32, 35, 36, 41, 61). Aspergillus nidulans CotA (the Ndr ortholog of ScCbk1) and MobB (the ortholog of ScMob2) have also been identified (72). Consistent with their functions in S. cerevisiae and the pathogenic fungi discussed below, these networks and genes all play roles in polarized growth and morphogenesis (57). Due to space limitations, however, a full discussion of this work is beyond the scope of this review.
ScAce2 regulation.
The best-characterized function of ScCbk1 and RAM is the phosphorylation and regulation of the zinc finger transcription factor ScAce2. ScAce2 was the first protein in this network to be described and is a paralog of ScSwi5 (8). ScAce2 is a daughter cell-specific transcription factor that is expressed during G2 phase of the cell cycle (15, 16, 70) and regulates the expression of genes such as the chitinase ScCTS1 (43) and the putative glucanase ScSCW11, which is involved in septum degradation and cell separation during mitotic exit (7, 8, 15, 16). In addition to cell separation genes, ScAce2 regulates cell wall genes and coregulates a subset of genes with ScSwi5, including the Cdk1 inhibitor ScSIC1 (15, 16).
Although ScAce2 is not essential in any fungus studied to date, deletion of ScACE2 leads to a dramatic defect in cell separation that is characterized by large clumps of cells that do not disperse when ultrasonicated (15). Consistent with the role of ScAce2 in orchestrating the degradation of the chitin-rich septum, the cells of Scace2Δ mutants remain attached at the chitinous septa (15). This phenotype is shared with all of the RAM pathway mutants in S. cerevisiae (65) and, as described below, is also observed in many pathogenic fungi containing mutations in the respective ACE2 orthologs. Deletion of ScACE2 in S. cerevisiae strain backgrounds that display filamentation and invasive growth (e.g., Σ1278b strain background) generates mutants that grow as rough, raised colonies on solid agar and produce high levels of pseudohyphae relative to wild-type strains (43); again, this phenotype is shared with ace2 mutants in the pathogenic yeast C. albicans.
An elegant series of recent studies has shed considerable light on the mechanism by which the mitotic exit network (MEN) controls ScCbk1 and ScAce2 function and, in turn, regulates daughter cell separation (7, 58, 59). As outlined in Fig. 1B, ScAce2 nuclear import is blocked by CDK-dependent phosphorylation until the metaphase-to-anaphase transition (58). The MEN-dependent release of ScCdc14 phosphatase is required to dephosphorylate the inhibitory CDK1 phosphorylation sites on ScAce2, leading to its nuclear import (6, 12, 58, 59, 65, 66, 70). ScCbk1 regulates the nuclear localization of ScAce2 by phosphorylating near a nuclear export sequence in ScAce2. This phosphorylation occurs specifically in daughter cell nuclei and blocks ScAce2 export, leading to its accumulation in the daughter cell nuclei (6, 59, 72). In the absence of S. cerevisiae RAM network signaling, ScAce2 fails to asymmetrically localize to daughter cell nuclei and does not promote the expression of cell separation genes (12, 65, 81). Thus, the asymmetric localization of ScCbk1 also controls the asymmetric localization and function of ScAce2.
ScCbk1 function and asymmetric ScAce2 localization are also dependent on phosphorylation of the ScCbk1 HM motif (67). Phosphorylation of the HM domain of ScCbk1 is dependent on the FEAR (Cdc-fourteen early anaphase release) pathway (7). Nevertheless, ScCbk1 HM phosphorylation alone does not appear to be sufficient for ScAce2 activation in vivo, because ScCbk1-mediated ScAce2 phosphorylation also requires MEN-dependent ScCdc14 phosphatase release. Thus, the asymmetric localization and activity of ScAce2 require the sequential interplay between CDK, FEAR, MEN, and RAM signaling pathways. This interplay is also observed in other fungi (24, 29).
ScSsd1 regulation.
Recent data indicate that the mRNA-binding protein ScSsd1 is also an important ScCbk1 substrate. ScSsd1 has been implicated in cell wall integrity (82) and mRNA processing (33, 37). The functional link between ScCbk1 and ScSsd1 was first implied by two-hybrid and genetic interactions (18, 39). Notably, the cell integrity and lysis defects of S. cerevisiae RAM pathway mutants are suppressed by loss-of-function alleles of ScSSD1 (18, 39). As described below, the mechanistic details underlying this genetic observation have begun to emerge recently.
ScSsd1 associates with a subset of mRNAs involved in cell wall biosynthesis and cell growth, including the transcripts of several genes known to be dosage suppressors of Sccbk1 mutants (37, 47, 48). ScCbk1 binds to and phosphorylates ScSsd1, thereby influencing the subcellular distribution and translation of ScSsd1-mRNA complexes (37, 48, 49). ScCbk1 inhibition causes ScSsd1-mRNA complexes to localize to P bodies and stress granules (49, 50), where the translation of the ScSsd1-targeted mRNAs is repressed. ScCbk1 inhibition also decreases the amount of ScSsd1-associated mRNA bound to polysomes (37). These data suggest that ScSsd1-associated mRNAs are constitutively repressed in S. cerevisiae RAM pathway mutants, leading to cell death.
Recent data also suggest that ScCbk1 directly regulates ScSsd1 localization (49, 50). For example, an S. cerevisiae ssd1 mutant lacking multiple ScCbk1 phosphorylation sites constitutively localizes to P bodies and, like Sccbk1 mutants, are toxic to cells (49). Conversely, phosphomimetic mutations of the ScCbk1 phosphorylation sites of ScSsd1 diminish its recruitment to P bodies and promote the localization of at least one mRNA to sites of polarized growth (49). These results suggest that ScCbk1 phosphorylation of ScSsd1 both prevents the recruitment of ScSsd1-mRNA complexes to sites of translational repression and promotes the localization of at least a subset of ScSsd1-mRNA complexes to sites of polarized growth where they are then translated.
The role of RAM in mRNA homeostasis may not be limited to modulation of ScSsd1. Genetic data suggest that ScCbk1 negatively regulates the pumilio family protein ScPuf5/Mpt5 (5), a conserved RNA-binding protein involved in yeast cell wall integrity, mating efficiency, and longevity. ScPuf5 contains putative ScCbk1 phosphorylation sites (5) and is genetically linked to ScSSD1, suggesting that ScCbk1 may regulate parallel processes in mRNA metabolism via ScSsd1 and ScPuf5.
Morphogenesis, polarized growth, secretion, and cell wall integrity.
As implied by its name, the RAM network plays a role in morphogenesis and polarized growth (3, 52, 65, 77). Indeed, all RAM pathway components have been shown to localize, at least in part, to sites of polarized growth, such as the new bud tip or the septum (3, 65, 69, 81). A lack of ScCbk1 activity results in random bud site selection rather than the normal axial or bipolar pattern displayed by S. cerevisiae (3, 65, 69) Accordingly, in the absence of functional ScSsd1, ramΔ cells are much rounder than corresponding RAM+/ssd1-d loss-of-function mutants (65, 81). In addition, RAM pathway mutants display defective mating projection formation and decreased mating efficiency (3, 5, 65, 81). Interestingly, these defects do not appear to be the result of dramatic changes to the actin cytoskeleton during polarized growth (81).
Several lines of evidence implicate the S. cerevisiae RAM pathway in the regulation of secretion and Golgi function during polarized growth. Notably, ScCbk1 binds to and phosphorylates ScSec2 (51), the GTPase exchange factor for the RAB GTPase and exocytosis regulator ScSec4 (78), and influences the polarity establishment of both proteins upon release from G0 block (51). In addition, conditional S. cerevisiae cbk1 mutants exhibit defects in protein glycosylation and are hypersensitive to hygromycin B, a characteristic phenotype of yeast cells with glycosylation defects (51). Moreover, ScCbk1 inhibition significantly disrupts the localization of a key Golgi mannosyltransferases and diminishes the accumulation of post-Golgi vesicles in secretion mutants. Overexpression of several mannosyltransferase genes (ScMNN1, ScMNN4, and ScMNN9) via multicopy plasmids also partially suppresses the lethality of conditional cbk1 mutants (51). Despite these findings, the ScCbk1 substrates that mediate its role in Golgi function are not yet known.
A final RAM-dependent process that is intimately related to morphogenesis, polarized growth, and secretion is the maintenance of cell wall integrity. Several lines of evidence suggest that the RAM pathway plays a role in the maintenance of cell wall integrity. For example, mutants of the RAM pathway (with the exception of ace2Δ) are inviable in the presence of a functional copy of ScSSD1 (ScSSD1-v) and undergo cell lysis (18, 39). The lethality of RAM mutants in the presence of ScSSD1-v is suppressed by overexpression of a variety of cell wall genes, suggesting that the lysis is due to defects in cell wall integrity (18, 48, 51). In addition, Sckic1Δ mutants have decreased levels of 1,6-β-glucan, a key component of the yeast cell wall (77). Finally, Sccbk1Δ and other RAM mutants are hypersensitive to Calcofluor white, a chitin-binding molecule that induces cell wall stress (46).
Recent work indicates that the RAM pathway affects cell wall integrity by at least two mechanisms. As outlined above, ScCbk1 regulates the expression of some cell wall proteins by preventing recruitment of ScSsd1-mRNA complexes to P bodies, where translation is repressed (49, 50, 67). In the absence of ScCbk1 activity, translation of ScSsd1-associated mRNAs is repressed (37). Recent data indicate that ScCbk1 also influences cell wall integrity during heat shock and cell wall stress via the cell wall integrity (CWI) signaling pathway (47). The CWI pathway is a MAP kinase cascade that regulates cell wall biosynthesis and cell wall stress responses (52). During heat shock and cell wall stress, ScCbk1 is critical for the activation but not the ScMkk1/Mkk2-dependent T190/Y192 phosphorylation of ScMpk1, the terminal MAP kinase of the CWI pathway (47). ScCbk1 also appears to regulate CWI pathway inactivation through the dual-specificity phosphatase ScSpd1 (52). Although the mechanism for the role of ScCbk1 in CWI pathway regulation is not understood, ScCbk1 appears to function via ScBck2, a component of the CWI pathway and putative ScCbk1 substrate. Interestingly, genetic experiments with Neurospora indicate that Ndr/COT1 is functionally linked to the MAPK pathways in filamentous fungi (55).
Although much remains to be learned regarding the underlying mechanisms through which the RAM pathway functions in fungi, the detailed studies in S. cerevisiae provide an impressive foundation upon which to base the study of the RAM pathway in pathogenic fungi. Undoubtedly, our understanding of the functions and mechanisms of the RAM pathway will increase as additional substrates are identified.
RAM PATHWAY IN PATHOGENIC FUNGI
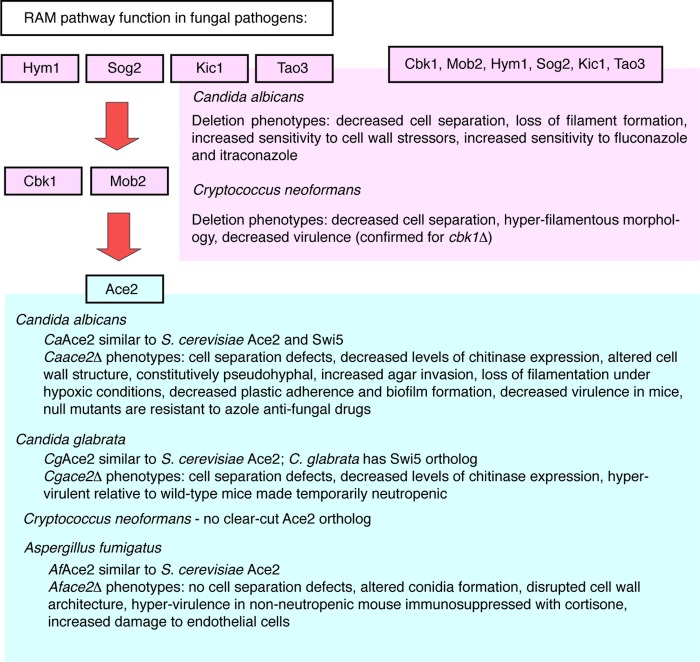
Orthologs of RAM pathway components are present in the major fungal pathogens. Among pathogenic fungi, RAM pathway function has been studied most extensively in Candida albicans, the most common human fungal pathogen; consequently, our discussion is primarily focused on this organism. A summary of the pathogenic yeasts for which functions of the RAM pathway have been identified is presented in Fig. 2. The first subsection focuses on the role of Ace2 orthologs, the second summarizes the functions of Cbk1 orthologs and its regulatory components, and the third discusses interactions between the RAM network and other signaling pathways.
Fig 2.
Summary of RAM pathway components and functions in pathogenic fungi.
Role of the RAM pathway-regulated transcription factor Ace2 in pathogenic fungi.
C. albicans (CaAce2) and C. glabrata (CgAce2) Ace2 orthologs are among the most extensively studied components of the RAM pathway in pathogenic fungi. C. glabrata is the Candida sp. most closely related to S. cerevisiae and has both Swi5 and Ace2 homologs (40). In contrast, the more distantly related C. albicans has a single gene, CaACE2, with homology to both ScACE2 (24% identity) and ScSWI5 (21% identity). Although the overall protein sequence identity between CaAce2 and ScAce2 is low, the zinc finger regions of these two proteins are highly conserved (42). CaAce2 contains a sequence that matches the consensus ScCbk1 phosphorylation motif and is presumed to be a substrate of CaCbk1, although this has not been confirmed experimentally. Consistent with its role as a daughter cell-specific transcription factor, multiple groups have shown that CaAce2 localizes to daughter cell nuclei in yeast-phase cells and in filamentous cells generated under a variety of inducing conditions (2, 42, 80). Although deletion of ACE2 does not affect vegetative growth in either C. glabrata or C. albicans, the strains display cell separation defects and decreased levels of chitinase expression (11, 40, 42, 73, 74).
The role of CaAce2 in gene expression has been characterized by transcriptional profiling (64). In addition to regulating the expression of cell separation and cell wall genes, CaAce2 induces the expression of glycolytic genes and represses, either directly or indirectly, genes involved in respiration (64). Recently, Cantero and Ernst found that CaAce2 also regulates the basal expression of protein-O-mannosyl transferases (9). Based on transcriptional profiling experiments, Mulhern et al. formulated AACCAGC as a candidate consensus binding site for CaAce2 (64). In addition to being dependent on other components of the RAM pathway (73), the expression of CaAce2 targets has also been shown to be modulated by both CaCdc28, a cyclin-dependent kinase (80), and CaCdc14, a cyclin-dependent phosphatase homologous to the mitotic exit regulator ScCdc14 (11). Wang et al. have also shown that CaCdc28 phosphorylates the transcriptional modulator CaEfg1, which in turn represses CaAce2-mediated expression of cell separation genes early in the cell cycle/hypha transition (80). CaCdc14 is required for the nuclear localization of CaAce2 and, consequently, for expression of CaAce2 targets (11). ScCdc14 has also been shown to influence ScCbk1 function during mitotic exit.
As in S. cerevisiae and consistent with its role in the expression of cell wall-related proteins, C. albicans ace2Δ/Δ mutants exhibit phenotypes indicative of altered cell wall structure, including hypersensitivity to selective inhibitors of PMT and glycosylation (9). Interestingly, the Caace2Δ/Δ mutant is less sensitive to the chitin-binding molecule Calcofluor white (34). Similarly, deletion of CaACE2 leads to increased resistance to the azole class of clinically used antifungal drugs (34). The mechanisms behind these interesting phenotypes are not clear, and their elucidation could provide interesting insights into both the function of the RAM pathway and the effects of this important class of antifungal drugs on C. albicans.
The transition between yeast and filamentous forms of C. albicans is associated with pathogenesis, and accordingly, the role of CaAce2 in morphogenesis is of interest (34, 42). Caace2Δ/Δ strains constitutively form pseudohyphae under growth conditions that do not induce filamentation in wild-type cells (e.g., rich yeast peptone dextrose medium [YPD] at 30°C). In addition, Caace2Δ/Δ cells display increased agar invasion, consistent with the phenotypes displayed in S. cerevisiae (32, 42, 44) Interestingly, Caace2Δ/Δ mutants form normal hyphae in the presence of serum, indicating that polarized growth is not affected by loss of CaAce2 function (42) and suggesting that CaAce2 is dispensable under these conditions. This is in stark contrast to other RAM pathway mutants, which are unable to form filaments under any conditions (11, 60, 73). A recent study suggested that the constitutive pseudohyphal phenotype of the Caace2Δ/Δ strains may be related to increased activity of the PKA pathway as a compensatory response to decreased CaRAM-mediated transcription (2).
The complex role that CaAce2 plays in filamentation is further highlighted by the fact that CaAce2 is required for hypha formation under oxygen-limited conditions or by yeast embedded in agar medium (64). Mulhern et al. propose that filamentation under low oxygen conditions is induced by decreased respiration (64). As mentioned above, CaAce2 is required for full expression of glycolytic genes and for decreased expression of respiratory genes. Consistent with these transcriptional effects, C. albicans mutants lacking CaACE2 have increased respiratory activity and fail to form filaments under hypoxic conditions. Similarly, Caace2Δ/Δ strains are resistant to the growth-inhibitory properties of antimycin A, a drug that inhibits cytochrome function and, consequently, respiration (64). Accordingly, antimycin A treatment induces filaments in wild-type cells but not in Caace2Δ/Δ cells. Thus, CaAce2 plays an important role in the hypoxic adaptation of C. albicans and is required for filamentation under these conditions.
CaAce2 is also important for the ability of C. albicans to adhere to plastic and to form biofilms (42). For example, the number of Caace2Δ/Δ cells that adhere to 96-well plates during short-term incubation is reduced relative to that of the wild type. Similarly, Caace2Δ/Δ biofilms matured for 24 h were much less dense than wild-type biofilms, as determined by dry cell mass measurements (42). Recently, the Mitchell laboratory found that CaAce2 is a key regulator of biofilm formation, particularly with respect to adherence (22). Indeed, overexpression of CaACE2 suppresses the defects in biofilm formation displayed by strains lacking CaSNF5, a component of the SWI/SNF complex. These data suggest that CaACE2 is a key target of CaSnf5 (22).
The effect of ACE2 deletion on the virulence of C. albicans and C. glabrata has also been examined in mouse models of disseminated candidiasis. For example, immunocompetent mice infected with a Caace2Δ/Δ strain survived more than 30 days, while the median survival of wild-type-infected mice was 3.7 days (42). In temporarily neutropenic mice, a minor but statistically significant decrease in virulence was observed in Caace2Δ/Δ strains. This suggests that the immune state of the host modulates the effect of CaACE2 on C. albicans virulence. This effect is more dramatic in C. glabrata. Cgace2Δ/Δ cells are hypervirulent relative to wild-type C. glabrata cells in a neutropenic mouse model of disseminated candidiasis (40). This difference was not due to vascular occlusion secondary to the highly clumped phenotype of Cgace2Δ/Δ but rather was associated with dramatically increased levels of interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α) relative to those in mice infected with wild-type C. glabrata. Further studies revealed that the hypervirulence of a Cgace2Δ/Δ strain was limited to neutropenic mice and that Cgace2Δ/Δ strains were slightly less virulent in immunologically intact BALB/c and DBA/2 mice (54). The mechanistic basis for this host-mutant interaction remains unclear.
Interestingly, disruption of the ACE2 ortholog in the pathogenic mold Aspergillus fumigatus also leads to increased virulence in a mouse model employing cortisone immunosuppression (20). However, the virulence of the Aface2Δ mutant was the same as that of the wild type in a neutropenic mouse model. The Aface2Δ strain also causes more damage to endothelial cells in vitro than wild type and, like Cgace2Δ mutants (40), induces increased levels of host inflammation. Aface2Δ mutants show a variety of other phenotypes, including altered conidium formation, disrupted cell wall architecture, and decreased pigment production (20). Unlike with all other fungal ACE2 mutants, there was no evidence of cell separation defects, such as cell clumping or aggregation. On the other hand, as with C. albicans, the germlings of Aface2Δ mutants are more resistant to oxidative stress than the wild type (20, 40). Although it is tempting to suggest that this resistance to oxidative stress may contribute to the increased virulence of Aface2Δ strains, it generally correlates poorly with virulence in these models.
Role of Cbk1 and upstream regulators in pathogenic fungi.
In pathogenic fungi, the upstream components of the RAM pathway have not been studied as extensively as Ace2 orthologs. CaCbk1 was first cloned by McNemar and Fonzi in 2002 (60). As in S. cerevisiae, loss of CaCbk1 profoundly affects cell separation, leading to aggregates of cells that are not disrupted by ultrasonication (60). Accordingly, the expression of cell separation genes such as CHT2 and CHT3 is also decreased in Cacbk1Δ/Δ strains, suggesting that CaCbk1 regulates the activity of CaAce2. Under a variety of hypha-inducing conditions, Cacbk1Δ/Δ cells fail to form filaments (60, 73), indicating that the RAM pathway also plays a role in C. albicans polarized growth. The heterozygote also shows decreased radial hyphae on solid medium. Consistent with this finding, Uhl et al. also isolated a Cacbk1Δ/CaCBK1 strain in a screen for mutants showing haploinsufficiency during filamentation (76). In liquid medium, the heterozygote also shows increased proportions of pseudohyphae relative to the wild type (2).
These results indicate that the general functions of the RAM pathway are conserved between S. cerevisiae and C. albicans. Further supporting this idea are the results of a recent genetic analysis of the components of the RAM pathway in C. albicans (73). With respect to morphogenesis and cell separation, Camob2Δ/Δ, Cakic1Δ/Δ, Capag1Δ/Δ, Cahym1Δ/Δ, and Casog2Δ/Δ strains show phenotypes similar to those of Cacbk1Δ/Δ strains, and most of the heterozygotes show haploinsufficiency as well (73). Consistent with these findings, the expression of Ace2-regulated cell separation genes as well as a number of hypha-specific genes is decreased in Camob2Δ/Δ, although the effects of mutations in other upstream RAM components on Ace2-mediated gene expression have not yet been examined (73).
Cacbk1Δ/Δ strains are hypersensitive to cell wall stressors such as Calcofluor white, Congo red, sodium dodecyl sulfate (SDS), and hygromycin B (73). Consistent with S. cerevisiae, the cell wall phenotypes of Cacbk1Δ/Δ and Cakic1Δ/Δ strains were suppressed by deletion of CaSSD1 (73). Although CaSsd1 has not been experimentally confirmed as a CaCbk1 substrate, it contains sequences that match the consensus ScCbk1 phosphorylation motif (D. J. Krysan, unpublished observations). An important distinction between S. cerevisiae and C. albicans RAM pathway mutants is that loss of RAM pathway activity in the presence of a functional SSD1 allele is lethal in S. cerevisiae (18, 39) but not in C. albicans (25, 60, 73). Interestingly, deletion of CaSSD1 leads to hypovirulence in a mouse model of disseminated candidiasis (23), while the Scssd1Δ mutants are hypervirulent in mouse models (82). The latter phenotype was linked to alterations in S. cerevisiae cell wall construction (82). However, Cassd1Δ/Δ mutants also have cell wall defects, suggesting that other factors are likely to be responsible for the differences in the virulence phenotypes. Thus, an important question in the study of the RAM pathway in pathogenic fungi is related to the nature of the relationship between Ssd1 and the RAM pathway.
Deletion of components of the C. albicans RAM pathway upstream of CaAce2 renders cells hypersensitive to the azole antifungal molecules fluconazole and itraconazole (73). Azoles inhibit ergosterol biosynthesis, and the expression of some ergosterol genes is decreased in Caace2Δ/Δ and Cacbk1Δ/Δ cells compared to that in wild-type cells (64, 73). However, the role of the RAM pathway in directly regulating ergosterol gene expression appears to be more complex. Deletion of RAM components upstream of CaAce2 appears to decrease ergosterol gene expression (73) and cause azole hypersensitivity, while transcriptional profiling indicates that the expression of many of these same ergosterol genes either is unaffected by deletion of CaAce2 or is actually increased in yeast (64). Additionally, Homann et al. found that Caace2Δ/Δ cells are actually resistant to azoles such as fluconazole (34), further illustrating the fact that the phenotypes of CaCBK1 and CaACE2 mutants are not always concordant. Elucidating the role of the RAM pathway in the regulation of ergosterol and resistance to antifungal drugs should provide interesting insights into these processes.
The RAM pathway has also been identified and studied in the basidiomycetous fungus Cryptococcus neoformans (79), an important cause of meningoencephalitis in immunocompromised patients. However, BLAST searches have not identified a clear-cut CnAce2 homolog. The C. neoformans gene most similar to the one encoding Ace2 actually has closer homology to the ScCrz1 and ScSwi5 genes than to the ScAce2 gene (79). Despite the lack of an obvious CnACE2 homolog, the RAM pathway mutants all display defects in cell separation. However, additional functional analysis of the RAM pathway mutants in C. neoformans has revealed some striking distinctions compared to S. cerevisiae and Candida spp. For example, deletion of the upstream components of the RAM pathway leads to a hyperfilamentous morphology rather than the filamentation defects displayed by mutants of CaRAM upstream components. These data indicate that the RAM pathway function in C. neoformans may have diverged dramatically from that in ascomycetes such as S. cerevisiae and C. albicans. Despite these significant functional divergences, the protein-protein complex interactions between the components of the CnRAM pathway are conserved with those of S. cerevisiae and C. albicans based on the fact that CnCbk1, CnKic1, and CnMob2 all physically interact by two-hybrid analysis.
To date, no specific studies of the pathogenesis of CnRAM pathway mutants have been reported. However, since most the mutants showed decreased growth at 37°C (but remained viable), which is the body temperature of mammalian hosts and a virulence factor for C. neoformans, they would be expected to show decreased virulence. Consistent with these expectations, a systematic virulence analysis of a large collection of C. neoformans mutants found that cbk1Δ cells were significantly less virulent than wild-type cells in a mouse model of pulmonary infection and that this defect was likely due to the strain's slow growth at 37°C (53). One of the other important virulence factors for C. neoformans is the ability to produce its polysaccharide capsule; however, this appears to be normal in CnRAM pathway mutants (53).
Finally, a CBK1 homologue has been cloned from rat-pathogenic Pneumocystis, a model for human-pathogenic Pneumocystis, an important cause of life-threatening pneumonia in immunocompromised patients (44). PnCBK1 was identified as a gene that is strongly expressed in lung epithelial cells exposed to Pneumocystis. The expression of PnCBK1 is repressed by low pH and high ionic strength but increased by high osmotic stress, suggesting that it is regulated by environmental conditions. PnCBK1 complements a number of Sccbk1Δ phenotypes, including cell separation defects, mating projection formation, and expression of the ScAce2-regulated transcript CTS1. These data indicate that a significant number of PcCbk1 functions may be conserved between S. cerevisiae and Pneumocystis. However, these functions can only be inferred, since Pneumocystis is not cultivatable outside the host and no system for its genetic manipulation is currently available.
INTERACTIONS OF THE RAM PATHWAY WITH OTHER SIGNALING NETWORKS
The RAM pathway regulates a variety of fundamental processes, and as such, its activity would be expected to correlate with other major signaling pathways in the cell. In S. cerevisiae (71), deletion of mob2 and cbk1 in a ras2Δ background causes synthetic growth defects relative to the single mutations. Similarly, the bud site selection defects displayed by mob2Δ strains are exacerbated by deletion of RAS2. The Ras2/PKA pathway plays an important role in coupling nutrient sensing to cell growth and cell cycle progression. The growth defects of the Scmob2Δ ras2Δ and Sccbk1Δ ras2Δ strains were suppressed by overexpression of the PKA catalytic subunit ScTpk1 but were not exacerbated by deletion of ScACE2 (71). These data suggest that the PKA pathway acts in parallel with the ScAce2-independent arm of the S. cerevisiae RAM pathway to regulate polarized growth.
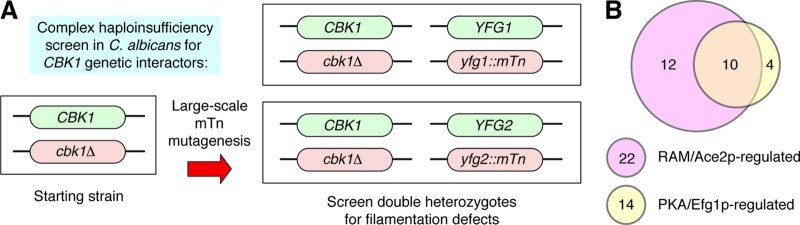
Recently, a large-scale genetic screen for genes and pathways that interact with CaCbk1 during hyphal morphogenesis identified a number of PKA pathway targets (2), indicating that the CaRAM and CaPKA pathways also interact during C. albicans morphogenesis (Fig. 3). Consistent with this hypothesis, strains containing mutations in both cbk1 and tpk1 mutations showed synthetic defects in morphogenesis (2). In addition, CaRAM pathway mutants (e.g., Cacbk1Δ/Δ and Caace2Δ/Δ) display increased EFG1 expression and PKA pathway activity relative to wild-type cells. The Ernst laboratory has shown that elevated or ectopic EFG1 expression leads to increased amounts of pseudohyphae. This same phenotype of increased pseudohyphae is displayed by Caace2Δ/Δ and is suppressed by chemical inhibition of PKA, supporting the notion that CaRAM pathway mutants have elevated levels of PKA activity (2).
Fig 3.
A complex haploinsufficiency-based screen identifies interactions between RAM and cAMP/PKA pathways in C. albicans. (A) Schematic of transposon-mediated mutagenesis of a heterozygous cbk1Δ CBK1 mutant, followed by screening for defects in filamentation relative to the parental strain. (B) Venn diagram of CBK1 interactors associated with links to the RAM and PKA pathways (2).
These data suggest that the CaPKA pathway may act to compensate for decreased CaRAM pathway function (2, 71). Further supporting this model, transcriptional profiling studies suggest that CaAce2 and CaEfg1 may coregulate a common set of genes. Consistent with that notion, >300 genes contain putative binding sites for both the PKA transcription factor Efg1 and the RAM pathway transcription factor CaAce2 (2). It therefore appears that the PKA and RAM pathways coregulate a common set of genes. Microscopy-based nuclear localization experiments indicate that Efg1 is present in the nucleus at the initiation of hyphal morphogenesis but is nearly absent from hyphal nuclei (2). In contrast, CaAce2 primarily localizes to the daughter cell nuclei of hyphae, and its expression is increased late in hyphal morphogenesis (2, 42, 80).
Taken together, these results suggest a model in which Efg1 and CaAce2 coregulate a common set of genes at different stages of hyphal development: Efg1 at the initiation of germ tube formation and CaAce2 once daughter cell nuclei accumulate within the hyphae. Further supporting this view, Wang et al. showed that Efg1 (80), which can act as either a transcriptional activator or a repressor, suppresses the expression of CaAce2-regulated cell separation genes early in morphogenesis (0 to 2 h). At later time points (3 h after hyphal induction), the repression is relieved, and thus the timing of Efg1-mediated CaAce2 suppression correlates well with the changes in nuclear localization of these two transcriptional modulators.
Wang et al. also found that Efg1 repression of CaAce2 targets such as CaCTS1 was dependent upon CaCdc28-mediated phosphorylation of Efg1 at T179; introduction of a nonphosphorylatable alanine at this position abolished suppression (80). The role of CaCdc28 in mediating the correlation of Efg1 and CaAce2 transcriptional modulation is also highlighted by the fact that CaCdc14 is also required for expression of CaAce2-regulated cell separation genes (11), an effect consistent with the fact that it is thought to dephosphorylate CaCdc28 targets.
Recently, a role for CaCdc28 in the regulation of the RAM pathway was further strengthened by the finding that CaCdc28 directly phosphorylates CaMob2 (25). This phosphorylation appears to occur relatively early in the course of hyphal development. C. albicans mob2-4A, a mutant containing alanine residues at the consensus CaCdc28 phosphorylation sites, is defective for polarized growth and hyphal development. Interestingly, expression of cell separation genes such as CaCHT3 and CaSCW11 is increased in C. albicans mob2-4A relative to that in the wild type (25). This observation suggests that CaCdc28-phosphorylated CaMob2 negatively regulates the ability of CaCbk1 to activate CaAce2-mediated transcription early in morphogenesis. Thus, it appears that CaCdc28 negatively regulates the activity of CaAce2 through both Efg1 and CaMob2 (25).
As a model for the interaction between CaCdc28, the PKA, and the RAM pathway (Fig. 4), we suggest that phosphorylation of Efg1 and CaMob2 by CaCdc28 prevents inappropriate CaAce2-mediated gene expression early in morphogenesis. Later in morphogenesis, CaAce2 localizes to the daughter cell nuclei within the growing hyphae, Efg1 nuclear localization decreases, and the CaCdc28 sites of CaMob2 are no longer phosphorylated. As a result, the repression of CaAce2-mediated transcription is relieved. Consistent with this model, Wang et al. (80) have shown that CaCHT3 expression increases late in morphogenesis. Since CaCdc14 is also required for CaAce2-mediated gene expression, it is possible that this phosphatase is responsible for dephosphorylating Efg1 and/or CaMob2 and, thereby, plays a role in alleviating CaCdc28-mediated repression of Ace2 transcription. Finally, it is likely that many levels of regulation are involved in the interplay between Efg1 and CaAce2 during morphogenesis, and hence, additional work will be required to fully understand this interaction.
Fig 4.
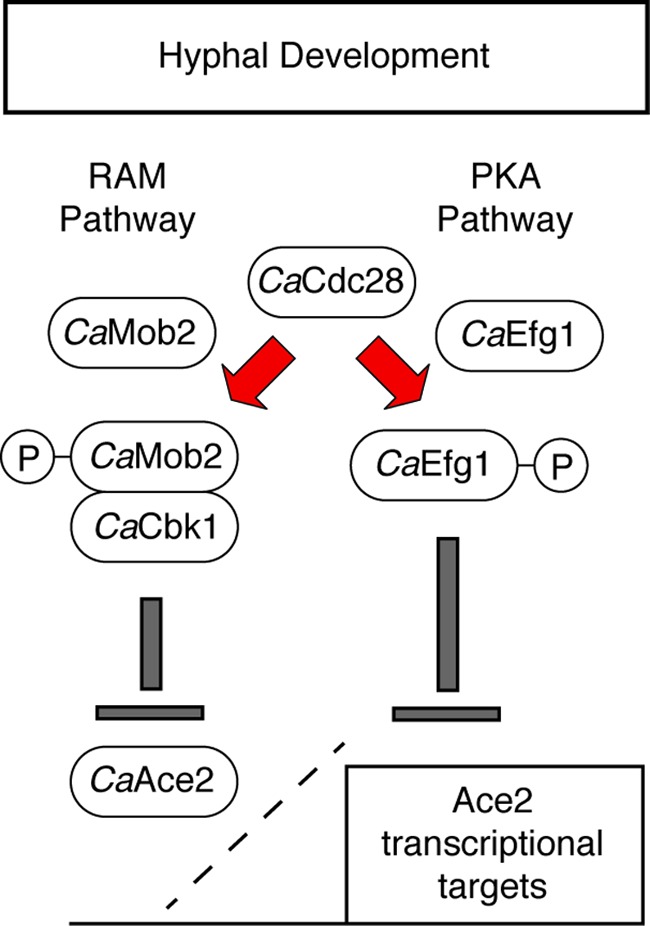
Model for CaCdc28-mediated modulation of components of the RAM and PKA pathways during hyphal morphogenesis.
An apparently unique feature of the RAM pathway in C. neoformans is that it functions in parallel with the calmodulin-calcineurin pathway. Walton et al. found that the CnRAM pathway mutants were all hypersensitive to the calcineurin inhibitors cyclosporine and FK506 (79). In addition, they were unable to isolate double Cncbk1Δ Cncna1Δ mutants. None of the RAM pathway mutants in S. cerevisiae were hypersensitive to calcineurin inhibitors, suggesting that this parallel function represents a divergent feature of the RAM pathway in C. neoformans. At this time, there are two interesting questions to be answered: (i) what is the identity of the essential functions shared by the RAM and calcineurin pathways, and (ii) do any other fungi display an interaction between these two important signaling pathways?
In summary, the RAM pathway is a well-conserved signaling network in eukaryotic cells that regulates a variety of important cellular functions in pathogenic yeasts. Consistent with its importance to fungal physiology, further studies on the mechanisms of RAM pathway function in pathogenic fungi are expected to provide insights into pathogenesis and drug resistance.
ACKNOWLEDGMENTS
Because of space constraints, not all relevant work could be cited in this article.
Work in our laboratories was supported by National Institute of Allergy and Infectious Diseases grants 5R21AI084539 (D.J.K. and A.K.), 1R01AI098450-1A1 (D.J.K.), and 5T32AI007464 (Y.C.-R.) and by March of Dimes grant 1-FY11-403 (A.K.).
Footnotes
Published ahead of print 27 April 2012
REFERENCES
- 1. Baxter BK, DiDone L, Oga D, Schor S, Krysan DJ. 2011. Identification, in vitro activity, and mode of action of phosphoinositide-dependent-1 kinase inhibitors as antifungal molecules. ACS Chem. Biol. 6:502–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bharucha N, et al. 2011. A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis. PLoS Genet. 7:e1002058 doi:10.1371/journal.pgen.1002058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bidlingmaier S, Weiss EL, Seidel C, Drubin DG, Snyder M. 2001. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:2449–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bogomolnaya LM, Pathak R, Guo J, Polymenis M. 2006. Roles of the RAM signaling network in cell cycle progression in Saccharomyces cerevisiae. Curr. Genet. 49:384–392 [DOI] [PubMed] [Google Scholar]
- 5. Bourens M, et al. 2009. Mutations in the Saccharomyces cerevisiae kinase Cbk1p lead to a fertility defect that can be suppressed by the absence of Brr1p or Mpt5p (Puf5p), proteins involved in RNA metabolism. Genetics 183:161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bourens M, et al. 2008. Mutations in a small region of the exportin Crm1p disrupt the daughter cell specific nuclear localization of the transcription factor Ace2p in Saccharomyces cerevisiae. Biol. Cell 100:343–354 [DOI] [PubMed] [Google Scholar]
- 7. Brace J, Hsu J, Weiss EL. 2011. Mitotic exit control of the Saccharomyces cerevisiae Ndr/LATS kinase Cbk1 regulates daughter cell separation after cytokinesis. Mol. Cell. Biol. 31:721–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler G, Thiele D. 1991. ACE2, an activator of yeast metallothionein expression, is homologous to SWI5. Mol. Cell. Biol. 11:476–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cantero PD, Ernst J. 2011. Damage to the glycoshield activates PMT-directed O-mannosylation via the Msb2-Cek1 pathway in Candida albicans. Mol. Microbiol. 80:715–725 [DOI] [PubMed] [Google Scholar]
- 10. Chen RE, Thorner J. 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773:1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clemente-Blanco A, et al. 2006. The Cdc14p phosphatase affects late cell-cycle events and morphogenesis in Candida albicans. J. Cell Sci. 119:1130–1143 [DOI] [PubMed] [Google Scholar]
- 12. Colman-Lerner A, Chin TE, Brent R. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739–750 [DOI] [PubMed] [Google Scholar]
- 13. Das M, Wiley DJ, Chen X, Shah K, Verde F. 2009. The conserved NDR kinase Orb6 controls polarized growth by spatial regulation of the small GTPase Cdc42. Curr. Biol. 19:1314–1319 [DOI] [PubMed] [Google Scholar]
- 14. Denning DW, Hope WW. 2010. Therapy for fungal diseases: opportunities and priorities. Trends Microbiol. 18:195–204 [DOI] [PubMed] [Google Scholar]
- 15. Dohrmann PR, et al. 1992. Parallel pathways of gene regulation: homologous regulators of SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 6:93–104 [DOI] [PubMed] [Google Scholar]
- 16. Doolin MT, Johnson AL, Johnston LH, Butler G. 2001. Overlapping and distinct roles of the duplicated yeast transcriptional factors Ace2p and Swi5p. Mol. Microbiol. 40:422–432 [DOI] [PubMed] [Google Scholar]
- 17. Dorland S, Meegenaars ML, Stillman DJ. 2000. Roles for the Saccharomyces cerevisiae SDS3, CBK1, and HYM1 genes in transcriptional repression by SIN3. Genetics 154:573–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du LL, Novick P. 2002. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell 13:503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eglen RM, Reisine T. 2009. The current status of drug discovery against the human kinome. Assay Drug Dev. Technol. 7:22–43 [DOI] [PubMed] [Google Scholar]
- 20. Ejzykowicz DE, et al. 2009. The Aspergillus fumigatus transcription factor Ace2p governs pigment production, conidiation, and virulence. Mol. Microbiol. 72:155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emoto K, et al. 2004. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell 119:245–256 [DOI] [PubMed] [Google Scholar]
- 22. Finkel JS, et al. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog. 8:e1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gank KD, et al. 2008. SSD1 is integral to host defense peptide resistance in Candida albicans. Eukaryot. Cell 7:1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gupta S, McCollum D. 2011. Crosstalk between NDR kinase pathways coordinates cell cycle dependent actin rearrangements. Cell Div. 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gutierrez-Escribano P, et al. 2011. CDK-dependent phosphorylation of Mob2 is essential for hyphal development in Candida albicans. Mol. Biol. Cell 22:2458–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hall RA, Cottier F, Mulschlegel FA. 2009. Molecular networks in the fungal pathogen Candida albicans. Adv. Appl. Microbiol. 67:191–212 [DOI] [PubMed] [Google Scholar]
- 27. Hao Chun YA, Cheung K, Rashidi B, Yang X. 2008. Tumor suppressor LATS1 is a negative regulator of YAP. J. Biol. Chem. 283:5496–5509 [DOI] [PubMed] [Google Scholar]
- 28. Hartwell LH. 2004. Yeast and cancer. Biosci. Rep. 24:523–544 [DOI] [PubMed] [Google Scholar]
- 29. Hergovich A, Stegert MR, Schmitz D, Hemmings BA. 2006. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell. Biol. 7:253–264 [DOI] [PubMed] [Google Scholar]
- 30. Hergovich A, Cornils H, Hemmings BA. 2008. Mammalian NDR protein kinases: from regulation to a role in centrosome duplication. Biochim. Biophys. Acta 1784:3–15 [DOI] [PubMed] [Google Scholar]
- 31. Hergovich A, Hemmings BA. 2009. Mammalian NDR/LATS protein kinases in hippo tumor suppressor signaling. Biofactors 35:338–345 [DOI] [PubMed] [Google Scholar]
- 32. Hirata D, et al. 2002. Fission yeast Mor2/Cps12, a protein similar to Drosophila Furry, is essential for cell morphogenesis and its mutation induces Wee1-dependent G(2) delay. EMBO J. 21:4863–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6:e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5:e1000783 doi:10.1371/journal.pgen.1000783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hou MC, Wiley DJ, Verde F, McCollum D. 2003. Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J. Cell Sci. 116:125–1335 [DOI] [PubMed] [Google Scholar]
- 36. Huang TY, Markley NA, Young D. 2003. Nak1, an essential germinal center (GC) kinase regulates cell morphology and growth in Schizosaccharomyces pombe. J. Biol. Chem. 278:991–997 [DOI] [PubMed] [Google Scholar]
- 37. Jansen JM, Wanless AG, Seidel CW, Weiss EL. 2009. Cbk1 regulation of the RNA binding protein Ssd1 integrates cell fate with translational control. Curr. Biol. 19:2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jansen JM, Barry MF, Yoo CK, Weiss EL. 2006. Phosphoregulation of Cbk1 is critical for RAM network control of transcription and morphogenesis. J. Cell Biol. 175:755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jorgensen PB, et al. 2002. High-resolution mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics 162:1091–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kamran M, et al. 2004. Inactivation of transcription factor gene ACE2 in the fungal pathogen Candida glabrata results in hypervirulence. Eukaryot. Cell 3:546–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kanai M, et al. 2005. Fission yeast MO25 protein is localized at SPB and septum and is essential for cell morphogenesis. EMBO J. 24:3012–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kelly MT, et al. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence, and virulence. Mol. Microbiol. 53:969–983 [DOI] [PubMed] [Google Scholar]
- 43. King L, Butler G. 1998. Ace2p, a regulator of CTS1 (chitinase) expression, affects pseudohyphal production in Saccharomyces cerevisiae. Curr. Genet. 34:183–191 [DOI] [PubMed] [Google Scholar]
- 44. Kottom TJ, Limper AH. 2003. Pneumocystis carinii cell wall biosynthesis kinase gene CBK1 is an environmentally responsive gene that complements cell wall defects of cbk1-deficient yeast. Infect. Immun. 71:6463–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kozubowski L, Lee SC, Heitman J. 2009. Signalling pathways in the pathogenesis of Cryptococcus neoformans. Cell Microbiol. 11:370–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS. 2005. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot. Cell 4:1364–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kuravi V, Kurischko C, Puri M, Luca FC. 2011. Cbk1 kinase and Bck2 control MAP kinase activation and inactivation during heat shock. Mol. Biol. Cell 22:4892–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kurischko C, Weiss G, Ottey M, Luca FC. 2005. A role for the Saccharomyces cerevisiae regulation of Ace2 and polarized morphogenesis signaling network in cell integrity. Genetics 171:443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kurischko C, Kim HK, Kuravi VK, Pratzka J, Luca FC. 2011. The yeast Cbk1 kinase regulates mRNA localization via the mRNA-binding protein Ssd1. J. Cell Biol. 192:583–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kurischko C, Kuravi VK, Herbert CJ, Luca FC. 2011. Nucleocytoplasmic shuttling of Ssd1 defines the destiny of its bound mRNAs. Mol. Microbiol. 81:831–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kurischko C, et al. 2008. The yeast LATS/Ndr kinase Cbk1 regulates growth via Golgi-dependent glycosylation and secretion. Mol. Biol. Cell 19:5559–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Levin DE. 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 189:1145–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu OW, et al. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135:174–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. MacCallum DM, et al. 2006. Different consequences of ACE2 and SWI5 gene disruptions for virulence in pathogenic and nonpathogenic yeasts. Infect. Immun. 74:5244–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maerz S, et al. 2008. The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion, and sexual development in Neurospora crassa. Genetics 179:1313–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Maerz S, et al. 2009. Two NDR kinase-MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa. Mol. Microbiol. 74:707–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maerz S, Seiler S. 2010. Tales of RAM and MOR: NDR kinase signaling in fungal morphogenesis. Curr. Opin. Microbiol. 13:663–671 [DOI] [PubMed] [Google Scholar]
- 58. Mazanka E, Weiss EL. 2010. Sequential counteracting kinases restrict an asymmetric gene expression program to early G1. Mol. Biol. Cell 21:2809–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mazanka E, et al. 2008. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 6:e203 doi:10.1371/journal.pbio.0060203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McNemar MD, Fonzi WA. 2002. Conserved serine and threonine kinase encoded by CBK1 regulates expression of several hypha-associated transcripts and genes encoding cell wall proteins in Candida albicans. J. Bacteriol. 184:2058–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mendoza M, Redemann S, Brunner D. 2005. The fission yeast MO25 protein functions in polar growth and cell separation. Eur. J. Cell Biol. 84:915–926 [DOI] [PubMed] [Google Scholar]
- 62. Millward TA, Hess D, Hemmings BA. 1999. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J. Biol. Chem. 274:33847–33850 [DOI] [PubMed] [Google Scholar]
- 63. Miyamoto H, Matsushiro A, Nozaki M. 1993. Molecular cloning of a novel mRNA sequence expressed in cleavage stage embryos. Reprod. Dev. 34:1–7 [DOI] [PubMed] [Google Scholar]
- 64. Mulhern SM, Logue ME, Butler G. 2006. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 5:2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nelson B, et al. 2003. RAM: a conserved signaling network that regulates Ace2 transcriptional activity and polarized growth. Mol. Biol. Cell 14:3782–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. O'Conallain C, Doolin MT, Taggart C, Thornton F, Butler G. 1999. Regulated nuclear localization of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 262:275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Panozzo C, Bourens M, Nowacka A, Herbert CJ. 2010. Mutations in the C-terminus of the conserved NDR kinase, Cbk1p of Saccharomyces cerevisiae, make the protein independent of its upstream activators. Mol. Genet. Genomics 283:111–122 [DOI] [PubMed] [Google Scholar]
- 68. Perfect JR, Casadevall A. 2006. Fungal molecular pathogenesis: what can it do and why do we need it? p3–13 In Heitman J, Filler SG, Edwards JE, Jr, Mitchell AP. (ed), Molecular principles of fungal pathogenesis. ASM Press, Washington, DC [Google Scholar]
- 69. Racki WJ, Becam A-M, Nasr F, Herbert CJ. 2000. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 19:4524–4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sbia M, et al. 2008. Regulation of the yeast Ace2 transcription factor during the cell cycle. J. Biol. Chem. 283:11135–11145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schneper L, Kraus A, Miyamoto R, Fang S, Broach JR. 2004. The Ras/protein kinase A pathway acts in parallel with the Mob2/Cbk1 pathway to effect cell cycle progression and proper bud site selection. Eukaryot. Cell 3:108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shi J, et al. 2008. Depletion of MoB and CotA complexes in Aspergillus nidulans causes defects in polarity maintenance that can be suppressed by environmental stress. Fungal Genet. Biol. 45:1570–1581 [DOI] [PubMed] [Google Scholar]
- 73. Song Y, et al. 2008. Role of the RAM network in cell polarity and hyphal morphogenesis in Candida albicans. Mol. Biol. Cell 19:5456–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stead DA, et al. 2010. Impact of the transcriptional regulator, Ace2, on the Candida glabrata secretome. Proteomics 10:212–223 [DOI] [PubMed] [Google Scholar]
- 75. Tamaskovic R, Bichsel SJ, Hemmings BA. 2003. NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Lett. 546:73–80 [DOI] [PubMed] [Google Scholar]
- 76. Uhl MA, Biery M, Craig N, Johnson AD. 2003. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen Candida albicans. EMBO J. 22:2668–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vink E, et al. 2002. The protein kinase Kic1 affects 1,6-β-glucan levels in the cell wall of Saccharomyces cerevisiae. Microbiology 148:4035–4048 [DOI] [PubMed] [Google Scholar]
- 78. Walch-Solimena C, Collins RN, Novick PJ. 1997. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 137:1495–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Walton FJ, Heitman J, Idnurm A. 2006. Conserved elements of the RAM signaling pathway establish cell polarity in the basidiomycete Cryptococcus neoformans in a divergent fashion from other fungi. Mol. Biol. Cell 17:3768–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang A, Raniga PP, Lane S, Lu Y, Liu H. 2009. Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol. Cell. Biol. 29:4406–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Weiss EL, et al. 2002. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell specific localization of Ace2p transcription factor. J. Cell Biol. 158:885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wheeler RT, Kupiec M, Magnelli P, Abejion C, Fink GR. 2003. A Saccharomyces cerevisiae mutant with increased virulence. Proc. Nat. Acad. Sci. U. S. A. 100:2766–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yarden OM, Pamann DJ, Ebbole Yanofsky C. 1992. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 11:2159–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]