Abstract
blaCTX-M beta-lactamases confer resistance to critically important cephalosporin drugs. Recovered from both hospital- and community-acquired infections, blaCTX-M was first reported in U.S. livestock in 2010. It has been hypothesized that veterinary use of cephalosporins in livestock populations may lead to the dissemination of beta-lactamase-encoding genes. Therefore, our objectives were to estimate the frequency and distribution of coliform bacteria harboring blaCTX-M in the fecal flora of Ohio dairy cattle populations. In addition, we characterized the CTX-M alleles carried by the isolates, their plasmidic contexts, and the genetic diversity of the bacterial isolates themselves. We also evaluated the association between ceftiofur use and the likelihood of recovering cephalosporinase-producing bacteria. Thirty fresh fecal samples and owner-reported ceftiofur use data were collected from each of 25 Ohio dairy farms. Fecal samples (n = 747) yielded 70 blaCTX-M-positive Escherichia coli isolates from 5/25 herds, 715 blaCMY-2 E. coli isolates from 25/25 herds, and 274 Salmonella spp. from 20/25 herds. The within-herd prevalence among blaCTX-M-positive herds ranged from 3.3 to 100% of samples. Multiple pulsed-field gel electrophoresis (PFGE) patterns, plasmid replicon types, and CTX-M genes were detected. Plasmids with CTX-M-1, -15, and -14 alleles were clonal by restriction fragment length polymorphism (RFLP) within herds, and specific plasmid incompatibility group markers were consistently associated with each blaCTX-M allele. PFGE of total bacterial DNA showed similar within-herd clustering, with the exception of one herd, which revealed at least 6 different PFGE signatures. We were unable to detect an association between owner-reported ceftiofur use and the probability of recovering E. coli carrying blaCTX-M or blaCMY-2.
INTRODUCTION
A powerful class of plasmid-encoded extended-spectrum beta-lactamases (ESBLs) of emerging importance is the cefotaximases (CTX-M). As their name suggests, CTX-M enzymes exhibit heightened activity against cefotaxime compared to that against other cephalosporins and in particular are characterized by their ability to hydrolytically inactivate and thus convey resistance to extended-spectrum cephalosporin drugs, including cefepime. Since emerging in the late 1980s, blaCTX-M alleles have rapidly become the most common genes to confer the ESBL phenotype among Enterobacteriaceae recovered from human health care settings (26). CTX-M-bearing isolates are also readily recovered from community-acquired infections in humans (35, 38) as well as from companion (34) and food (46, 49) animals in many parts of the world. The CTX-M genes are primarily associated with conjugative plasmids recovered from Escherichia coli, Salmonella spp., and Klebsiella pneumoniae (48). Although there is little information to account for the rapid dissemination of these genes, conjugal transfer of blaCTX-M-bearing plasmids has been reported to be highly efficient (40). The global dissemination of this resistance genotype has been compared to that of the broad-spectrum TEM beta-lactamase seen in the 1960s in addition to the proliferation of blaCMY-2 in the 1990s and 2000s (40).
Although only recently reported in U.S. livestock (52), blaCTX-M has been reported in food animal populations (17, 46, 49), food products (11), and both domestic (34) and wild (9, 36) animal species throughout the world. While the zoonotic food-borne transmission of bacteria harboring blaCTX-M has not yet been reported in the United States, a sampling of 20 packages of fresh retail meat from 7 Pittsburgh, PA, area supermarkets yielded an E. coli isolate carrying blaCTX-M-1 with a pulsed-field gel electrophoresis (PFGE) pattern identical to that of a hospital-associated isolate identified during the same study period (11). In addition, the international spread of the highly virulent extraintestinal pathogenic E. coli (ExPEC) B2-O25:H4-ST131 is associated with the pandemic dissemination of blaCTX-M-14 and blaCTX-M-15 (20, 21, 44). E. coli O25:H4 ST131 has been linked to both community-acquired and nosocomial infections and reported in North America, Europe, Asia, Africa, and the Middle East, from multiple sources, including humans, their companion animals, and fresh meat products (22).
Extended-spectrum cephalosporin drugs are considered to be “critically important” to human medicine by the World Health Organization (53). It has been hypothesized that veterinary use of ceftiofur and cefquinome in livestock populations may provide selection pressure contributing to the dissemination of blaCTX-M (23, 46). The presence of extended-cephalosporin-resistant organisms in food animal populations could serve as a reservoir of resistance genes for their food-borne transmission in fresh meat products (11, 31). However, the extent of the association of veterinary cephalosporin use in livestock with the emergence or dissemination of blaCTX-M has not been established.
We previously reported the presence of blaCTX-M in fecal E. coli isolates recovered from cattle in Ohio (52). However, basic epidemiologic information regarding the frequency, distribution, and predisposing factors for this important resistance gene in U.S. livestock populations is unknown. Therefore, our objectives were to estimate the frequency and distribution of Salmonella spp. and coliform species harboring blaCTX-M among dairy cattle. In addition, we characterized the CTX-M alleles carried by the isolates, their plasmidic contexts, and the genetic diversity of the bacterial isolates themselves via PFGE. We additionally sought to evaluate the association between herd-level ceftiofur use and the likelihood of recovering blaCTX-M-bearing bacteria.
MATERIALS AND METHODS
Source of the isolates.
A convenience sample of 25 Ohio dairy farms was recruited for voluntary participation in this cross-sectional study. At each farm, 30 fresh fecal samples were collected and placed in individual sterile 50-ml conical tubes. Samples were freshly voided feces that appeared to be from a single animal and were collected from free-stall barn and alleyway floors throughout each facility to minimize the possibility of repeated samples from the same animal. Samples were transported at ambient temperature to our research laboratory, where they were stored overnight at 4°C. The day following their collection, fecal samples were divided into duplicate 4-g aliquots for the culture and isolation of extended-spectrum-cephalosporin-resistant E. coli and Salmonella spp. Cow inventory and ceftiofur use data for each farm, including the number of cows treated therapeutically with ceftiofur (injectable or intramammary) during the previous month and the previous 6 months, were obtained from the owner or manager of each herd.
Bacterial culture and antimicrobial susceptibility testing.
For the recovery of E. coli resistant to extended-spectrum cephalosporins, 4-g fecal aliquots were homogenized into 36 ml nutrient broth containing 2 μg/ml cefotaxime. After overnight incubation, this broth was streaked onto MacConkey agar containing 4 μg/ml cefepime to identify isolates with a blaCTX-M phenotype and onto MacConkey agar containing 4 μg/ml cefoxitin to identify isolates with a blaCMY-2 phenotype. Characteristic lactose-positive and indole-positive isolates were confirmed as E. coli by PCR (1). We have previously used these methods to successfully recover fecal E. coli isolates harboring blaCTX-M and blaCMY-2 (52).
The remaining 4-g fecal aliquot was used for isolation of Salmonella spp. We used a two-phase enrichment in supplemented tetrathionate (TTB) and Rappaport-Vassiliadis R10 (RV) broths followed by differential selection on xylose-lysine-Tergitol 4 agar (XLT-4) (16, 31). From each sample, bacteria from a single black colony on XLT-4 were isolated on MacConkey agar and confirmed as Salmonella by using standard biochemical reactions, including triple sugar iron (TSI) agar, urea broth, and polyvalent antisera testing. Confirmed Salmonella sp. isolates were screened for extended-spectrum-cephalosporin resistance by inoculation onto MacConkey agar plates containing 4 μg/ml cefepime or 4 μg/ml cefoxitin with overnight incubation at 37°C.
Isolates with the expected phenotypes on selective media were further characterized to fully describe their resistance phenotypes. MICs of a panel of 26 antimicrobial drugs important to human and veterinary medicine were generated using a semiautomated broth microdilution system (CMV1AGNF and ESB1F MIC plates, TREK Diagnostic Systems, Cleveland, OH) following Clinical and Laboratory Standards Institute guidelines (8).
Isolate characterization.
PFGE genotyping (CHEF-DRIII; Bio-Rad Laboratories, Hercules, CA) was performed on total genomic DNA by using SpeI (New England BioLabs, Ipswich, MA) following CDC-recommended procedures (7, 37). The genetic similarities of strains were compared by examining banding patterns after electrophoresis and applying generally accepted criteria to assign levels of similarity (45). In addition, the isolates were clustered into genotypic groups by using the Dice coefficient similarity index with clustering settings of 1.00% optimization and 1.00% band position tolerance via Bionumerics software (Applied Maths, Kortrijik, Belgium). To determine if isolates with the expected resistance genotype (i.e., blaCTX-M-14 or blaCTX-M-15) belonged to the B2-O25:H4-ST131 strain, sequencing (51) was performed on a subset of isolates by using adk (adenylate kinase), fumC (fumarate hydratase), gyrB (DNA gyrase), and mdh (malate dehydrogenase) housekeeping genes.
Plasmid characterization.
The plasmid content of each isolate was visualized by electrophoresis using a standard procedure (24). Conjugation experiments (14) to establish the transmissibility of plasmids harboring blaCTX-M utilized wild-type E. coli donors with a rifampin- and nalidixic acid-resistant derivative of E. coli K-12 MG1655 as the recipient strain. Recipient acquisition of the expected plasmids and resistance genes was established with additional plasmid profiling, blaCTX-M PCR, and sequencing of the resulting amplicons recovered from transconjugants. Individual plasmids were classified according to a PCR-based replicon typing procedure (PRT) that detects 18 replicon types based on incompatibility group loci (4, 5) by using boiled lysate as template DNA. Plasmids were characterized using restriction fragment analysis by digestion of 10 μl of extracted plasmid DNA (39) overnight with 1 μl of AccI (New England BioLabs) at 37°C.
Discrimination of blaCTX-M alleles.
PCR, utilizing previously reported primer sets, was used to detect blaCTX-M and to screen for other classes of beta-lactamase resistance genes, including CMY-2, TEM, SHV, and OXA (25, 26, 29, 43). The blaCTX-M genes were sequenced using sequencing primers for CTX-M groups A1 and 9 (Table 1), which were designed in our laboratory (PrimerQuest; Integrated DNA Technologies, Coralville, IA). Amplicons were sequenced bidirectionally using the CEQ 8000 capillary electrophoresis system (Beckman Coulter, Palo Alto, CA) and analyzed using a BLAST search (http://blast.ncbi.nlm.nih.gov/). Additional plasmid-mediated beta-lactamase resistance gene sequencing was accomplished using the corresponding PCR amplification primers (GENEWIZ, South Plainfield, NJ) and analyzed using BLAST (http://blast.ncbi.nlm.nih.gov/).
Table 1.
Primers used for sequencing beta-lactamase genes from Escherichia coli isolates and their transconjugants
| Target | Primer name | Oligonucleotide sequence (5′–3′) |
Product size (bp) | Reference | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| blaCTX–M group A1 | CTX–M–1 upstream | ATGTTGTTGTTAATTCGTCTC | CGTTATCGCTGTACTGTAG | 446 | 52 |
| CTX–M–1 downstream | TTAACTATAATCCGATTGCG | TTTCTGCCTTAGGTTGAG | 507 | 52 | |
| blaCTX–M group 9 | CTX–M–4 upstream | ATTCCGCTGCTGCTGGGCAG | GCGGCGTGGTGGTGTCTCTC | 511 | This study |
| CTX–M–4 downstream | CGGGAGGCGTGACGGCTTTT | TTGCCACGGAACGGTCTGCG | 520 | This study | |
| blaCMY–2 | AmpC | ATGATGAAAAAATCGTTATGC | TTGCAGCTTTTCAAGAATGCGC | 1,143 | 25 |
| blaTEM | TEM | ATAAAATTCTTGAAGACGAAA | GACAGTTACCAATGCTTAATCA | 1,074 | 29 |
Localization of blaCTX-M genes to plasmid DNA.
Southern hybridization was used for the detection of blaCTX-M on plasmid fragments following AccI restriction fragment length polymorphism (RFLP). DNA was transferred from agarose gels to positively charged nylon membranes following standard procedures (41). Digoxigenin (DIG)-labeled probes were synthesized using previously reported CTX-M PCR primer sets with a panspecific primer used to detect group 1 blaCTX-M and a group 9-specific primer to detect blaCTX-M-14 by using the PCR DIG kit (Roche, Basel, Switzerland) (26). Following probe hybridization and stringent washing, anti-DIG antibody conjugated to alkaline phosphatase was colorimetrically visualized with nitroblue tetrazolium (NBT)/BCIP (5-bromo-4-chloro-3-indolylphosphate) by using the DIG High Prime kit (Roche), following the manufacturer's instructions.
Data analysis.
The relationship between owner-reported ceftiofur use and the probability of recovery of fecal bacteria carrying blaCTX-M or blaCMY-2 was investigated using multivariable logistic regression procedures (Proc GENMOD in SAS version 9.2 [2008]; SAS Institute Inc.) with the events/trial syntax. Generalized estimating equations were utilized to account for expected clustering within herds. Ceftiofur use in each herd was expressed as the proportion of cows that had been treated with ceftiofur during the 1-month and 6-month periods just prior to our sampling. These variables were included in independent models that also included herd size as a fixed effect so that the recent and longer-term ceftiofur uses could both be evaluated.
RESULTS
The 25 study herds were made up predominantly of Holsteins and ranged in size from 40 to 475 lactating cows, with a mean herd size of 150 lactating cows. Except for the single herd producing milk to organic standards, owners/managers of all other herds (96%) reported that it was their policy to include ceftiofur as one of their treatment options for sick cows. These herds had mean ceftiofur treatment rates of 3.6% (median, 2.3%; range, 0 to 10.9% within individual herds) over the previous month and 16.4% (median, 10.7%; range, 0 to 72.5% within individual herds) over the previous 6 months immediately prior to our sampling.
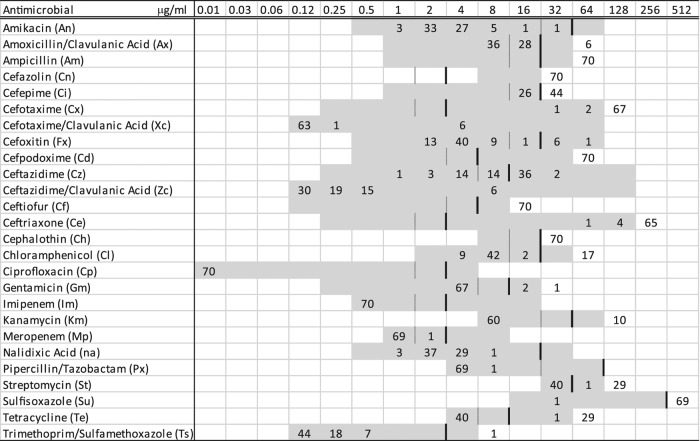
We collected the expected 30 fecal samples from each of 23 of the 25 study herds, but 29 samples were obtained from herd 21 and 28 samples from herd 25. E. coli isolates with blaCTX-M were recovered from 70 samples among the 747 collected (9.4%), representing 5 blaCTX-M-positive herds (20%). Susceptibility profiles revealed the characteristic blaCTX-M phenotype with resistance or reduced susceptibility to all penicillins and cephalosporins except cefoxitin, as well as variable resistance to other drugs, including tetracycline, streptomycin, and chloramphenicol (Fig. 1). Within-herd recovery of E. coli with blaCTX-M among the 5 positive herds ranged from 1 (3%) to 30 (100%) positive fecal samples (Table 2). In addition, 711 samples (94.8%), representing all 25 herds, yielded E. coli carrying blaCMY-2, with a minimum within-herd recovery of 18 (60%) positive fecal samples. Salmonella spp. were recovered from 284 (37.9%) fecal samples, representing 19 (76%) of the 25 study herds. Within-herd Salmonella recovery for the 19 positive herds ranged from 1 (3%) to 30 (100%) positive samples. Salmonella isolates were susceptible to both third- and fourth-generation cephalosporins.
Fig 1.
MICs of 26 antimicrobial agents for 70 Escherichia coli isolates containing blaCTX-M recovered from Ohio dairy cattle fecal samples (numbers of isolates are shown in the body of the table). Broken lines represent susceptible breakpoints and solid lines represent resistant breakpoints where available. Corresponding to the concentration listed at the top of each column (μg/ml), the included range of each antimicrobial is shown in gray.
Table 2.
Summary of E. coli isolates with blaCTX-M recovered from the fecal flora of Ohio dairy cattle including specific CTX-M gene, additional β-lactamase genes carried by the isolates, plasmid incompatibility group, and replicon type of additional plasmids
| Herd | No. of isolates | CTX-M gene type | Additional β-lactamase | CTX-M plasmida | Additional plasmid(s) |
|---|---|---|---|---|---|
| 5 | 24 | CTX-M-15 | I1 | F | |
| 6 | CTX-M-15 | TEM–1 | I1 | F | |
| 6 | 4 | CTX-M-14 | F | ||
| 2 | CTX-M-14 | * | |||
| 7 | 1 | CTX-M-14 | F | ||
| 17 | 6 | CTX-M-1 | CMY–2 | N | |
| 1 | CTX-M-1 | N | A/C | ||
| 19 | 6 | CTX-M-1 | N | Y | |
| 6 | CTX-M-1 | TEM–1 | N | Y | |
| 7 | CTX–M-1 | N | |||
| 6 | CTX-M-1 | N | FIB | ||
| 1 | CTX-M-1 | * |
*, plasmid could not be typed using our incompatibility group typing procedure.
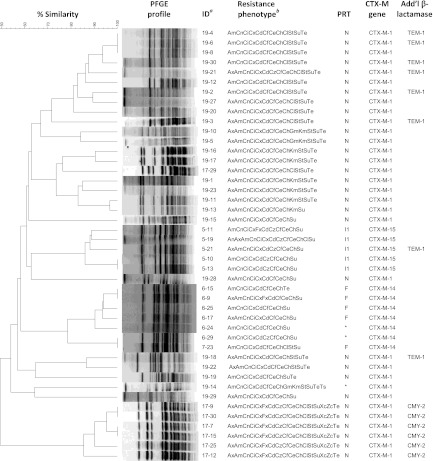
All 30 fecal samples from herd 5 yielded an E. coli isolate with blaCTX-M. PFGE of a subset of these 30 isolates indicated that they represented a single E. coli strain (Fig. 2), and plasmid analysis of every isolate indicated that all carried both an IncI1 plasmid of approximately 95 kb, bearing blaCTX-M-15, and an IncF plasmid of approximately 60 kb in size. In addition, a subset of 6 of these isolates also carried blaTEM-1, although we did not determine if this gene was localized to a specific plasmid. All fecal samples from herd 5 also yielded an E. coli isolate harboring blaCMY-2. Plasmid replicon typing found that 5 of the E. coli isolates with blaCMY-2 carried only an IncI1 plasmid, while another 13 of the E. coli isolates with blaCMY-2 carried an IncI1 plasmid in addition to IncA/C and IncFIB plasmids. One additional E. coli isolate with blaCMY-2 carried only IncI1 and IncA/C plasmids, while plasmids from the remaining E. coli isolates with blaCMY-2 from this herd were not typeable using our procedures. Restriction digestion with AccI yielded multiple bands among the blaCTX-M-positive IncI1 plasmids, while blaCMY-2-positive IncI1 plasmids recovered from the same samples yielded no multiband pattern, indicating that these plasmids, while belonging to the same incompatibility group, are polymorphic at the crude single-enzyme RFLP level. Screening of a subset of these isolates by using mdh indicated that isolates from this herd were not the pandemic B2-O25:H4-ST131 strain.
Fig 2.
Dendrographic analysis and SpeI pulsed-field gel electrophoresis (PFGE) profiles for a subset of 44 Escherichia coli isolates containing blaCTX-M recovered from dairy cattle fecal samples on 5 Ohio farms. The isolate identification (ID), antimicrobial resistance phenotype, plasmid replicon type (PRT), CTX-M gene, and additional β-lactamase genes of each isolate are indicated. The dendrogram was assembled by using Dice coefficients and the unweighted-pair group method using arithmetic averages. Default clustering settings of 1.00% optimization and 1.00% band position tolerance were used. a, isolate IDs are identified as herd number-isolate number. b, resistance phenotype indicates antimicrobial resistance or decreased susceptibility. Antimicrobial names with corresponding abbreviations are presented in Fig. 1.
E. coli isolates with blaCTX-M were recovered from 6 (20%) of the herd 6 fecal samples, all representing a single clonal strain (Fig. 2). The only E. coli isolate with blaCTX-M recovered from herd 7 samples produced a PFGE banding pattern identical to that of the isolates from herd 6. All 7 of these isolates from herds 6 and 7 carried blaCTX-M-14. Four of these 6 E. coli isolates from herd 6 and the herd 7 isolate each harbored blaCTX-M-14 on an IncF plasmid approximately 80 kb in size. The remaining 2 E. coli isolates with blaCTX-M from herd 6 also carried an 80-kb plasmid with blaCTX-M-14, but it was not typeable using our plasmid replicon typing procedure. All 7 isolates also carried a second plasmid of approximately 30 kb that was not typeable. No additional beta-lactamase genes were detected among these 7 E. coli isolates from herds 6 and 7. The fecal samples from herds 6 and 7 each produced an E. coli isolate with blaCMY-2; two samples from each of these two herds were evaluated for plasmid replicon type. Both isolates from herd 6 carried blaCMY-2 on IncA/C plasmids, while only one of the herd 7 isolates carried blaCMY-2 on an IncA/C plasmid and the second carried it on an IncFIA plasmid. Isolates from these herds were not the pandemic B2-O25:H4-ST131 strain based on screening of the mdh sequencing results from a subset of isolates.
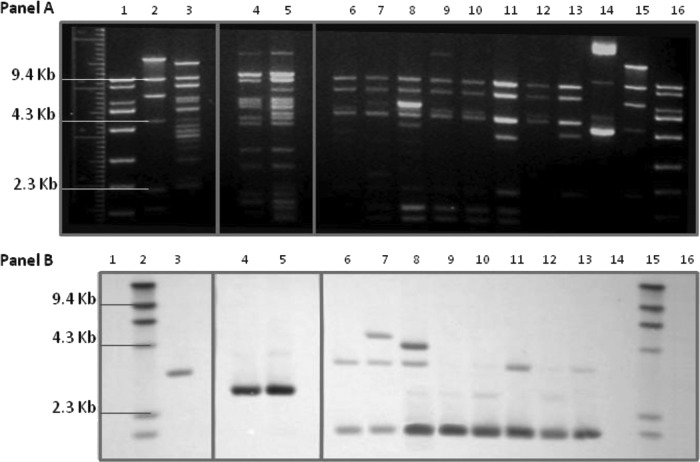
Two distinct E. coli strains each carrying blaCTX-M-1 were observed in herd 17 (Fig. 2). Six of the 7 isolates containing blaCTX-M-1 recovered from herd 17 shared an identical PFGE banding pattern. This subset of isolates expressed the blaCMY-2 phenotype in addition to the blaCTX-M phenotype, including resistance or reduced susceptibility to cefoxitin and the beta-lactamase inhibitors (Fig. 1). Carriage of blaCMY-2 by these 6 isolates was confirmed by PCR, although blaCMY-2 was not present on either the transconjugants or transformants of these isolates, suggesting that the blaCMY-2 was located either on the bacterial chromosome or on a plasmid that we were unable to isolate in our laboratory recipient E. coli. All 7 E. coli isolates from herd 17 carried blaCTX-M-1 on an IncN plasmid of approximately 40 kb, although the IncN plasmid of the unique strain appeared to be slightly larger, at approximately 55 kb. Interestingly, the single E. coli isolate with a unique PFGE pulsotype was the only isolate that did not also carry blaCMY-2, but it was the only isolate to also harbor an IncA/C plasmid (Table 2), the replicon type that is commonly associated with blaCMY-2 (32). Restriction digests of the IncN plasmids carrying blaCTX-M-1 resulted in similar banding patterns (Fig. 3). Southern hybridization of the restriction digests indicated that blaCTX-M was located on common fragments of all plasmids (Fig. 3). In addition, because blaCTX-M alleles contain no internal AccI restriction sites, the observed hybridization of multiple fragments indicates gene duplication or partial duplication within the plasmids, similar to what has been reported for blaCMY-2 carried on Inc A/C plasmids (2).
Fig 3.
(A) AccI restriction analysis of IncN plasmid DNA isolated from Escherichia coli transconjugants containing blaCTX-M recovered from Ohio dairy cattle fecal samples in two herds. Lanes: 1, Fisher exACTGene 1kb Plus DNA ladder; 2, Roche Applied Science DNA molecular weight marker II (DIG labeled); 3, 17-7; 4, 17-12; 5, 19-4; 6, 19-7; 7, 19-10; 8, 19-12; 9, 19-13; 10, 19-15; 11, 19-17; 12, 19-19; 13, 19-29; 14, negative-control CMY-2 plasmid DNA; 15, Roche Applied Science DNA molecular weight marker II (DIG labeled); 16, Fisher exACTGene 1kb Plus DNA ladder (lanes 3 to 13 are identified as herd number-isolate number). (B) Southern blot hybridization using CTX-M probe of AccI restriction analysis of IncN plasmid DNA isolated from Escherichia coli transconjugants containing blaCTX-M recovered from Ohio dairy cattle fecal samples in two herds. Lanes: 1, Fisher exACTGene 1kb Plus DNA ladder; 2, Roche Applied Science DNA molecular weight marker II (DIG labeled); 3, 17-7; 4, 17-12; 5, 19-4; 6, 19-7; 7, 19-10; 8, 19-12; 9, 19-13; 10, 19-15; 11, 19-17; 12, 19-19; 13, 19-29; 14, negative-control CMY-2 plasmid DNA; 15, Roche Applied Science DNA molecular weight marker II (DIG labeled); 16, Fisher exACTGene 1kb Plus DNA ladder (lanes 3 to 13 are identified as herd number-isolate number).
While isolates from the other 4 blaCTX-M-positive study herds exhibited a high degree of within-herd clonality, the 26 blaCTX-M-carrying E. coli isolates from herd 19 produced 6 different bacterial fingerprints on PFGE analysis (Fig. 4). We detected blaCTX-M-1 on IncN plasmids ranging in size from approximately 30 to 60 kb for 25 of the 26 isolates. The remaining isolate carried blaCTX-M-1 on a plasmid of similar size that could not be typed using our replicon typing procedure. Twelve of the 26 blaCTX-M-carrying E. coli isolates also carried an IncY plasmid, and 6 other isolates contained an IncFIB plasmid. A subset of 6 isolates carried blaTEM-1 in addition to blaCTX-M-1 (Table 2). Restriction analysis and southern hybridization of AccI restriction digests of plasmid DNA from a subset of transconjugants containing only a single plasmid found that IncN plasmids harboring blaCTX-M-1 produced nearly identical banding patterns with the blaCTX-M-1 on a common fragment, suggesting a clonal IncN plasmid present in multiple herds and in multiple E. coli strains within the same herd (Fig. 5). Restriction digest banding patterns and fragments hybridizing the CTX-M probe were unique for plasmids of different incompatibility groups (Fig. 5).
Fig 4.
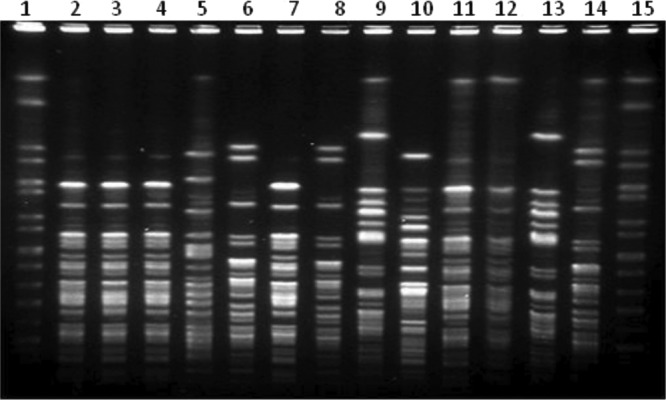
SpeI PFGE of 13 Escherichia coli isolates containing blaCTX-M-1 on IncN plasmids recovered from dairy cattle fecal samples in a single Ohio dairy herd (herd 19). Lanes: 1, Salmonella enterica serovar Braenderup H9812; 2, 19-4; 3, 19-6; 4, 19-8; 5, 19-10; 6, 19-11; 7, 19-12; 8, 19-13; 9, 19-18; 10, 19-19; 11, 19-20; 12, 19-21; 13, 19-22; 14, 19-23; 15, Salmonella Braenderup H9812 (lanes 2 to 14 are identified as herd number-isolate number).
Fig 5.
(A) AccI restriction analysis of plasmid DNA from Escherichia coli transconjugants that contained a single plasmid carrying blaCTX-M representing 5 Ohio dairy herds. Lanes: 1, Fisher exACTGene 1kb Plus DNA ladder; 2, Roche Applied Science DNA molecular weight marker II (DIG labeled); 3, 5-5; 4, 6-17; 5, 7-23; 6, 17-9; 7, 17-29; 8, 19-1; 9, 19-2; 10, 19-7; 11, 19-10; 12, 19-14; 13, 19-18; 14, negative-control CMY-2 plasmid DNA; 15, Roche Applied Science DNA molecular weight marker II (DIG labeled); 16, Fisher exACTGene 1kb Plus DNA ladder (lanes 3 to 13 are identified as herd number-isolate number). (B) Southern blot hybridization using CTX-M probe of AccI restriction analysis of plasmid DNA from a subset of Escherichia coli transconjugants that contained a single plasmid carrying blaCTX-M representing 5 Ohio dairy herds. Lanes: 1, Fisher exACTGene 1kb Plus DNA ladder; 2, Roche Applied Science DNA molecular weight marker II (DIG labeled); 3, 5-5; 4, 6-17; 5, 7-23; 6, 17-9; 7, 17-29; 8, 19-1; 9, 19-2; 10, 19-7; 11, 19-10; 12, 19-14; 13, 19-18; 14, negative-control CMY-2 plasmid DNA; 15, Roche Applied Science DNA molecular weight marker II (DIG labeled); 16, Fisher exACTGene 1kb Plus DNA ladder (lanes 3 to 13 are identified as herd number-isolate number).
All blaCTX-M gene-harboring plasmids were transferred in conjugation experiments. PCR for blaCTX-M performed on DNA isolated from MG1655 transconjugants yielded amplicons of the expected size of 544 bp (26, 29, 43). Subsequent plasmid replicon typing and sequencing confirmed carriage of the expected replicons and blaCTX-M alleles that were identified in their respective parental donor strains. SHV and OXA genes were not detected in this group of E. coli isolates with blaCTX-M.
Mean owner-reported herd-level ceftiofur treatment rates for blaCTX-M-positive herds were 3.3% and 13.5% for the previous 1 and 6 months, respectively. Ceftiofur treatment rates of 3.6% and 17.2% for blaCTX-M-negative herds were reported over the same 2 time periods. Neither ceftiofur treatment rate for the prior 1 month or 6 months was associated with the probability of recovery of E. coli harboring blaCTX-M or blaCMY-2 from fecal samples in these herds by using multivariable logistic regression models that adjusted for herd size as a continuous independent variable.
DISCUSSION
We recovered E. coli carrying blaCTX-M from 70 (9.4%) of the 747 dairy cattle fecal samples collected in this study. blaCTX-M-carrying isolates were present in 5 (20%) of 25 participating herds, with recovery rates within herds ranging from 3 to 100% of samples. Bovine fecal E. coli isolates harboring blaCTX-M were first reported in U.S. livestock in 2010 (52). While the prevalence of blaCTX-M-positive E. coli in the enteric flora of U.S. food animals has been previously unaddressed, studies from both Europe and Asia have shown the recovery of blaCTX-M from livestock with increasing frequency (27, 46). Our results suggest that the prevalence of blaCTX-M in U.S. food animal populations may be similar to those seen in Europe and Asia.
We found no association between the owner-reported therapeutic use of ceftiofur and the recovery of blaCTX-M-positive E. coli from bovine fecal flora. It is important to note that all herds, with the exception of the single organic dairy, reported ceftiofur as part of their treatment policy options. Mean ceftiofur treatment rates of 3.6% and 16.4% were reported for the previous 30 days and 6 months prior to sampling, respectively, in these herds. Ceftiofur use in swine has been associated with higher recovery of fecal coliform bacteria with blaCTX-M from individual treated animals than from untreated controls (6), but similar data for cattle are not available. At the herd level, there is a paucity of information regarding a potential association between ceftiofur use and the recovery of blaCTX-M-mediated resistance. Amplification of another extended-spectrum-cephalosporin resistance gene, blaCMY-2, in fecal E. coli has been associated with ceftiofur use in livestock (19, 28, 47). However, others have reported no association of ceftiofur use with extended-spectrum-cephalosporin resistance or simply a reduction in susceptible bacteria allowing for these rare resistant phenotypes to be detected (10, 16, 30, 42). The frequent use of ceftiofur in dairy herds may make the detection of a true association between ceftiofur administration and the recovery of blaCTX-M-positive fecal E. coli difficult. Our characterization of herd ceftiofur use as the proportion of treated cows also may not be an appropriate measure to fully capture the complex interactions of management, environment, and antimicrobial selection pressure in the dissemination of bacterial resistance genes.
PFGE analysis of the 70 blaCTX-M-positive E. coli isolates indicated that the clonal dissemination of a single or predominant strain was common within herds 5, 6, and 17. Of note, the PFGE pulsotype of the single herd 7 CTX-M-producing E. coli isolate was identical to those from herd 6, which is located in the same county in Ohio but is otherwise epidemiologically unrelated. This suggests between-herd transmission of this resistant strain, which could result from the movement of people, domestic animals, and wildlife between farms. The fact that these two herds were also sampled for this research project on the same day suggests the possibility of cross-contamination of samples at the farm or in the laboratory by the investigators. The total plasmid content of the single herd 7 isolate was also identical to that of the herd 6 isolates.
While the E. coli isolates harboring blaCTX-M in the other herds displayed notable within-herd homogeneity, the E. coli isolates from herd 19 revealed 6 different bacterial fingerprints on PFGE. Multiple pulsotypes of blaCTX-M-positive E. coli isolates from individual dairy herds have been previously reported in Europe (12, 27). Differences in the environment, management, or antimicrobial use practices that might have resulted in the exchange of a single plasmid among multiple E. coli strains are not clear. The owner of this herd did not report unusual ceftiofur use compared to other herds in the study, as 1.7% and 22% of cows were treated with ceftiofur in the previous 1 month and 6 months, respectively.
We found that the blaCTX-M gene was carried by plasmids of incompatibility groups I1, F, and N with a high degree of herd-level clonality (Table 2). In herd 5, the CTX-M-15 gene was located on an IncI1 plasmid; the CTX-M-14 gene of herds 6 and 7 was carried on an IncF plasmid, and the CTX-M-1 gene from herds 17 and 19 was located on an IncN plasmid. Both IncI1 and IncN plasmids have been suggested as the animal reservoir for the blaCTX-M-1 gene, with a high prevalence reported in livestock and poultry as well as in retail meat products (33). The IncF plasmid family is often carried by the enteric floras of both humans and animals worldwide and has been reported to harbor various resistance genes (3). These 3 major plasmid families have been previously associated with specific blaCTX-M genes, illustrating that the dissemination of this resistance gene is strongly linked to its association with a plasmid rather than an epidemic bacterial strain (13).
Individual CTX-M-1, -14, and -15 alleles were clonally conserved within herds. CTX-M-1 and -15 of group A1 and the CTX-M-14 of group 9 are among the most common of the CTX-Ms, often cultured from both humans and animals globally, and have all been identified with highly virulent multidrug-resistant (MDR) E. coli ST131-O25:H4-B2 (33). Our recovery of these three specific alleles from dairy cattle corresponds with the range of blaCTX-M genes reported from human isolates in North America (15). The global emergence of this mobile resistance determinant has been compared to that of the broad-spectrum TEM beta-lactamase seen in the 1960s (18) and the proliferation of the blaCMY-2 enzyme in the 1990s, with animal populations serving as a potential genetic reservoir (50).
In addition to recovering blaCTX-M, we found that 100% of herds and 95% of fecal samples had E. coli carrying blaCMY-2, with owner-reported ceftiofur use on 24 (96%) of the 25 farms. We previously reported that ceftiofur was used by 61% of Ohio dairy farms in 2006 (47) and 88% of such farms in 2009 (16), suggesting that ceftiofur use by dairy producers may have increased. Over the same time period, we recovered E. coli with blaCMY-2 from 67% of herds and 34% of individual cow fecal samples in 2006 (47) and from 92% of herds and 70% of cows in 2009 (16). With on-farm use of ceftiofur potentially contributing to the emergence, maintenance, and dissemination of extended-spectrum-cephalosporin resistance genes within dairy herds, our observed increase in blaCMY-2 prevalence in dairy populations concurrent with increasing ceftiofur use may portend a similar dissemination of blaCTX-M in U.S. livestock.
Footnotes
Published ahead of print 27 April 2012
REFERENCES
- 1. Bayardelle P, Zafarullah M. 2002. Development of oligonucleotide primers for the specific PCR-based detection of the most frequent Enterobacteriaceae species DNA using wec gene templates. Can. J. Microbiol. 48:113–122 [DOI] [PubMed] [Google Scholar]
- 2. Call DR, et al. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 5. Carattoli A, et al. 2006. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg. Infect. Dis. 12:1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cavaco LM, Abatih E, Aarestrup FM, Guardabassi L. 2008. Selection and persistence of CTX-M-producing Escherichia coli in the intestinal flora of pigs treated with amoxicillin, ceftiofur, or cefquinome. Antimicrob. Agents Chemother. 52:3612–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention (ed) 1996. (updated 2000) Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis: a manual. National Center for Infectious Diseases, Atlanta, GA [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed CLSI document M07-A8. CLSI, Wayne, PA [Google Scholar]
- 9. Costa D, et al. 2006. Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 58:1311–1312 [DOI] [PubMed] [Google Scholar]
- 10. Daniels JB, et al. 2009. Role of ceftiofur in selection and dissemination of blaCMY-2-mediated cephalosporin resistance in Salmonella enterica and commensal Escherichia coli isolates from cattle. Appl. Environ. Microbiol. 75:3648–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doi Y, et al. 2010. Extended-spectrum and CMY-type beta-lactamase-producing Escherichia coli in clinical samples and retail meat from Pittsburgh, USA and Seville, Spain. Clin. Microbiol. Infect. 16:33–38 [DOI] [PubMed] [Google Scholar]
- 12. Dolejska M, et al. 2011. IncN plasmids carrying blaCTX-M-1 in Escherichia coli isolates on a dairy farm. Vet. Microbiol. 149:513–516 [DOI] [PubMed] [Google Scholar]
- 13. GarcíA-Fernández A, et al. 2011. Multilocus sequence typing of IncN plasmids. J. Antimicrob. Chemother. 66:1987–1991 [DOI] [PubMed] [Google Scholar]
- 14. Gebreyes WA, Thakur S. 2005. Multidrug-resistant Salmonella enterica serovar Muenchen from pigs and humans and potential interserovar transfer of antimicrobial resistance. Antimicrob. Agents Chemother. 49:503–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hawkey PM, Jones AM. 2009. The changing epidemiology of resistance. J. Antimicrob. Chemother. 64:i3–i10 [DOI] [PubMed] [Google Scholar]
- 16. Heider LC, et al. 2009. Identification of Escherichia coli and Salmonella enterica organisms with reduced susceptibility to ceftriaxone from fecal samples of cows in dairy herds. Am. J. Vet. Res. 70:389–393 [DOI] [PubMed] [Google Scholar]
- 17. Horton R, et al. 2011. Faecal carriage and shedding density of CTX-M ESBL Escherichia coli in cattle, chickens, and pigs: implications for environmental contamination and food production. Appl. Environ. Microbiol. 77:3715–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jacoby GA, Archer GL. 1991. New mechanisms of bacterial resistance to antimicrobial agents. N. Engl. J. Med. 324:601–612 [DOI] [PubMed] [Google Scholar]
- 19. Jiang XP, Yang H, Dettman B, Doyle MP. 2006. Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodborne Pathog. Dis. 3:355–365 [DOI] [PubMed] [Google Scholar]
- 20. Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294 [DOI] [PubMed] [Google Scholar]
- 21. Johnson JR, Kuskowski MA, Gajewski A, Sahm DF, Karlowsky JA. 2004. Virulence characteristics and phylogenetic background of multidrug-resistant and antimicrobial-susceptible clinical isolates of Escherichia coli from across the United States, 2000–2001. J. Infect. Dis. 190:1739–1744 [DOI] [PubMed] [Google Scholar]
- 22. Johnson JR, Miller S, Johnston B, Clabots C, Debroy C. 2009. Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and urovirulent E. coli strains among dogs and cats within a household. J. Clin. Microbiol. 47:3721–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jørgensen CJ, Cavaco LM, Hasman H, Emborg HD, Guardabassi L. 2007. Occurrence of CTX-M-1-producing Escherichia coli in pigs treated with ceftiofur. J. Antimicrob. Chemother. 59:1040–1042 [DOI] [PubMed] [Google Scholar]
- 24. Kado C, Liu S. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koeck JL, et al. 1997. A plasmid-mediated CMY-2 β-lactamase from an Algerian clinical isolate of Salmonella senftenberg. FEMS Microbiol. Lett. 152:255–260 [DOI] [PubMed] [Google Scholar]
- 26. Lewis JS, II, Herrera M, Wickes B, Patterson JE, Jorgensen JH. 2007. First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob. Agents Chemother. 51:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liebana E, et al. 2006. Longitudinal farm study of extended-spectrum beta-lactamase-mediated resistance. J. Clin. Microbiol. 44:1630–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lowrance TC, et al. 2007. Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. Am. J. Vet. Res. 68:501–507 [DOI] [PubMed] [Google Scholar]
- 29. Ma L, et al. 2005. Variety of TEM-, SHV-, and CTX-M-type beta-lactamases present in recent clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae from Taiwan. Microb. Drug Resist. 11:31–39 [DOI] [PubMed] [Google Scholar]
- 30. Mann S, Siler JD, Jordan D, Warnick LD. 2011. Antimicrobial susceptibility of fecal Escherichia coli isolates in dairy cows following systemic treatment with ceftiofur or penicillin. Foodborne Pathog. Dis. 8:861–867 [DOI] [PubMed] [Google Scholar]
- 31. Mollenkopf DF, Kleinhenz KE, Funk JA, Gebreyes WA, Wittum TE. 2011. Salmonella enterica and Escherichia coli harboring blaCMY in retail beef and pork products. Foodborne Pathog. Dis. 8:333–336 [DOI] [PubMed] [Google Scholar]
- 32. Mulvey MR, Susky E, McCracken M, Morck DW, Read RR. 2009. Similar cefoxitin-resistance plasmids circulating in Escherichia coli from human and animal sources. Vet. Microbiol. 134:279–287 [DOI] [PubMed] [Google Scholar]
- 33. Naseer U, Sundsfjord A. 2011. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 17:83–97 [DOI] [PubMed] [Google Scholar]
- 34. O'Keefe A, Hutton TA, Schifferli DM, Rankin SC. 2010. First detection of CTX-M and SHV extended-spectrum β-lactamases in Escherichia coli urinary tract isolates from dogs and cats in the United States. Antimicrob. Agents Chemother. 54:3489–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pitout JDD, Gregson DB, Campbell L, Laupland KB. 2009. Molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob. Agents Chemother. 53:2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Radhouani H, et al. 2010. Detection of Escherichia coli harbouring extended-spectrum β-lactamases of the CTX-M classes in faecal samples of common buzzards (Buteo buteo). J. Antimicrob. Chemother. 65:171–173 [DOI] [PubMed] [Google Scholar]
- 37. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 38. Rodríguez-Baño J, et al. 2009. Escherichia coli producing SHV-type extended-spectrum β-lactamase is a significant cause of community-acquired infection. J. Antimicrob. Chemother. 63:781–784 [DOI] [PubMed] [Google Scholar]
- 39. Rondon MR, Raffel SJ, Goodman RM, Handelsman J. 1999. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc. Natl. Acad. Sci. U. S. A. 96:6451–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rossolini GM, D'Andrea MM, Mugnaioli C. 2008. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14(Suppl 1):33–41 [DOI] [PubMed] [Google Scholar]
- 41. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed, p 2.82–2.98 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42. Singer RS, Patterson SK, Wallace RL. 2008. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl. Environ. Microbiol. 74:6956–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siu L, et al. 2000. Beta-lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like beta-lactamase, OXA-30. Antimicrob. Agents Chemother. 44:2034–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smet A, et al. 2008. Diversity of extended-spectrum β-lactamases and class C β-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob. Agents Chemother. 52:1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tian GB, et al. 2009. Detection of CTX-M-15, CTX-M-22, and SHV-2 extended-spectrum beta-lactamases (ESBLs) in Escherichia coli fecal-sample isolates from pig farms in China. Foodborne Pathog. Dis. 6:297–304 [DOI] [PubMed] [Google Scholar]
- 47. Tragesser LA, Wittum TE, Funk JA, Winokur PL, Rajala-Schultz PJ. 2006. Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am. J. Vet. Res. 67:1696–1700 [DOI] [PubMed] [Google Scholar]
- 48. Walther-Rasmussen J, Hoiby N. 2004. Cefotaximases (CTX-M-ases), an expanding family of extended-spectrum beta-lactamases. Can. J. Microbiol. 50:137–165 [DOI] [PubMed] [Google Scholar]
- 49. Watson E, et al. 2012. Epidemiology of extended spectrum beta-lactamase E. coli (CTX-M-15) on a commercial dairy farm. Vet. Microbiol. 154:339–346 [DOI] [PubMed] [Google Scholar]
- 50. Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV. 2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wirth T, et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wittum TE, et al. 2010. CTX-M-type extended-spectrum β-lactamases present in Escherichia coli from the feces of cattle in Ohio, United States. Foodborne Pathog. Dis. 7:1575–1579 [DOI] [PubMed] [Google Scholar]
- 53. World Health Organization 2007. Critically important antimicrobials for human medicine: categorization for the development of risk management strategies to contain antimicrobial resistance due to non-human antimicrobial use. Report of the 2nd WHO Expert Meeting (Copenhagen, Denmark), 29 to 31 May 2007 WHO, Geneva, Switzerland [Google Scholar]