Abstract
The bacterium Mycoplasma agalactiae is responsible for contagious agalactia (CA) in small domestic ruminants, a syndrome listed by the World Organization for Animal Health and responsible for severe damage to the dairy industry. Recently, we frequently isolated this pathogen from lung lesions of ibexes during a mortality episode in the French Alps. This situation was unusual in terms of host specificity and tissue tropism, raising the question of M. agalactiae emergence in wildlife. To address this issue, the ibex isolates were characterized using a combination of approaches that included antigenic profiles, molecular typing, optical mapping, and whole-genome sequencing. Genome analyses showed the presence of a new, large prophage containing 35 coding sequences (CDS) that was detected in most but not all ibex strains and has a homolog in Mycoplasma conjunctivae, a species causing keratoconjunctivitis in wild ungulates. This and the presence in all strains of large integrated conjugative elements suggested highly dynamic genomes. Nevertheless, M. agalactiae strains circulating in the ibex population were shown to be highly related, most likely originating from a single parental clone that has also spread to another wild ungulate species of the same geographical area, the chamois. These strains clearly differ from strains described in Europe so far, including those found nearby, before CA eradication a few years ago. While M. agalactiae pathogenicity in ibexes remains unclear, our data showed the emergence of atypical strains in Alpine wild ungulates, raising the question of a role for the wild fauna as a potential reservoir of pathogenic mycoplasmas.
INTRODUCTION
Mycoplasma agalactiae is a wall-less bacterium responsible for contagious agalactia (CA) in small ruminants, a syndrome that causes important economic losses to the dairy industry and thus is listed as a notifiable disease by the World Organization for Animal Health (OIE). CA has been reported from many countries worldwide and has been documented frequently in Mediterranean countries (4, 8). While M. agalactiae is the historical etiological agent of CA, three other mycoplasma taxa are also responsible for this syndrome: Mycoplasma mycoides subsp. capri and Mycoplasma capricolum subsp. capricolum, two species that belong to the “M. mycoides” cluster, and Mycoplasma putrefaciens (4, 6, 8).
CA has three main, typical clinical signs—mastitis, arthritis, and keratoconjunctivitis—but others, such as pneumonia and septicemia in kids and lambs, or abortion, have been reported from various outbreaks (8). In France, CA of domestic goats is associated mostly with M. mycoides subsp. capri or M. capricolum subsp. capricolum (6), with the exception of the last dramatic episode reported in the 1990s, which was due to M. agalactiae and occurred in the French Savoy, a district located in the northwestern part of the Alps. In this area, the disease was considered enzootic until it was eradicated in 2002 after a long period of drastic sanitary measures, including herd slaughtering. In contrast, CA in sheep is due mainly to M. agalactiae and has been endemic in a restricted area of southern France (Western Department of the Pyrénées Atlantiques) for years (20). Prior to the current study, no isolation of M. agalactiae from wild Caprinae in France had been reported.
The Alps are known to shelter native populations of Alpine ibex (Capra ibex ibex), a wild ungulate endemic to Europe that is protected by European or national legislation in most European Union countries. In these populations, several keratoconjunctivitis outbreaks that were associated with Mycoplasma conjunctivae in Switzerland (31), or with other Mycoplasma species in Italy (10), were reported, but never any associated with M. agalactiae. In contrast, free-ranging ibexes of Spain (Capra pyrenaica) were shown to harbor M. agalactiae in their ear canals or eyes with no associated clinical signs (11, 33). Recently, an outbreak that could be regarded as CA, with mainly keratoconjunctivitis and arthritis but no pneumonia lesions, was reported in wild Caprinae of the Spanish Sierra Nevada region (32).
Since there is no satisfactory preventive or therapeutic treatment for CA, one main concern in controlling this infectious disease is to understand the origin of the pathogen and its mode of propagation within and among herds. Recent genome sequencing of two M. agalactiae strains, the type strain PG2 (27) and strain 5632 (22), has boosted the understanding of the evolutionary history of this species and the development of typing tools based on variable-number tandem repeat (VNTR) or multilocus sequence typing (MLST) analyses (17, 18). Furthermore, these genomic data have allowed the detection of horizontal gene transfer (HGT) between M. agalactiae and members of the phylogenetically distant “M. mycoides” cluster by comparative in silico analyses (27), suggesting that mycoplasma genomes are more dynamic than previously thought (26).
In recent years, repetitive isolations of M. agalactiae from the lungs of Alpine ibexes with severe pneumonia lesions by our group raised the question of the emergence and circulation of new M. agalactiae strains among protected wildlife. To address this issue, we undertook fine characterization of these ibex strains using a number of approaches, including genome sequencing of a representative strain, optical mapping, and molecular typing. Taken together, the new genome data and molecular typing results indicated that M. agalactiae strains isolated from wild ungulates of the French Alps have a highly dynamic genome but a common, unique parental origin, distinct from that of strains associated with previous CA episodes. Genome analyses also led to the discovery in ibex strains of a new, large mobile genetic element that displays phage features and is absent from the M. agalactiae genomes published so far.
MATERIALS AND METHODS
Sampling campaigns, isolation, and identification of M. agalactiae strains from ibexes.
Samplings were performed in the French Savoy region (between 45.2 and 45.5° latitude) in a restricted area delimited by the Italian frontier and the Gran Paradiso park (east) and by a line drawn between Modane, Champagny en Vanoise, and Peisey-Nancroix (west). Between 2003 and 2010, 60 ibex carcasses were collected, and necropsies were conducted at the Veterinary Laboratory of the Savoy Department (LDAV73). Three chamois carcasses found in the winter of 2009 to 2010 were also examined. At necropsy, tissues from the lungs, uteruses, and testicles and swabs from the eyes, nares, and ear canals were subjected to bacteriological analyses. In 2010, the Vanoise National Park (PNV) and the National Hunting and Wildlife Agency (ONCFS) organized a live-capture campaign consisting of the capture of approximately 100 ibexes from which two swabs from the ear canal, two from the nares, and four from the eyes were collected.
Bacteriological analyses were conducted by the LDAV73 using standard procedures, with particular attention to the isolation and culture of mycoplasmas (19). Detection of M. conjunctivae, a species known to be fastidious in culture, was conducted by PCR directly on eye swabs, in parallel with cultivation (34). Mycoplasma isolates were further identified by dot immunoblotting on a filtration membrane (MF-dot) as reported previously (23). Briefly, each isolate was tested using specific hyperimmune sera prepared against reference strains of the most commonly isolated ruminant mycoplasmas. MF-dot results obtained with sera prepared against the M. mycoides subsp. capri type strain PG3 or the M. agalactiae type strain PG2 (Table 1) were ambiguous, and strains were further identified using PCR assays specific for Mycoplasma species commonly found in ruminants as described previously (14, 16). More specifically, a PCR assay targeting the polC gene (16) was used for M. agalactiae isolates.
Table 1.
M. agalactiae isolates from wild fauna included in the study, isolation details, and characterization by MF-dot, mobilome composition, and VNTR profile
| Source and isolate no. | Description | Isolation |
Antigenic signatured |
Mobilome |
VNTR profilee |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date (mo/yr) | Sourcea | Other bacteria isolated from lungs | Host age (yr)b | Probable cause of deathc | M. mycoides subsp. capri PG3 | M. agalactiae PG2 | Phage | ICE | 5 | 14 | 17 | 19 | St | ||
| Capra ibex | |||||||||||||||
| 13387 | 07/03 | L | Mannheimia haemolytica, Arcanobacterium pyogenes | 14 | P, K | ++ | +/− | − | + | p0 | p0 | p0 | p2 | St02 | |
| 13501 | 09/03 | L | None | 3 | P | +/− | +/− | − | + | p0 | p0 | p0 | p2 | St02 | |
| 14628 | 03/06 | L | M.haemolytica, A. pyogenes | 11 | P | +++ | + | + | + | p0 | p0 | p0 | p2 | St02 | |
| 14797 | 05/07 | L | M. haemolytica, A. pyogenes | 5 | P, K | ++ | + | + | + | p0 | p0 | p0 | p2 | St02 | |
| 14934 | 01/08 | L | Pasteurella multocida, Streptococcus spp. | NK | P, K | − | +++ | + | + | p0 | p0 | p0 | p2 | St02 | |
| 14944 | 02/08 | L | M. haemolytica | NK | P, K | − | + | + | + | p0 | p0 | p0 | p2 | St02 | |
| 14980 | 03/08 | EC | P. multocida | 1 | P | +++ | ++ | − | + | p0 | p0 | p0 | p2 | St02 | |
| 14989 | 04/08 | EC | P.multocida, A. pyogenes | NK | P | ++ | + | + | + | p0 | p0 | p0 | p2 | St02 | |
| 15009 | 04/08 | L | P. multocida | 11 | P | + | +++ | − | + | p0 | p0 | p0 | p2 | St02 | |
| 15027 | 05/08 | EC | None | 2 | Capture only | − | − | + | + | p0 | p0 | p0 | p2 | St02 | |
| 15044 | 05/08 | Uterus | P. multocida, Streptococcus spp. | 5 | P | +/− | + | + | + | p0 | p0 | p0 | p2 | St02 | |
| 15179 | 12/08 | L | None | 3 | K | +++ | ++ | − | + | p0 | p0 | p0 | p2 | St02 | |
| 15196 | 01/09 | L, N | P. multocida, A. pyogenes | 6 | P | +/− | ++ | + | + | p0 | p0 | p0 | p2 | St02 | |
| 15201 | 02/09 | N | P. multocida | 14 | P | ++ | + | − | + | p0 | p0 | p0 | p2 | St02 | |
| 15261 | 04/09 | L | A. pyogenes | 11 | Enterotoxemia | +++ | + | − | + | p0 | p0 | p0 | p2 | St02 | |
| 15310 | 06/09 | L | Streptococcus spp. | 7 | P | − | ++ | − | + | p0 | p0 | p0 | p2 | St02 | |
| 15406 | 04/10 | L, N | P. multocida, A. pyogenes, Micrococcus spp. | NK | P | +++ | +/− | − | + | p0 | p0 | p0 | p2 | St02 | |
| 15409 | 04/10 | EC | Staphylococcus spp., Corynebacterium spp. | NK | Capture only | +++ | +/− | + | + | p0 | p0 | p0 | p2 | St02 | |
| Chamois | |||||||||||||||
| 15341 | 12/09 | N | M. haemolytica, P. multocida | Young | P, K | − | +++ | − | + | p0 | p0 | p0 | p2 | St02 | |
| 15379 | 03/10 | L, N | P. multocida, M. haemolytica | Adult | P, K | − | +++ | − | + | p0 | p0 | p0 | p2 | St02 | |
| Reference strains | |||||||||||||||
| PG2 | Type strain (goat, 1952, Spain) | − | +++ | − | − | p1 | p1 | p1 | p1 | St06 | |||||
| 5632 | Sequenced strain (goat joint, before 1991, Spain) | − | ++ | − | + | p1 | p0 | p5 | p1 | St13 | |||||
| 4908 | Reference for the historical CA in the Savoys (goat milk, before 1990) | − | ++ | − | − | p1 | p1 | p1 | p0 | St05 | |||||
| 4206 | Reference for the area of CA endemicity in the Pyrénées Atlantiques (ovine milk, 1981) | − | ++ | − | − | p1 | p1 | p3 | p1 | St10 | |||||
L, lung; EC, ear canal; N, nares.
NK, not known.
P, pneumonia; K, keratoconjunctivitis.
Antigenic profiles were determined by MF-dot using antisera prepared against the type strain of M. mycoides subsp. capri (PG3) or of M. agalactiae (PG2). The intensities of the MF-dot reaction were noted as follows: −, negative; +/−, doubtful; +, weak; ++, average; +++, strong.
VNTR profiles and share types (St) are numbered as proposed by L.-X. Nouvel et al. (20).
A total of 20 M. agalactiae strains from ibexes or chamois were included in the present study (Table 1). Other strains from the VIGIMYC/Anses collection (6) were also used as representatives of M. agalactiae intraspecies diversity or of specific outbreaks (Table 1; see also Table S1 in the supplemental material). M. agalactiae type strain PG2, M. agalactiae strain 5632, and Mycoplasma bovis type strain PG45 were used as reference strains because of the availability of their fully sequenced genomes (GenBank accession numbers CU179680, FP671138, and CP002188, respectively).
DNA extraction and PCR assays.
All PCR assays described in this study were conducted using purified genomic DNA except for the detection of M. conjunctivae, which was performed directly on swabs. Genomic DNA was extracted from mycoplasma culture in stationary-growth phase by using a standard phenol-chloroform procedure (7, 24). PCR assays were performed using an iCycler thermocycler (Bio-Rad, Marnes-La-Coquette, France) and GoTaq polymerase with a reaction buffer from Promega (Charbonnières, France). Real-time PCR assays for M. conjunctivae were performed on an ABI 7500 platform using TaqMan Universal PCR Master Mix (Life Technologies SAS, Villebon sur Yvette, France).
Strain typing by PFGE, VNTR analysis, and sequence analysis of the housekeeping gene polC.
Pulsed-field gel electrophoresis (PFGE) analyses were performed as described previously (30) with a slight modification of the migration conditions (6 V/cm; included angle, 120°C; pulse time from 5 to 40 s over 24 h with a linear ramping factor). Three restriction endonucleases (SmaI, MluI, and XhoI) were used. MluI-digested DNA was subsequently transferred to a Hybond N+ membrane (GE Healthcare, Chalfont St. Giles, United Kingdom) to be hybridized as described previously (14) using PCR products labeled with the enhanced chemiluminescence direct nucleic acid labeling system (GE Healthcare, Chalfont St. Giles, United Kingdom).
Strains were also characterized by their VNTR profiles as described previously (17, 20).
The polC PCR products generated for the purpose of strain identification were further sequenced by Beckman Coulter Genomics (Grenoble, France). The sequences were aligned using SeaView (http://pbil.univ-lyon1.fr) (12), and a maximum-likelihood-based tree was generated using MEGA5 (28) with a 216-nucleotide (nt) subsequence corresponding to nt 3711 to 3926 of the MAG0650 gene of M. agalactiae PG2.
Whole-genome sequencing and analysis.
The whole-genome sequence of M. agalactiae 14628 was obtained using a combination of new-generation sequencing technologies. A single (library A) and a mate-paired (insert size, 8 kb) (library B) 454 library were constructed using mycoplasma DNA purified as described previously (22). The sequencing of 35-fold coverage of GS FLX reads (issued from library A) was combined with 26-fold coverage of Titanium reads (issued from library B) and was assembled using Newbler, version 2.3 (Roche). For quality improvement, approximately 220-fold coverage of Illumina reads (36 bp) was mapped onto the whole-genome sequence by using SOAP (http://soap.genomics.org.cn) as described previously (1).
Annotation was conducted using a customized version of the CAAT-BOX platform (9) with automatic preannotation for coding sequences (CDSs) showing high similarity to PG2 or 5632, followed by expert validation as described previously (22). Genome analysis and comparisons were conducted mainly using tools provided by the MolliGen (version 3.0) platform (http://www.molligen.org) (2).
CDSs that are potential candidates for HGT in the 14628 genome were detected as described previously (27). Briefly, prediction resulted from a combination of best-BLAST-hit (BBH) analysis (using a BLASTP threshold E value of 10−8 in MolliGen, version 3.0), pairwise alignments of proteins from different phylogenetic groups, and construction of protein phylogeny trees (using the maximum-likelihood or distance/neighbor-joining methods and the complete-deletion option for gaps). When supported by significant bootstrap values in the calculated trees, incongruence between protein and species phylogenies was understood as a sign of potential HGT.
Optical mapping.
Optical maps were generated by OpGen (OpGen Technologies Inc., Madison, WI) as described previously (35). Briefly, mycoplasma cells were embedded in low-melting-point agar and were gently lysed. High-molecular-mass genomic DNA molecules were spread and immobilized onto derivatized glass slides and were digested with BglII. This restriction enzyme was selected to generate DNA fragments compatible with the technique (number and size distribution of fragments) using the in silico maps of available M. agalactiae genome sequences of strains 5632 and PG2. The DNA digests were stained with a fluorescent dye, and the pattern was recorded using a fluorescence microscope interfaced with a digital camera. Multiple scans were assembled to produce whole-chromosome-ordered optical maps using image analysis software. The MapSolver program (version 3.1; OpGen Technologies Inc.) was used to compare maps for different strains. Three experimental optical maps were generated, one each for M. agalactiae strain 5632 and M. agalactiae Alpine isolates 14628 and 15341. The accuracy and reliability of the technique were assessed by comparing in silico and experimental maps of strain 5632.
Prophage and ICE detection.
All M. agalactiae strains from wild ungulates were screened for the presence of a prophage similar to that detected in the 14628 genome and for the presence of integrative conjugative elements (ICEs), as detected in the genomes of both 5632 and 14628, by PCRs. Three sets of primers were designed for the detection of the prophage. They targeted (i) putative conserved regions, one that encodes a phage prohead protein (MAGb_3220) and one that encodes the phage terminase (MAGb_3240), and (ii) the extrachromosomal intermediate of the phage by using complementary reverse primers located at each end of the element (MAGb_3270 and MAGb_2930). The corresponding primers were, respectively, MAGb_3220-F (5′-ACCAACAAGAAACACAAACA-3′) and MAGb_3220-R (5′-AGGAATATATACGGCTTTCG-3′; MAGb_3240-F (5′-TGAAGCACGGAAACAATGAA-3′) and MAGb_3240-R (5′-TGTTCCCTTTTGTGGTGTCA-3′); and Circ-ph F (5′-CAACATTCCACTATCTGCAA-3′) and Circ-ph R (5′-TTTATCTGCGTCTGTTAGGG-3′). These three PCRs were run with the same annealing temperature of 53°C.
The presence of an ICE was analyzed by a PCR targeting the CDS22 element using primers cds22for3 (5′-TTTATGCTTTGAGACCAG-3′) and cds22rev3 (5′-GTAGTAATAACTTTAGCTCCA-3′), which are specific to M. agalactiae 5632 and give no amplification with M. agalactiae PG2. The annealing temperature was 52°C. The extrachromosomal form of the 5632-like ICE was also amplified as described previously (15).
Nucleotide sequence accession number.
The Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession AJPR00000000. The version described in this paper is the first version, AJPR01000000.
RESULTS
Recurrent isolation of atypical M. agalactiae strains from Alpine ibexes during an abnormal mortality episode.
In the course of the winter of 2007 to 2008, we observed a decrease in the ibex population of the Vanoise National Park, France, and collected, in the wild, 21 ibex carcasses that repetitively showed the presence of atypical lung lesions, such as interstitial pneumonia. Bacteriological analyses of the corresponding lung tissues often resulted in the isolation both of Pasteurella spp. and of mycoplasmas. Keratoconjunctivitis lesions were also observed, but to a lesser extent, and no M. conjunctivae was detected by PCR from eye swabs. This situation was reminiscent of four previous sporadic cases that had occurred between 2003 and 2007 and for which mycoplasma identification at the species level was at first ambiguous by use of the standard MF-dot assay. Indeed, the isolated mycoplasmas gave strongly and weakly positive reactions with the M. mycoides subsp. capri-specific and M. agalactiae-specific sera, respectively (Table 1). The four isolated strains (strains 13387, 13501, 14628, and 14797) were then unambiguously identified as belonging to the species M. agalactiae by using the specific polC PCR assay (16). The mortality episode that was particularly severe in 2008 declined in 2009 and 2010, while during the same period, 2008 to 2010, the ratio of carcasses with M. agalactiae remained constant, representing one-third of the total carcasses. Three chamois carcasses found in the same geographical area were also examined, and two were shown to present pneumonia lesions associated with M. agalactiae isolation (strains 15341 and 15379).
In 2010, the presence of mycoplasmas in the ear canals of 100 healthy live-captured ibexes was assessed. The results showed that 40 animals carried mycoplasmas of the “M. mycoides” cluster, while only 2 hosted M. agalactiae (isolates 15027 and 15409). This contrasted with the previous finding showing an association of 57% of the 60 carcasses collected since 2003 with mycoplasmas, half of which were identified as M. agalactiae recovered mostly from lung or naris samples.
Whole-genome analysis of an ibex M. agalactiae strain: an important set of mobile elements, including a new large prophage.
Preliminary typing analyses (see below) suggested that the ibex M. agalactiae strains were rather different from those isolated from domestic ruminants in France and Europe. To define their particular features, the genome of one ibex strain, strain 14628, was sequenced. This strain was chosen because it was isolated at the beginning of the mortality episode from the lung of an ibex carcass with pneumonia lesions and because of its strong reactivity with the M. mycoides subsp. capri-specific hyperimmune serum (Table 1).
A total-DNA sequence of 940,298 bp was obtained, with 99.3% of the sequence consisting of a single scaffold. Of the 806 annotated open reading frames (ORFs), 719 were predicted to be coding sequences (CDSs) and 53 to be pseudogenes or truncated CDSs (Table 2). Gap closing was impaired by the presence of repeated sequences previously characterized in the two fully sequenced genomes of M. agalactiae strains PG2 and 5632 (22, 27). These repeated sequences corresponded to (i) the vpma locus, a gene family involved in high-frequency surface variation that is composed of closely related sequences repeated among and within vpma genes (21), and (ii) mobile genetic elements, such as the integrative conjugative element (ICE) identified in 5632 (15). Indeed, 23 CDSs related to the ICE were identified, indicating that 14628 possesses at least one entire copy of this element as well as some vestiges of another one corresponding to the 3′ end. These features were confirmed by Southern blot data (data not shown).
Table 2.
General properties of M. agalactiae strains and genomesa
| Characteristic | PG2 | 5632 | 14628 |
|---|---|---|---|
| Date of isolation | 1952 | Before 1991 | 2006 |
| Country | Spain | Spain | France |
| Source | Unknown | Articulation | Lung |
| Host | Caprine | Caprine | Ibex |
| Genome size (bp) | 877,438 | 1,006,702 | 934,310 (scaffold 1)b |
| G+C content (%) | 29.70 | 29.62 | 29.87 |
| Gene density (%) | 88.5 | 88.7 | 86.9 |
| Total no. of CDSs | 713 | 815 | 719 |
| No. of (conserved) hypothetical proteins | 314 | 297 | 287 |
| No. of CDSs with predicted functions | 399 | 518 | 443 |
| No. of predicted lipoproteins | 66 | 102 | 81 |
| No. of pseudogenesc | 69 | 14 | 53 |
| No. of rRNA sets | 2 | 2 | 2 |
| No. of tRNAs | 34 | 34 | 34 |
| GenBank accession no. | CU179680 | FP671138 | AJPR01000000 |
| No. of ICEs | 1 vestigial | 3 (+ 2 vestigial) | At least 1 (+ 1 vestigial) |
| No. of transposasesd | 1 (+ 2 pseudogenes) | 15 (+ 3 pseudogenes) | 0 (+ 2 pseudogenes) |
| RMSe | |||
| R | 5 (+ 3) | 9 (+ 1) | 10 (+ 3) |
| M | 1 (+ 2) | 6 | 5 (+ 3) |
| S | 3 | 3 | 1 |
| Type I RMS | 0 (+ 2) | 1 (+ 1) | 0 (+ 2) |
| Type II RMS | 0 (+ 5) | 3 (+ 3) | 2 (+ 6) |
| Type III RMS | 1 | 1 (+ 1) | 1 (+ 2) |
| Genomic DNA digestionf by: | |||
| DpnII (sensitive to Dam methylation) | Yes | No | No |
| DpnI (needs Dam to cut) | No | Yes | Yes |
All data were calculated using the MolliGen database, version 3.0.
Sequencing and assembly (without genome circularization) resulted in the definition of 1 main scaffold of 934,310 bp (14 contigs) and 8 small contigs of 516, 547, 560, 663, 697, 853, 1,032, and 1,120 bp, respectively.
Includes pseudogenes and truncated genes. Each part of a pseudogene was counted individually, although a pseudogene could correspond to several parts of the same gene (e.g., C-terminal and N-terminal parts).
Numbers include transposases annotated as IS30-like protein.
RMS, restriction-modification systems. Shown are the numbers of DNA methyltransferases (M), restriction endonucleases (R), specificity subunits (S), or different types of RMS predicted in M. agalactiae 5632, PG2, and 14628. Numbers in parentheses indicate truncated genes or partial RMS (those in which at least one subunit is lacking).
Experimental data.
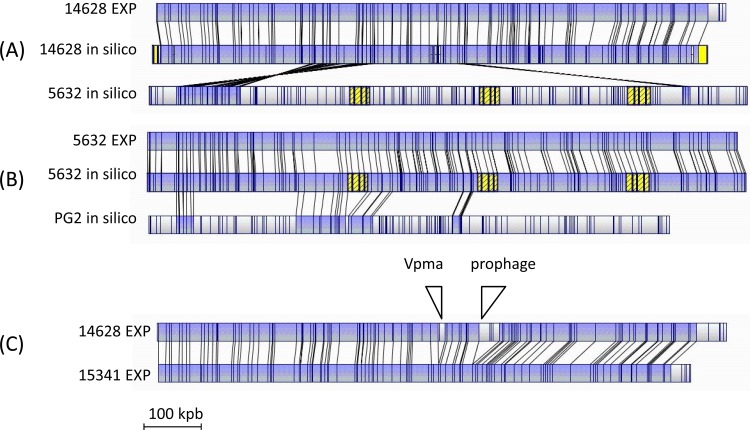
Correct assembly of the main scaffold of 14628 was confirmed by optical mapping using BglII, a restriction enzyme that generates size fragments compatible with the technique. As shown in Fig. 1A, a fine alignment was obtained between the experimental map and that generated in silico using the main scaffold of 14628. Controls included the comparison of (i) the optical and in silico BglII maps of strain 5632 (Fig. 1B), which matched almost perfectly, with only some fragments smaller than 2 kbp not detected by optical mapping, and (ii) the in silico BglII maps of strains 5632 and PG2, which, as expected, did not align well (Fig. 1B). Finally, comparison of the 14628 optical map with that generated with 5632 (Fig. 1A) or with PG2 (not shown) clearly indicated that the ibex strain is different, and this was supported by whole-genome alignment of the three strains.
Fig 1.
Alignments of M. agalactiae BglII optical maps generated using the BglII restriction enzyme. (A) Comparison of the 14628 map generated in silico from the main scaffold obtained by 454 MP genome sequencing with those obtained experimentally (EXP) after BglII digestion of the 14628 genomic DNA or generated in silico with the 5632 genome sequence. Using the same stringency parameters, in silico maps of PG2 and 14628 did not align (not shown). (B) Comparison of the 5632 map generated in silico from the genome sequence (GenBank accession number FP671138) with those obtained experimentally with 5632 total DNA or generated in silico with the PG2 genome sequence (GenBank accession number CU179680). (C) Comparison of experimental optical maps of strains 14628 and 15341. The positions of the Vpma locus and the prophage are shown. Maps were compared two by two using the default parameters of MapSolver, version 3.1. Lines between maps indicate the positions of identical restriction patterns. The blue background highlights single alignment. Blocks in yellow indicate the positions of ICEs or components of ICEs when known. The 5′ end of the dnaA gene was used as the +1 nucleotide.
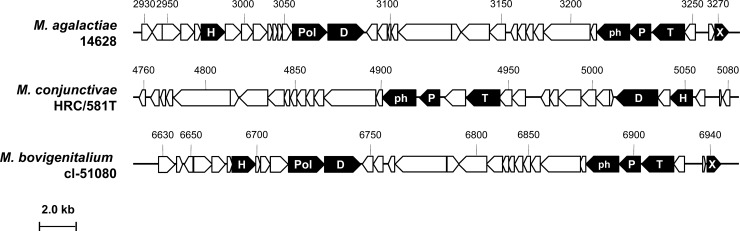
A rapid survey of the 14628 genome revealed a striking feature: the presence of a 34-kbp region that contains 35 CDSs and is totally absent from the M. agalactiae strains previously sequenced. Best-BLAST-hit (BBH) analyses indicate that this region corresponds to a prophage, because (i) it encodes a number of proteins with similarities to phage components found in other bacterial species or with known phage-related domains and (ii) it displays a gene content and organization similar to those of a putative prophage of M. conjunctivae strain HRC/581T (about 40% of the 35 CDSs encoded by the M. agalactiae putative prophage show 35.8 to 64.7% overall similarities with their M. conjunctivae homologs) (see Table S2 in the supplemental material). As illustrated in Fig. 2, major common phage features, such as the prohead, portal, and terminase coding sequences, are shared by the two Mycoplasma species. The 14628 prophage is inserted within an AT-rich region located at the beginning of a putative CDS, MAGb_3280 (see Fig. S1 in the supplemental material). In PG2 and 5632, homologs to MAGb_3280 (MAG6500 and MAGa7480, respectively) were annotated as hypothetical proteins with unknown functions and were not previously detected by whole-proteomic analyses (22). These CDSs all have an AT-rich region in their 5′ end, the length of which differs between the three strains. Whether these differences reflect excision-insertion of the 14628 prophage, and whether they are responsible for the lack of expression of the nearby CDS, is not known. A PCR assay using phage-specific outward (back-to-back) primers detected the presence of a free circular intermediate, suggesting that excision of the prophage is occurring in 14628. Finally, the 14628 prophage is inserted in a region that may have undergone horizontal gene transfer with members of the “M. mycoides” cluster (27), suggesting that the prophage may be directly and indirectly associated with genome dynamics. Recently, we found that another ruminant mycoplasma species, Mycoplasma bovigenitalium, whose genome is currently being sequenced by our consortium (project EVOLMYCO, ANR-07-GMGE-001), displays a similar prophage (Fig. 2; see also Table S2 in the supplemental material).
Fig 2.
Genome organization of the prophage identified in strains M. agalactiae 14628, M. conjunctivae HRC/581T, and M. bovigenitalium cl-51080. The locations, sizes, and orientations of the CDSs identified in each prophage are indicated by arrows. CDS numbers refer to the mnemonic codification used for each mycoplasma in the databases (see Table S2 in the supplemental material). CDSs encoding common phage products are highlighted in black using the following letter code: H, helicase; Pol, DNA polymerase; D, DNA primase; ph, prohead protein; P, portal; T, terminase; X, Xer. The overall organization of the prophages is similar, with some differences in M. conjunctivae HRC/581T, including the inversion of the region from MCJ_005050 to MCJ_005030, the absence of a DNA polymerase gene, and the absence of a recombinase gene (xer).
Searching for 14628 strain-specific genes resulted in only 23 CDSs (including 11 pseudogenes) that gave neither BLASTP nor TBLASTN hits with either of the two sequenced M. agalactiae strains (see Table S3 in the supplemental material) in addition to the 35 CDSs included in the prophage (see above). Of the 23 CDSs, 12 (52%) encode restriction-modification (RM) systems that have homologs and conserved synteny in another ruminant Mycoplasma species, M. bovis. Most likely, these RM genes have undergone HGT, as suggested by their BBHs outside the M. agalactiae and M. bovis species, which were obtained mainly with members of the “M. mycoides” cluster (see Table S3 in the supplemental material). Of the remaining 11 strain-specific CDSs of 14628, 3 (MAGb_8010, -8020, and -8030) encode hypothetical products of unknown function and have significant best-BLAST hits with M. bovis and Mycoplasma leachii, two bovine pathogens with lung tropism. These three CDSs are clustered on the chromosome between CDS14 and CDSF, which are parts of an ICE in strain 5632. Other strain-specific CDSs were mainly annotated as pseudogenes and have no particular features. The data related to 14628 strain-specific CDSs indicate that most may have undergone HGT among ruminant Mycoplasma species, and we addressed this question further at the genome level using a methodology described previously (27). A total of 163 CDSs (including pseudogenes) were predicted to have been exchanged with mycoplasmas outside the M. bovis/M. agalactiae group (see Table S4 in the supplemental material); 126 of these CDSs had BBHs with organisms from the “M. mycoides” cluster and no significant similarity outside this cluster. Of the 163 CDSs, 28 were shown to have their BBHs with mycoplasmas from the Mycoplasma hominis phylogenetic group (exclusive of M. bovis and M. agalactiae), and 25 of these correspond to the phage whose counterpart is found in M. conjunctivae and M. bovigenitalium, two members of this group. In contrast, very few CDSs were predicted to be exchanged with the Mycoplasma pneumoniae group (2 CDSs) or with organisms that do not belong to the Mollicutes (7 CDSs).
Overall, strain 14628 appears to be well equipped with large mobile elements, such as ICEs and prophage, that together account for at least 5% of the genome (60 kbp).
Emergence of M. agalactiae in ibexes: dissemination of a clonal lineage, distinct from strains circulating in domestic ruminants.
To further comprehend the dissemination of M. agalactiae in the wild ungulate population of the French Alps, molecular features of representative isolates were defined and compared to those of strains that had been isolated from domestic ruminants and characterized previously. For this purpose, 20 M. agalactiae isolates, 18 from ibexes and 2 from chamois, were chosen; these represent the mortality episode observed across the years and include 2 isolates (15027 and 15409) collected from healthy animals during capture (Table 1).
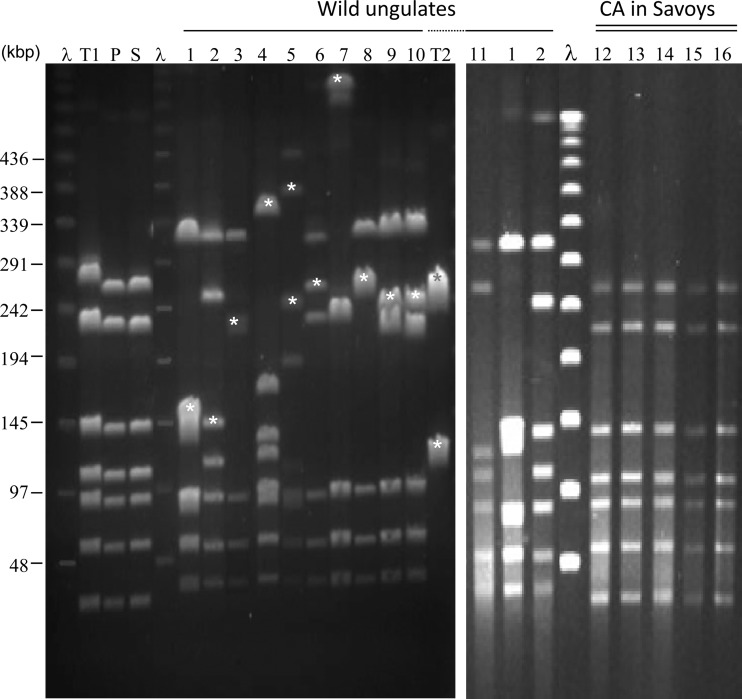
PFGE profiles indicated that wild-ungulate isolates differ (i) from strains that circulated in domestic goats of the neighboring Savoy region before CA was eradicated in 2002, (ii) from strains currently present in an area of endemicity for ovines in southern France (20), and (iii) from strains PG2 and 5632, which have been fully sequenced previously and have marked genetic differences (Fig. 3). The two chamois isolates (15341 and 15379) shared an identical pulsotype, but each ibex isolate yielded a unique restriction profile, raising the question of the circulation of several strains versus a single, highly dynamic strain. Southern blot analyses showed that PFGE polymorphisms are indeed associated with ICEs, which are mobile elements per se, and thus might reflect only ICE movements in one strain rather than several different strains (Fig. 3). A PCR assay targeting a conserved part of the M. agalactiae ICE, CDS22, confirmed the presence of this element in all ibex strains (Table 1), while it was shown to be absent from strains collected in Savoy or the Pyrénées-Atlantiques (Fig. 3; see also Table S1 in the supplemental material). This indicates that all ibex strains display ICEs or ICE components, the genomic locations of which differ among the isolates. The detection of the free, extrachromosomal ICE intermediate and the genomic organization within the ICE module were also variable among ibex strains (data not shown), suggesting that the ICEs might also differ in their functionality. The occurrence of the 14628 prophage in the other strains was also assessed by a PCR assay designed to detect two prophage CDSs (encoding, respectively, the putative phage prohead and terminase) and the circular extrachromosomal form of the phage. The data indicated the presence of the prophage in 50% of the ibex isolates (Table 1), with no correlation between the presence of 14628 prophage elements and the year of isolation. As well, there was no link between the prophage and the presumed higher virulence of M. agalactiae in ibexes, since the prophage was detected in two strains isolated from healthy captured animals. Finally, the relationships among M. agalactiae strains from wild ungulates were addressed by VNTR analyses, and all isolates were shown to display a unique common shared type (ST02 according to the classification system of reference 20) (Table 1).
Fig 3.
Representative PFGE patterns of M. agalactiae isolates following digestion of their chromosomal DNA by MluI. Lanes 1 to 11, M. agalactiae strains from wild fauna (strains 13387, 14628, 14797, 15009, 15044, 15196, 15261, 15406, 15341, 15379, and 13501, respectively, with lanes 1 and 2 represented twice). For history, see Table 1. Lane T1, M. agalactiae PG2; lane T2, M. agalactiae 5632; lane P, strain 4206, used as representative of the clonal population currently circulating in the Pyrénées-Atlantiques region. Lanes 12 to 16 represent several isolates collected in Savoy from domestic small ruminants during the historical episode of CA. Strain 4908 (lane S) was chosen as a representative. Stars indicate fragments that were detected by Southern blotting with a probe targeting an ICE (CDS22) of M. agalactiae strain 5632. Lane λ contained lambda DNA concatemers for which the molecular sizes are indicated on the right.
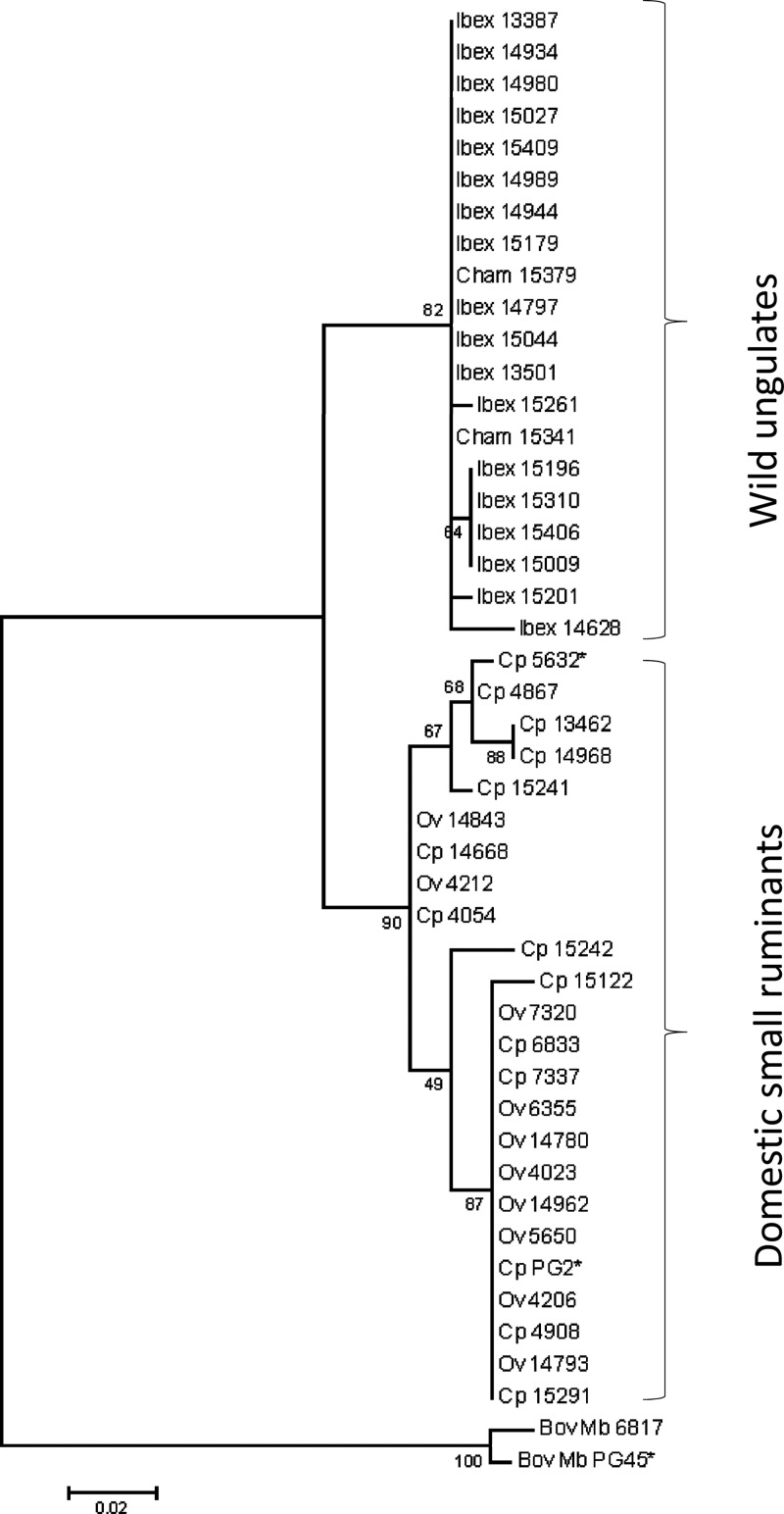
The data argued that the wild-ungulate isolates were genetically highly homogeneous apart from their mobilomes (Table 1) and different from strains isolated in Savoy or in southwestern France from domestic ruminants. To further strengthen these findings, we amplified a polC region of 216 nt from 44 strains isolated from ibexes, chamois, and small ruminants (Fig. 4 and Table 1; see also Table S1 in the supplemental material). Subsequent sequencing of the amplicons revealed 56 variable positions, of which 46 were informative for the construction of a maximum-likelihood tree that clustered all ibex and chamois isolates in one branch. The other main branch was composed of isolates from domestic ruminants that grouped with PG2 or 5632, two strains that are considered representative of each end of the genetic spectrum of the species (22).
Fig 4.
Clustering of M. agalactiae strains isolated from different hosts using the maximum-likelihood method. A 216-nt portion of the polC gene was sequenced, and 46 variable positions were used to construct a maximum-likelihood tree. The tree with the highest log likelihood is shown, and the percentage of trees in which the associated strains clustered together is indicated at each node. The branch lengths indicate the number of substitutions per site. The tree was outrooted using two M. bovis strains. Strains are designated by a number preceded by “Ibex,” “Cham,” “Ov,” or “Cp,” referring to the host origin (ibex, chamois, ovine, or caprine, respectively).
Comparison of the partial polC sequences and of the molecular typing data indicated that the ibex and chamois strains are highly related. This observation was further supported by optical mapping of genomes from strains 15341 and 14628, which were isolated 4 years apart from a chamois and an ibex, respectively, from geographically close areas. Their optical maps were nearly identical (Fig. 1C), with only two major differences, which corresponded to the prophage and to the Vpma locus of 14628, two regions expected to differ greatly even within clonal populations.
DISCUSSION
In this study, M. agalactiae was isolated for the first time from Alpine wild ungulates, both ibexes and chamois, during a severe mortality episode. These isolates were recovered mainly from animals with atypical lung lesions, while healthy ibexes of the same geographical area were found to harbor M. mycoides subsp. capri in their ear canals frequently and M. agalactiae very rarely. In small ruminants, M. agalactiae displays a predilection for the mammary gland, the joint, and the eye but is rarely found in the lung (8). In contrast, M. mycoides subsp. capri strains are often found in goats, either in the ear canals of asymptomatic carriers or associated with the lower respiratory tract, where they cause important lesions (8). Thus, the situation observed in Alpine ibexes suggests that the associated M. agalactiae strains are atypical in their tissue tropism and virulence, yet the direct role of this pathogen in lung lesions observed during the mortality episode cannot be experimentally addressed in this protected species.
To identify specific molecular features of these strains isolated from ibexes, the genome of strain 14628, which was recovered at the beginning of the mortality episode, was sequenced. Genomic and proteomic data were already available for two M. agalactiae strains, PG2 and 5632, both of which were isolated from small ruminants with CA. These two strains were considered to stand at each end of the genetic spectrum encountered in the species and were shown to differ mainly by the presence in 5632 of an important mobile gene set composed of both insertion sequences (IS) and integrative conjugative elements (ICEs) (22). In comparison, the 14628 genome showed only a small number of strain-specific CDSs, many of which had homologs in M. bovis, a phylogenetically closely related species responsible for severe infections in cattle. Several CDSs correspond to RM systems, but a cluster of three CDSs (MAGb_8010, -8020, and -8030) with no predicted function might be potential candidates for virulence factors, because they have homologs in M. leachii, a Mycoplasma species of the “M. mycoides” cluster that has been associated with acute arthritis, mastitis, and pneumonia in cattle (29). Interestingly, the 14628 genome revealed the presence of a large prophage that is very similar to that described in the type strain of M. conjunctivae, a species causing keratoconjunctivitis in wild ungulates (5, 31). This finding was striking because no phage had ever been described in the species M. agalactiae, but also because the occurrence of phages in Mycoplasma species has rarely been reported. The circulation of the 14628 phage in ruminant mycoplasmas might not be restricted to M. agalactiae and M. conjunctivae, because the genome of M. bovigenitalium strain 51080 also displays a similar prophage. This Mycoplasma species, which has been documented mainly in genital tract disorders of domestic ruminants, has not yet attracted much interest, and its occurrence in wildlife cannot be ruled out. One interesting feature of the prophage is the presence of a sequence coding for an integrase-recombinase that is found in M. agalactiae and M. bovigenitalium but not in M. conjunctivae and could play a role either in prophage excision or in the reorganization of phage modules, as shown for some viruses (13). These findings suggest that the 14628 prophage has been transmitted horizontally across diverse Mycoplasma species that shared a ruminant host. Prophage sequences were found in 50% of M. agalactiae strains isolated from ibexes, including two carriage strains isolated from the ear canals of healthy animals. However, the presence of phage variants not detected by our assay cannot be ruled out. A circular, nonchromosomal intermediate of the phage was evidenced in several strains, strongly suggesting that it may be functional, at least for excision. Yet many CDSs carried by the prophage remain hypothetical, without any associated functions, and a putative role of the 14628 phage or related variants in M. agalactiae virulence has yet to be addressed.
Strain 14628, as well as other ibex strains. displays two phenotypic features that are usually ascribed to members of the “M. mycoides” cluster: a preferential lung tropism and a number of antigens that are recognized by M. mycoides-specific sera. As mentioned above, several Mycoplasma species cohabit in the domestic or wild ruminant host, and the likelihood that ibex M. agalactiae strains have horizontally acquired genetic material from the “M. mycoides” cluster was evaluated by in silico analyses of the 14628 genome. While HGT prediction clearly identified large mobile genetic elements, such as the prophage or the ICE initially identified in strain 5632, genes that were exchanged specifically with the “M. mycoides” cluster were very similar in number and functions to those previously predicted in strain PG2 or 5632 (22, 27). More-detailed, functional studies are needed to identify the genetic factors responsible for the M. mycoides-like features of the M. agalactiae strains from ibexes. In this effort, analysis of the M. mycoides subsp. capri strains that were isolated from the same ibex populations may be key.
Genomic analyses and molecular typing showed that M. agalactiae strains isolated from Alpine ibexes are very distinct from strains circulating in France and in Europe and, more specifically, from strains that had been collected from goats in the nearby Savoy area during previous CA outbreaks. Molecular data also clearly showed that the 20 wild-ungulate isolates studied here form a homogeneous group and differ from each other mainly by the contents and genomic locations of mobile genetic elements (or mobilomes) such as ICEs, suggesting an important plasticity of their genomes. Overall, our findings point toward the introduction of one strain, of yet unknown origin, that has adapted to the ibex host and has subsequently spread in the wild-ungulate population, resulting in a current M. agalactiae clonal lineage composed of isolates displaying a variety of mobilomes. As well, the data indicate that M. agalactiae isolates from two wild Caprinae species, ibex and chamois, are genetically closely related and are probably derived from a unique parent strain. Such interspecies transmission had already been observed with M. conjunctivae, a Mycoplasma species known to induce keratoconjunctivitis in Alpine wild ungulates (3). From an epidemiological perspective, our study shows the benefit of old and new molecular typing tools that can now be used to trace the M. agalactiae strains within the ibex population but also elsewhere. Optical mapping had never been used for mycoplasmas before except once, to validate the sequence assembly of the Mycoplasma haemofelis genome (25). In the present work, we confirmed the usefulness of the technique for this type of purpose and showed, in addition, that optical mapping is a good alternative to sequencing for defining the genetic relationship between two Mycoplasma isolates.
Overall, our study provides evidence for the emergence and dissemination in Alpine wild ungulates of atypical M. agalactiae strains that may be highly pathogenic in the ibex host. This finding further raises the question of the importance of wild fauna as reservoirs for bacterial pathogens and, more specifically, for pathogens, such as Mycoplasma species, listed by the OIE as representing a threat to livestock.
Supplementary Material
ACKNOWLEDGMENTS
Financial support was provided by the EVOLMYCO project (ANR-07-GMGE-001) from ANR to A. Blanchard (principal investigator [PI]), F. Thiaucourt (co-PI), F. Poumarat (co-PI), and C. Citti (co-PI) and by the MYCOIBEX project from Parc National de la Vanoise, Office National de la Chasse et de la Faune Sauvage, DDCSPP73, and Fédération Nationale de la Chasse, to F. Poumarat and F. Tardy (PIs).
We thank the biologists, veterinarians, game wardens, and park rangers who helped collect the samples. We also thank the VIGIMYC's partner laboratories and the technical team at Anses, Laboratoire de Lyon, for the supply and identification, respectively, of mycoplasma isolates from domestic ruminants. Finally, we are grateful to the “Sequencing Development” Group of the Genoscope (Centre National de Séquençage, Evry, France) for technical assistance in sequencing and assembly.
Footnotes
Published ahead of print 20 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aury JM, et al. 2008. High quality draft sequences for prokaryotic genomes using a mix of new sequencing technologies. BMC Genomics 9:603 doi:10.1186/1471-2164-9-603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barre A, de Daruvar A, Blanchard A. 2004. MolliGen, a database dedicated to the comparative genomics of Mollicutes. Nucleic Acids Res. 32:D307–D310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belloy L, et al. 2003. Molecular epidemiology of Mycoplasma conjunctivae in Caprinae: transmission across species in natural outbreaks. Appl. Environ. Microbiol. 69:1913–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bergonier D, Berthelot X, Poumarat F. 1997. Contagious agalactia of small ruminants: current knowledge concerning epidemiology, diagnosis and control. Rev. Sci. Tech. 16:848–873 [DOI] [PubMed] [Google Scholar]
- 5. Calderon-Copete SP, et al. 2009. The Mycoplasma conjunctivae genome sequencing, annotation and analysis. BMC Bioinformatics 10(Suppl. 6):S7 doi:10.1186/1471-2105-10-S6-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chazel M, Tardy F, Le Grand D, Calavas D, Poumarat F. 2010. Mycoplasmoses of ruminants in France: recent data from the national surveillance network. BMC Vet. Res. 6:32 doi:10.1186/1746-6148-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen WP, Kuo TT. 1993. A simple and rapid method for the preparation of gram negative bacterial genomic DNA. Nucleic Acids Res. 21:2260 doi:10.1093/nar/21.9.2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corrales JC, et al. 2007. Contagious agalactia in small ruminants. Small Ruminant Res. 68:154–166 [Google Scholar]
- 9. Frangeul L, et al. 2004. CAAT-Box, Contigs-Assembly and Annotation Tool-Box for genome sequencing projects. Bioinformatics 20:790–797 [DOI] [PubMed] [Google Scholar]
- 10. Giangaspero M, et al. 2010. Characterization of Mycoplasma isolated from an ibex (Capra ibex) suffering from keratoconjunctivitis in northern Italy. J. Wildl. Dis. 46:1070–1078 [DOI] [PubMed] [Google Scholar]
- 11. Gonzalez-Candela M, Verbisck-Bucker G, Martin-Atance P, Cubero-Pablo MJ, Leon-Vizcaino L. 2007. Mycoplasmas isolated from Spanish ibex (Capra pyrenaica hispanica): frequency and risk factors. Vet. Rec. 161:167–168 [DOI] [PubMed] [Google Scholar]
- 12. Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224 [DOI] [PubMed] [Google Scholar]
- 13. Groth AC, Calos MP. 2004. Phage integrases: biology and applications. J. Mol. Biol. 335:667–678 [DOI] [PubMed] [Google Scholar]
- 14. Maigre L, Citti C, Marenda M, Poumarat F, Tardy F. 2008. Suppression-subtractive hybridization as a strategy to identify taxon-specific sequences within the “M. mycoides” cluster: design and validation of a Mycoplasma capricolum subsp. capricolum-specific PCR assay. J. Clin. Microbiol. 46:1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marenda M, et al. 2006. A new integrative conjugative element occurs in Mycoplasma agalactiae as chromosomal and free circular forms. J. Bacteriol. 188:4137–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marenda MS, Sagne E, Poumarat F, Citti C. 2005. Suppression subtractive hybridization as a basis to assess Mycoplasma agalactiae and Mycoplasma bovis genomic diversity and species-specific sequences. Microbiology 151:475–489 [DOI] [PubMed] [Google Scholar]
- 17. McAuliffe L, et al. 2008. VNTR analysis reveals unexpected genetic diversity within Mycoplasma agalactiae, the main causative agent of contagious agalactia. BMC Microbiol. 8:193 doi:10.1186/1471-2180-8-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McAuliffe L, et al. 2011. Multilocus sequence typing of Mycoplasma agalactiae. J. Med. Microbiol. 60:803–811 [DOI] [PubMed] [Google Scholar]
- 19. Nicholas R, Ayling R, McAuliffe L. 2008. Mycoplasma diseases of ruminants, p 3–14 CABI, Wallingford, United Kingdom [Google Scholar]
- 20. Nouvel LX, et al. Molecular typing of Mycoplasma agalactiae: tracing European-wide genetic diversity and an endemic clonal population. Comp. Immunol. Microbiol. Infect. Dis., in press [DOI] [PubMed] [Google Scholar]
- 21. Nouvel LX, et al. 2009. Occurrence, plasticity, and evolution of the vpma gene family, a genetic system devoted to high-frequency surface variation in Mycoplasma agalactiae. J. Bacteriol. 191:4111–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nouvel LX, et al. 2010. Comparative genomic and proteomic analyses of two Mycoplasma agalactiae strains: clues to the macro- and micro-events that are shaping mycoplasma diversity. BMC Genomics 11:86 doi:10.1186/1471-2164-11-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poumarat F, Perrin B, Longchambon D. 1991. Identification of ruminant mycoplasma by dot-immunobinding on membrane filtration (MF dot). Vet. Microbiol. 29:329–338 [DOI] [PubMed] [Google Scholar]
- 24. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Santos AP, et al. 2011. Genome of Mycoplasma haemofelis, unraveling its strategies for survival and persistence. Vet. Res. 42:102 doi:10.1186/1297-9716-42-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sirand-Pugnet P, Citti C, Barre A, Blanchard A. 2007. Evolution of mollicutes: down a bumpy road with twists and turns. Res. Microbiol. 158:754–766 [DOI] [PubMed] [Google Scholar]
- 27. Sirand-Pugnet P, et al. 2007. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet. 3:e75 doi:10.1371/journal.pgen.0030075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tardy F, Maigre L, Poumarat F, Citti C. 2009. Identification and distribution of genetic markers in three closely related taxa of the Mycoplasma mycoides cluster: refining the relative position and frontiers of the Mycoplasma sp. bovine group 7 taxon (Mycoplasma leachii). Microbiology 155:3775–3787 [DOI] [PubMed] [Google Scholar]
- 30. Tardy F, Mercier P, Solsona M, Saras E, Poumarat F. 2007. Mycoplasma mycoides subsp. mycoides biotype large colony isolates from healthy and diseased goats: prevalence and typing. Vet. Microbiol. 121:268–277 [DOI] [PubMed] [Google Scholar]
- 31. Tschopp R, Frey J, Zimmermann L, Giacometti M. 2005. Outbreaks of infectious keratoconjunctivitis in alpine chamois and ibex in Switzerland between 2001 and 2003. Vet. Rec. 157:13–18 [DOI] [PubMed] [Google Scholar]
- 32. Verbisck G, et al. 2010. Mycoplasma agalactiae in Iberian ibex (Capra pyrenaica) in Spain. Vet. Rec. 167:425–426 [DOI] [PubMed] [Google Scholar]
- 33. Verbisck-Bucker G, et al. 2008. Epidemiology of Mycoplasma agalactiae infection in free-ranging Spanish ibex (Capra pyrenaica) in Andalusia, southern Spain. J. Wildl. Dis. 44:369–380 [DOI] [PubMed] [Google Scholar]
- 34. Vilei EM, et al. 2007. Validation and diagnostic efficacy of a TaqMan real-time PCR for the detection of Mycoplasma conjunctivae in the eyes of infected Caprinae. J. Microbiol. Methods 70:384–386 [DOI] [PubMed] [Google Scholar]
- 35. Zhou S, et al. 2004. Single-molecule approach to bacterial genomic comparisons via optical mapping. J. Bacteriol. 186:7773–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.