Abstract
We have previously characterized from Lactobacillus casei BL23 three α-l-fucosidases, AlfA, AlfB, and AlfC, which hydrolyze in vitro natural fucosyl-oligosaccharides. In this work, we have shown that L. casei is able to grow in the presence of fucosyl-α-1,3-N-acetylglucosamine (Fuc-α-1,3-GlcNAc) as a carbon source. Interestingly, L. casei excretes the l-fucose moiety during growth on Fuc-α-1,3-GlcNAc, indicating that only the N-acetylglucosamine moiety is being metabolized. Analysis of the genomic sequence of L. casei BL23 shows that downstream from alfB, which encodes the α-l-fucosidase AlfB, a gene, alfR, that encodes a transcriptional regulator is present. Divergently from alfB, three genes, alfEFG, that encode proteins with homology to the enzyme IIAB (EIIAB), EIIC, and EIID components of a mannose-class phosphoenolpyruvate:sugar phosphotransferase system (PTS) are present. Inactivation of either alfB or alfF abolishes the growth of L. casei on Fuc-α-1,3-GlcNAc. This proves that AlfB is involved in Fuc-α-1,3-GlcNAc metabolism and that the transporter encoded by alfEFG participates in the uptake of this disaccharide. A mutation in the PTS general component enzyme I does not eliminate the utilization of Fuc-α-1,3-GlcNAc, suggesting that the transport via the PTS encoded by alfEFG is not coupled to phosphorylation of the disaccharide. Transcriptional analysis with alfR and ccpA mutants shows that the two gene clusters alfBR and alfEFG are regulated by substrate-specific induction mediated by the inactivation of the transcriptional repressor AlfR and by carbon catabolite repression mediated by the catabolite control protein A (CcpA). This work reports for the first time the characterization of the physiological role of an α-l-fucosidase in lactic acid bacteria and the utilization of Fuc-α-1,3-GlcNAc as a carbon source for bacteria.
INTRODUCTION
Fucosyl-oligosaccharides (FUSs) are present in human milk, blood group antigens, mammalian cell surfaces, and intestinal mucin (5). They are involved in a variety of biological processes, such as tumor metastasis (21), inflammation (17), cell-cell adhesion (25), and colonization by symbiotic microbiota of the adult mammalian gut (10, 20). FUSs from human milk have been shown to exert antiadhesive activity toward pathogens, and they protected infants against diarrheal diseases (23). The adaptation of probiotic intestinal bacteria to the gastrointestinal tract of both infants and adults depends on several factors, including their ability to compete for the available substrates. Extensive research on the utilization of FUSs and other human milk oligosaccharides (HMOs) in Bifidobacterium species from the infant intestine has been performed (3, 38, 39). The HMOs 2′-fucosyllactose, 3′-fucosyllactose, and lacto-N-fucopentaose II are fermented by Bifidobacterium bifidum (3). The utilization of these FUSs is conditioned to the expression of two α-l-fucosidases, AfcA, which acts on α-1,2-linked fucoses, and AfcB, which hydrolyzes α-1,3- and α-1,4-fucosylated oligosaccharides (3). Moreover, the capacity to degrade intestinal mucin of two B. bifidum strains in vitro has been suggested to be related to the induction of two genes coding for extracellular glycosidases, afcA (1,2-α-l-fucosidase) and engBF (endo-α-N-acetylgalactosaminidase) in the presence of mucin (33). B. longum subsp. infantis carries gene clusters related to the utilization of glycans that encode the activities necessary for the hydrolysis of a variety of glycosidic linkages present in HMOs, including α-l-fucosidases (39).
Several species of Lactobacillus, another group of probiotic microorganisms, are common inhabitants of the human gastrointestinal tract (29) and have also been isolated from human milk (18). Although genome analysis of intestinal strains revealed the particular adaptation to this niche (4, 8, 13), their capacity to exploit the carbohydrate resources of the intestinal mucosa seems to be limited. The Lactobacillus casei species is widely used in the dairy industry as a starter culture, and some strains have been applied in functional foods as probiotics. L. casei strain BL23 showed immunomodulatory probiotic properties (11, 31) and is used extensively for physiological and genetic studies (15, 24, 30, 36). This strain utilized carbohydrates very efficiently by the phosphoenolpyruvate (PEP):sugar phosphotransferase system (PTS) (1, 44, 45). This consists of the general phosphotransferase proteins enzyme I (EI) and HPr (43) and carbohydrate-specific transporters (EII). EI autophosphorylates at a histidine residue by using PEP and then transfers the phosphoryl group to HPr, which becomes phosphorylated at the conserved histidine-15 residue (P-His-HPr). P-His-HPr functions as a phosphoryl donor for the different PTS transporters, which consist of three or four different proteins or domains: the cytoplasmic domains EIIA and EIIB and the transmembrane transporters EIIC and EIID (28).
We have recently identified and purified from L. casei BL23 three α-l-fucosidases (AlfA, AlfB, and AlfC) that cleaved α-linked l-fucose from natural FUSs (32). AlfA hydrolyzed only fucosyl-α-1,6-N-acetylglucosamine (Fuc-α-1,6-GlcNAc) with low specific activity, while AlfB and AlfC act on a few FUSs, showing the maximum activity on fucosyl-α-1,3-N-acetylglucosamine (Fuc-α-1,3-GlcNAc) and Fuc-α-1,6-GlcNAc, respectively. However, their real substrates were unknown. With the aim of studying the physiological role of the alf genes, we screened L. casei BL23 for its ability to use several FUSs. We found that Fuc-α-1,3-GlcNAc can be fermented by this strain. We have demonstrated that the clusters alfBR, encoding AlfB and the transcriptional repressor AlfR, and alfEFG, encoding the EII components of a mannose-class PTS transporter, are involved in the utilization of this disaccharide.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
The strains and plasmids used in this study are listed in Table 1. The Lactobacillus casei strains were routinely grown at 37°C under static conditions on MRS medium (Difco). L. casei wild type was also grown on MRS fermentation medium (Scharlau), which contains chlorophenol red as a pH indicator, supplemented with 0.5% l-fucose. Escherichia coli, which was used as the host in cloning experiments, was grown in Luria-Bertani medium at 37°C (Oxoid). The corresponding solid media were prepared by adding 1.8% agar. E. coli transformants were selected with ampicillin (100 μg/ml), and L. casei transformants were selected with erythromycin (5 μg/ml).
Table 1.
Strains and plasmids used in this studya
| Strain or plasmid | Relevant genotype or properties | Source or reference |
|---|---|---|
| Strains | ||
| Lactobacillus casei | ||
| BL23 | Wild type | CECT 5275 |
| BL126 | BL23 ptsI | 43 |
| BL71 | BL23 ccpA | 22 |
| BL370 | BL23 alfD::pRV300 Ermr | This work |
| BL371 | BL23 alfF::pRV300 Ermr | This work |
| BL372 | BL23 alfH::pRV300 Ermr | This work |
| BL373 | BL23 alfB (frameshift at SphI site) | This work |
| BL374 | BL23 alfR::pRV300 Ermr | This work |
| Escherichia coli DH10B | F− endA1 recA1 galE15 galK16 nupG rpsL ΔlacX74 ϕ80lacZΔM15 araD139 Δ(ara leu)7697mcrA Δ(mrr-hsdRMS-mcrBC) λ− | Invitrogen |
| Plasmids | ||
| pRV300 | Suicide vector carrying Ermr from pAMβ1 | 16 |
| pRValfD | pRV300 with a 0.6-kb alfD fragment | This work |
| pRValfF | pRV300 with a 0.45-kb alfF fragment | This work |
| pRValfH | pRV300 with a 0.6-kb alfH fragment | This work |
| pRValfB | pRV300 with a 0.9-kb alfB fragment | This work |
| pRValfBSphI | pRV300 with a frameshift at SphI site in alfB | This work |
| pRValfR | pRV300 with a 0.45-kb alfR fragment | This work |
CECT, Colección Española de Cultivos Tipo; Ermr, erythromycin resistance.
Vector pRV300 (16) was used for cloning experiments with E. coli and for insertional inactivation of genes in L. casei. E. coli and L. casei strains were transformed by electroporation with a Gene Pulser apparatus (Bio-Rad Laboratories) as recommended by the manufacturer (E. coli) or described earlier (L. casei) (27).
Culture of L. casei strains with fucosyl-oligosaccharides.
The L. casei strains were grown overnight at 37°C under static conditions on sugar-free MRS medium containing Bacto peptone (Difco), 10 g/liter; yeast extract (Pronadisa), 4 g/liter; sodium acetate, 5 g/liter; triammonium citrate, 2 g/liter; magnesium sulfate 7-hydrate, 0.2 g/liter; manganese sulfate monohydrate, 0.05 g/liter; and Tween 80, 1 ml/liter. Overnight cultures were diluted to an optical density at 550 nm (OD550) of 0.05 in 100 μl of MRS medium containing 2 mM fucosyl-oligosaccharides (antigen H disaccharide, antigen H type II trisaccharide, 2′-fucosyl-lactose, 3′-fucosyl-lactose, Lewis x trisaccharide, Lewis a trisaccharide, Lewis b trisaccharide, fucosyl-α-1,3-N-acetylglucosamine, fucosyl-α-1,4-N-acetylglucosamine, fucosyl-α-1,6-N-acetylglucosamine, or fucosyl-α-1,4-galactose). All these sugars were obtained from Carbosynth (Compton, Berkshire, United Kingdom). Bacterial growth was monitored over 22.5 h by spectrophotometric measurements every 30 min at 550 nm in 96-well plates at 37°C without shaking in a POLARstar Omega microplate reader (BMG Labtech, Offenburg, Germany). At least three independent biological replicates for each growth curve were obtained. Results were expressed as means ± standard deviations. The growth rates (μ) were calculated by using the Gompertz model (GraphPad Software, San Diego, CA).
DNA manipulation, oligonucleotides, and sequencing.
The oligonucleotides used in this study are listed in Table S1 in the supplemental material. Total DNA was isolated from L. casei BL23 as described before (27). Recombinant DNA techniques were performed by following standard procedures (34). All PCRs were performed with an Expand High Fidelity PCR system (Roche). DNA sequencing was carried out by the Central Service of Research Support of the University of Valencia (Spain). M13 universal and reverse primers or custom primers hybridizing within the appropriate DNA fragments were used for sequencing. Sequence analyses were carried out with DNAMAN (version 4.0) for Windows (Lynnon BioSoft), and sequence similarities were analyzed with the BLAST program (2).
Construction of recombinant strains.
Internal DNA fragments of alfD, alfF, alfH, and alfR were obtained by PCR using L. casei BL23 chromosomal DNA and the oligonucleotides pairs alfAper1/alfAper2, alfBmanC1/alfBmanC2, alfCper1/alfCper2, and regFucB1/regFucB2, respectively. The PCR products were cloned into pRV300 digested with EcoRV. The resulting plasmids, pRValfD, pRValfF, pRValfH, and pRValfR, respectively, were used to transform L. casei BL23, and single-crossover integrants were selected by resistance to erythromycin and confirmed by PCR. This was performed with primers pairs universal M13 reverse/qAlfArv (alfD), universal M13 reverse/qAlfBrv (alfF), universal M13 forward/qAlfCrv (alfH), and universal M13 reverse/qAlfBfw (alfR). One mutant with a mutation of each gene was selected, and the mutants were named BL370 (alfD), BL371 (alfF), BL372 (alfH), and BL374 (alfR), respectively. To construct an alfB mutant, a DNA fragment containing part of alfB was obtained by PCR using L. casei BL23 chromosomal DNA and the primers IIABfucB1 and fucB1. This DNA fragment was cloned into pRV300 digested with EcoRV, giving pRValfB. This plasmid was cleaved at the unique SphI restriction site present in the alfB coding region, treated with the Klenow fragment of DNA polymerase I, ligated, and transformed. One plasmid-containing clone in which a frameshift was introduced at the SphI site in alfB (pRValfBSphI) was selected. L. casei was transformed with this plasmid, and one erythromycin-resistant clone carrying the plasmid integrated by a single crossover was grown in MRS medium without erythromycin for 200 generations. Cells were plated on MRS medium and replica plated on MRS medium plus erythromycin. Antibiotic-sensitive clones were isolated, and among them, one clone (the BL373 strain) in which a second recombination event led to the excision of the plasmid, leaving a mutated alfB copy, as was confirmed by sequence analysis of appropriate PCR products, was selected.
Oligosaccharide and monosaccharide analysis.
To determine the carbohydrates present in the supernatants from the L. casei cultures, the cells were removed by centrifugation and the cultures were analyzed by high-pH anion-exchange chromatography with pulsed amperometric detection in an ICS3000 chromatographic system (Dionex) using a CarboPac PA100 column (Dionex). A combined gradient of 100 to 300 mM NaOH and 0 to 150 mM acetic acid was used for 20 min at a flow rate of 1 ml/min. Oligo- and monosaccharides were confirmed by comparison of their retention times with those of standards.
RNA isolation and RT-qPCR.
For RNA isolation, strains were grown at 37°C under static conditions in 100 μl MRS fermentation medium containing 2 mM different sugars until mid-log phase was reached. The inocula for these cultures (starting OD, 0.05) were cells pregrown in MRS fermentation medium without sugar added. Cells were collected by centrifugation, washed with 1 ml of 50 mM EDTA (pH 8), and resuspended in 1 ml of TRIzol (Gibco). One gram of 0.1-mm glass beads was added, and cells were broken with a Mini-BeadBeater apparatus (Biospec Products, Bartlesville, OK). RNA was isolated as described by the manufacturers of TRIzol. One hundred nanograms of RNA was digested with DNase I, RNase free (Fermentas), and 50 ng of digested RNA was retrotranscribed using a Maxima First Strand cDNA synthesis kit (Fermentas) following manufacturer instructions. The resulting cDNA was subjected to quantitative PCR for the genes alfR, alfB, alfE, alfF, alfG, alfA, and alfC. Reverse transcriptase quantitative PCR (RT-qPCR) was performed using a LightCycler (version 2.0) system (Roche) and LC Fast Start DNA Master SYBR green I (Roche). Primers were designed by using the Primer-BLAST service (http://www.ncbi.nlm.nih.gov/tools/primer-blast) in order to generate amplicons ranging from 70 to 100 bp in size (see Table S1 in the supplemental material). RT-qPCR was performed for each cDNA sample in triplicate and by using the primers pairs qRegFucBfw/qRegFucBrv (alfR), qAlfBfw/qAlfBrv (alfB), qIIABfucBfw/qIIABfucBrv (alfE), qIICfucBfw/qIICfucBrv (alfF), qIIDfucBfw/qIIDfucBrv (alfG), qAlfAfw/qAlfArv (alfA), and qAlfCfw/qAlfCrv (alfC). The reaction mixture (10 μl) contained 5 μl of 2× master mix, 0.5 μl of each primer (10 μM), and 1 μl of a 20-times-diluted sample from the cDNA synthesis reaction. Reaction mixtures without a template were run as controls. The cycling conditions were as follows: 95°C for 10 min, followed by 45 cycles of three steps consisting of denaturation at 95°C for 10 s, primer annealing at 60°C for 20 s, and primer extension at 72°C for 20 s. For each set of primers, the cycle threshold (crossing point [CP]) values were determined by the automated method implemented in the LightCycler software (version 4.0; Roche). The pyrG gene was selected as the reference gene on the basis of previous studies of the group (15). The relative expression, based on the expression ratio between the target genes and reference genes, was calculated using the software tool REST (relative expression software tool) (26). Linearity and amplification efficiency were determined for each primer pair. Every RT-qPCR was performed at least in triplicate with two biologically independent samples.
RESULTS
Growth of L. casei BL23 in media containing fucosyl-oligosaccharides.
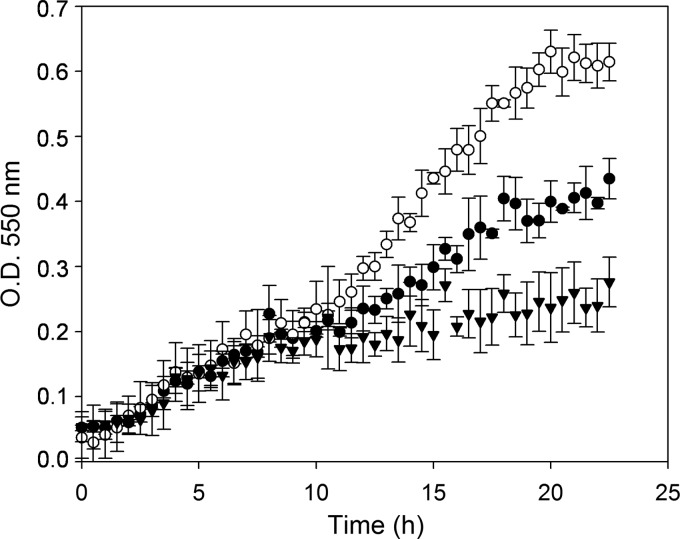
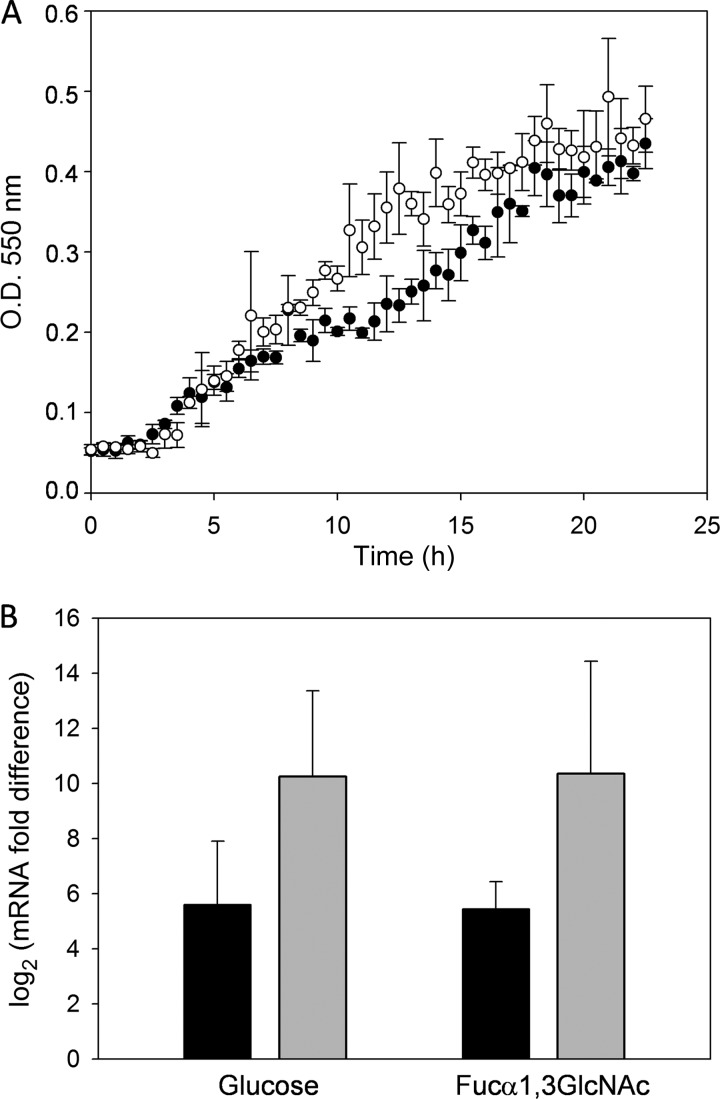
In order to determine the ability of L. casei BL23 to utilize FUSs as a carbon and energy source, the growth pattern in MRS medium supplemented with an FUS at 2 mM was followed. From the 11 FUS structures assayed (see Materials and Methods), L. casei BL23 was able to grow only in the presence of fucosyl-α-1,3-N-acetylglucosamine (Fuc-α-1,3-GlcNAc) (Fig. 1). The rest of the FUSs tested did not allow any significant growth compared with the growth in MRS medium without added carbohydrates (data not shown). To confirm that Fuc-α-1,3-GlcNAc was fermented by L. casei, the supernatants of the cultures were analyzed for sugar content. Surprisingly, while Fuc-α-1,3-GlcNAc was exhausted at the stationary phase of the bacterial growth curve, at this point, l-fucose was found in the supernatant at 2 mM. This suggested that the disaccharide was split by an α-l-fucosidase, and only the N-acetylglucosamine (GlcNAc) moiety was metabolized. This finding was also in agreement with the inability of L. casei BL23 to ferment l-fucose in MRS medium containing 0.5% of this sugar (data not shown). The three α-l-fucosidases seem to be present intracellularly since they do not have any predicted signal peptide. Then, the most plausible hypothesis was that Fuc-α-1,3-GlcNAc was taken up and hydrolyzed, and the resulting and nonmetabolizable l-fucose was excreted from the cells. The growth pattern of L. casei in the presence of lactose as a positive control is also shown in Fig. 1. The OD550 values reached by L. casei with lactose are higher than those reached with Fuc-α-1,3-GlcNAc, which is in accordance with the sole utilization of the GlcNAc moiety of Fuc-α-1,3-GlcNAc, while both galactose and glucose from lactose are metabolized by L. casei.
Fig 1.
Growth curves of Lactobacillus casei BL23 on MRS fermentation medium without a carbon source (▼) or with lactose (○) or fucosyl-α-1,3-N-acetylglucosamine (●) as the only carbon source. Data presented are mean values based on at least three replicates. Error bars indicate standard deviations.
Identification of the genes responsible for Fuc-α-1,3-GlcNAc utilization in L. casei BL23.
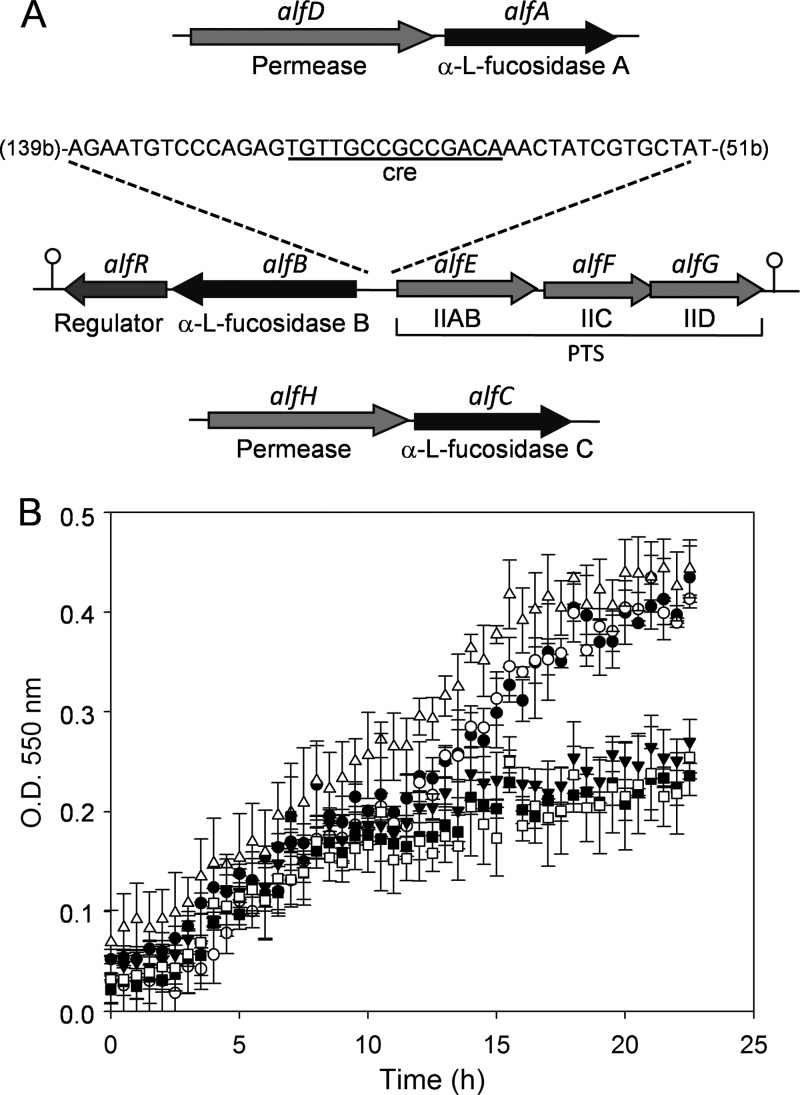
The capacity to hydrolyze natural FUSs in vitro has recently been shown for three α-l-fucosidases (AlfA, AlfB, and AlfC) cloned from L. casei BL23 (32). The chromosomal regions of the genes alfA, alfB, and alfC, encoding the three α-l-fucosidases, respectively, were analyzed using the genomic sequence of L. casei BL23 (GenBank accession no. FM177140) (19) (Fig. 2A). A gene (LCABL_20380), named here alfD, that encodes a protein homologous to putative carbohydrate symporters is present upstream from alfA. Downstream from alfB, alfR (LCAB_28260) is present. alfR encodes a protein with homology to putative transcriptional regulators, showing a highly conserved winged helix-turn-helix (WHTH) DNA-binding domain of the GntR family at the N terminus. Divergently from alfB, three genes (LCAB_28280, LCAB_28290, and LCAB_28300), designated alfEFG, encode proteins with homology to the EIIAB, EIIC, and EIID components of the mannose-class PTS transporters, respectively. Two putative rho-independent terminators were identified, one downstream from alfR (ΔG, −17.8 kcal/mol) and another downstream from alfG (ΔG, −25.3 kcal/mol). A gene (LCABL_29330), named alfH, that encodes a protein homologous to putative permeases of the major facilitator superfamily is present upstream from alfC. As the three α-l-fucosidases seem to be intracellular enzymes, the putative permeases present in the genetic surroundings of the genes coding for the α-l-fucosidases could have a role in the transport of Fuc-α-1,3-GlcNAc. To determine if these permeases are involved in the utilization of Fuc-α-1,3-GlcNAc in L. casei BL23, disruptions of alfD (BL370), alfF (BL371), and alfH (BL372) were introduced into BL23 by integration of the nonreplicative plasmid pRV300 (16) carrying an internal fragment of the respective genes. The growth patterns of the three mutants were compared to the pattern of the wild type by measuring OD550 values in MRS medium with Fuc-α-1,3-GlcNAc as the carbon source (Fig. 2B). The results showed that the growth patterns of BL370 (alfD) and BL372 (alfH) were similar to the pattern of the wild type; however, BL371 (alfF) exhibited a growth rate comparable to that of the control (nonsupplemented MRS medium). Sugar content analysis of the culture supernatants detected l-fucose in the supernatants from BL370 (alfD) and BL372 (alfH) cultures but detected Fuc-α-1,3-GlcNAc in the supernatants from BL371 (alfF). These results demonstrated that alfF, which encodes a specific EIIC domain of the PTS, participates in the uptake of Fuc-α-1,3-GlcNAc. In order to analyze the presumed involvement of the PTS from L. casei BL23 in the fermentation of this disaccharide, the growth pattern of BL126 (ptsI), a mutant deficient in the PTS general component enzyme I (43), was tested in Fuc-α-1,3-GlcNAc as the only carbon source. BL126 (ptsI) presented a growth rate similar to that of the wild type (data not shown), suggesting that the utilization of Fuc-α-1,3-GlcNAc is independent of enzyme I of the PTS. Lactose was used as a control for sugar uptake dependent on enzyme I (43), and it was observed that BL126 is not able to grow in the presence of this sugar (data not shown).
Fig 2.
(A) Schematic representation of the chromosomal regions of the genes alfA, alfB, and alfC encoding the three α-l-fucosidases of Lactobacillus casei BL23. The cre-like sequence is shown in the alfB-alfE intergenic region. Numbers in parentheses indicate distances (in nucleotides) from the alfB and alfE start codons. Hairpin loops indicate putative rho-independent transcriptional terminators. PTS, phosphoenolpyruvate:sugar phosphotransfrase system. (B) Growth curves of Lactobacillus casei strains BL23 (●), BL370 (alfD) (○), BL371 (alfF) (▼), BL372 (alfH) (Δ), and BL373 (alfB) (□) on MRS fermentation medium with fucosyl-α-1,3-N-acetylglucosamine as the only carbon source and BL371 (alfF) (■) on MRS fermentation medium without a carbon source. Data presented are mean values based on at least three replicates. Error bars indicate standard deviations.
As mentioned above, the alfEFG cluster is placed in the chromosome divergently from the alfBR cluster, suggesting that α-l-fucosidase B is involved in the hydrolysis of Fuc-α-1,3-GlcNAc by L. casei BL23. To prove this hypothesis, a mutant with a frameshift in alfB was constructed (strain BL373). In this strain, no plasmid remains integrated in the gene, and therefore, minor effects on the downstream alfR gene are expected. This mutant lost the ability to grow with Fuc-α-1,3-GlcNAc as the only carbon source (Fig. 2B), suggesting that α-l-fucosidase B is necessary for its hydrolysis, which would release the GlcNAc moiety required for obtaining energy.
Transcriptional analyses of the alf genes in L. casei.
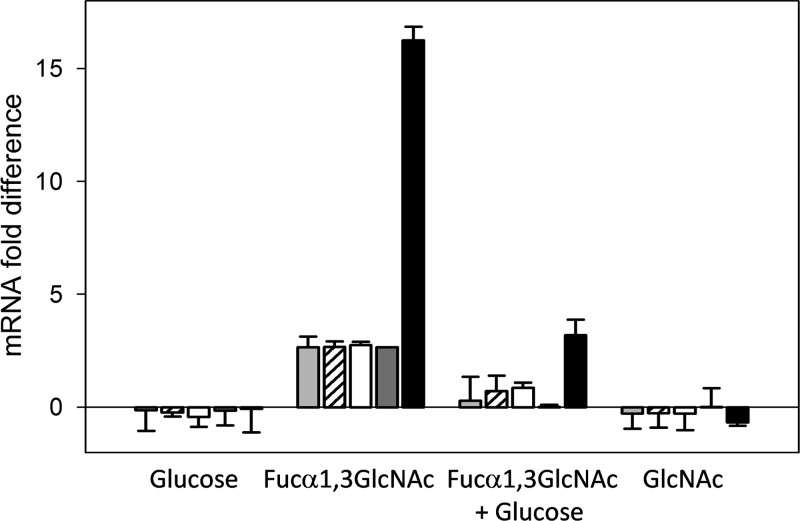
In order to determine whether the transcription of the alf genes was regulated by the carbon source, RNA was isolated from L. casei BL23 grown in MRS medium containing glucose, Fuc-α-1,3-GlcNAc, glucose plus Fuc-α-1,3-GlcNAc, or GlcNAc and used for RT-qPCR analyses (Fig. 3). Taking as a reference the transcript levels in cells grown in MRS medium without a carbon source, the alfB gene in L. casei BL23 is not expressed in the presence of glucose. However, its expression increased about 16-fold in the presence of Fuc-α-1,3-GlcNAc. This expression level was reduced in a mix of glucose and Fuc-α-1,3-GlcNAc. These results indicated that alfB is induced by this disaccharide and subjected to carbon catabolite repression (CCR) by glucose. The genes alfA and alfC, encoding the other two α-l-fucosidases present in the L. casei genome, were not induced in the presence of Fuc-α-1,3-GlcNAc (data not shown). Similar to alfB, the genes alfR, alfE, alfF, and alfG were upregulated by Fuc-α-1,3-GlcNAc and repressed by glucose, although the induction levels were lower (Fig. 3). In the presence of GlcNAc, none of the tested alf genes were upregulated, demonstrating that the disaccharide and not the monosaccharide resulting from the hydrolysis is responsible for the induction of alf genes.
Fig 3.
Transcriptional analysis by RT-qPCR of the expression of alfE (light gray bars), alfF (striped bars), alfG (white bars), alfR (dark gray bars), and alfB (black bars) in Lactobacillus casei BL23 grown in MRS fermentation medium containing glucose, fucosyl-α-1,3-N-acetylglucosamine (Fucα1,3GlcNAc), a mix of Fuc-α-1,3-GlcNAc and glucose, or N-acetylglucosamine (GlcNAc). Cells grown in MRS fermentation medium without a carbon source were used as a reference. Data presented are mean values based on three replicates of at least two biologically independent samples. Error bars indicate standard deviations.
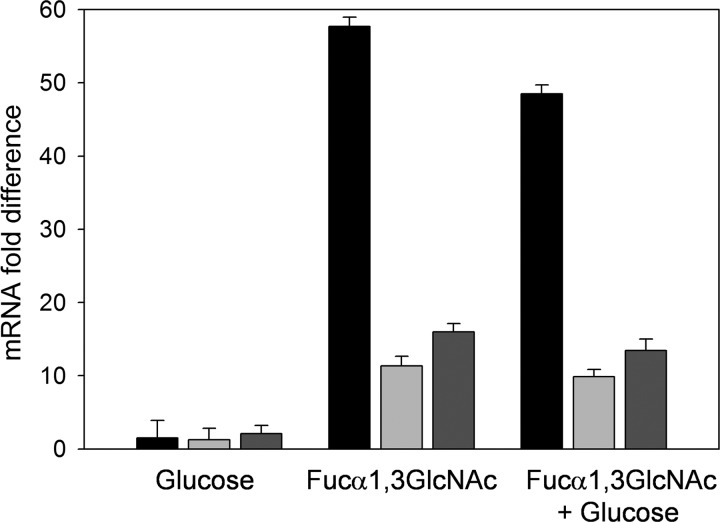
A DNA sequence (TGTTGCCGCCGACA) matching the consensus motif of a catabolite-responsive element (cre), the binding site of the CcpA carbon catabolite control protein in Gram-positive bacteria (41), was found in the alfB-alfE intergenic region (Fig. 2A), suggesting that CCR of the alfEFG cluster was mediated via the PTS/CcpA pathway (9). In order to test this, RNA was isolated from L. casei strain BL71, a mutant with inactivated ccpA (22). As shown in Fig. 4, the genes alfB, alfR, and alfE provided relief from CCR in this strain, with repression factors being close to 1 for the three genes when cells were grown in a mixture of glucose plus Fuc-α-1,3-GlcNAc. This demonstrated that the CCR effect on alfBR and alfEFG clusters occurs via CcpA.
Fig 4.
Transcriptional analysis by RT-qPCR of the expression of alfB (black bars), alfE (light gray bars), and alfR (dark gray bars) in Lactobacillus casei BL71 (ccpA) grown in MRS fermentation medium containing glucose, fucosyl-α-1,3-N-acetylglucosamine (Fucα1,3GlcNAc), or a mix of Fuc-α-1,3-GlcNAc and glucose. Cells grown in MRS fermentation medium without a carbon source were used as a reference. Data presented are mean values based on three replicates of at least two biologically independent samples. Error bars indicate standard deviations.
alfR codes for a transcriptional repressor.
To analyze the participation of AlfR in the regulation of Fuc-α-1,3-GlcNAc utilization in L. casei BL23, a mutant, BL374 (alfR), was constructed by insertional inactivation of the gene alfR with the plasmid pRV300. In MRS medium supplemented with Fuc-α-1,3-GlcNAc, this mutant showed a growth rate (0.031 h−1) higher than that of the wild type (0.017 h−1), although the final OD reached by both strains was similar (Fig. 5A). Taking as a reference the transcript levels in BL23 wild-type cells, transcription of alfB and alfE genes in the BL374 (alfR) mutant occurred at high levels in the presence of glucose and also in the presence of Fuc-α-1,3-GlcNAc (Fig. 5B), indicating that AlfR is a transcriptional repressor.
Fig 5.
(A) Growth curves of Lactobacillus casei BL23 (●) and BL374 (alfR) (○) on MRS fermentation medium with fucosyl-α-1,3-N-acetylglucosamine as the only carbon source. Data presented are mean values based on at least three replicates. Error bars indicate standard deviations. (B) Transcriptional analysis by RT-qPCR of the expression of alfB (black bars) and alfE (gray bars) in L. casei BL374 (alfR) grown in MRS fermentation medium containing glucose or fucosyl-α-1,3-N-acetylglucosamine (Fucα1,3GlcNAc). L. casei BL23 was used as the reference strain. Data presented are mean values based on three replicates of at least two biologically independent samples. Error bars indicate standard deviations.
DISCUSSION
The disaccharide Fuc-α-1,3-GlcNAc forms part of the Lewis x antigen core, which is found in many human glycoproteins at mucosal surfaces (14). In this work, we demonstrated that L. casei BL23 is able to grow in the presence of Fuc-α-1,3-GlcNAc as a carbon source, releasing the l-fucose moiety into the environment and metabolizing the GlcNAc. We have determined that the α-l-fucosidase AlfB is involved in the hydrolysis of Fuc-α-1,3-GlcNAc and that the PTS encoded by the adjacent alfEFG cluster is necessary for the metabolism of the disaccharide. The regulation of the alfBR and alfEFG clusters is dual and comprises induction by Fuc-α-1,3-GlcNAc mediated by the transcriptional repressor AlfR and CCR involving the CcpA global regulator (see Fig. S1 in the supplemental material). The fact that in an alfR mutant strain the induction levels of alfB and alfE were similar for glucose- and Fuc-α-1,3-GlcNAc-grown cells indicates that regulation by AlfR is superimposed onto CCR. The signal triggering AlfR inactivation is unknown, but it could probably be the presence of intracellular Fuc-α-1,3-GlcNAc, as growth on its derived metabolite (GlcNAc) did not result in gene induction. The effect of the carbohydrate source on expression of α-l-fucosidases has been investigated in B. longum subsp. infantis ATCC 15697. No FUS was identified to be a specific inducer, and only the enzyme encoded by Blon_2335 was induced 2-fold by growth with HMOs. In addition, for some of the B. longum subsp. infantis α-l-fucosidase genes (Blon_0248 and Blon_0426), a CCR effect that was probably activated by the most abundant HMOs, such as lacto-N-tetraose, was reported (39).
Despite the fact that numerous Lactobacillus species colonize the gastrointestinal tract and other human and vertebrate mucous membranes, analysis of the available genome sequences of lactobacilli revealed that genes encoding α-l-fucosidases are present only in the L. casei/Lactobacillus paracasei/Lactobacillus rhamnosus group. L. casei LC2W (GenBank accession no. CP002618), L. casei BD-II (GenBank accession no. CP002616), L. rhamnosus GG (GenBank accession no. FM179322), and L. rhamnosus HN001 (GenBank accession no. ABWJ00000000) contain homologues to alfB, alfR, alfE, alfF, and alfG which display the same cluster organization that they do in L. casei BL23. The predicted amino acid sequences of the five genes present in the LC2W and BD-II strains exhibit 100% identity to the BL23 sequence, whereas in those of L. rhamnosus strains, identity was between 75% and 95%. Thus, among probiotic microorganisms, only that group, together with some species of bifidobacteria (3, 39), appears to be adapted to metabolize FUSs. In the case of the BL23 strain, up to three α-l-fucosidases clustered with different transport systems can be found. The natural substrates for the two additional AlfA and AlfC α-l-fucosidases from BL23 remain to be determined. Even if AlfC can efficiently cleave Fuc-α-1,6-GlcNAc (32), the BL23 strain does not utilize this FUS under our experimental conditions, suggesting that the physiological substrate for this enzyme is different.
The α-l-fucosidases AfcA and AfcB from Bifidobacterium bifidum are extracellular membrane-bound enzymes (3). However, the five α-l-fucosidases encoded in the Bifidobacterium longum genome seem to be intracellular, since they lack a signal peptide at the N terminus (39). Similarly, all the evidence suggests that AlfB acts intracellularly in L. casei. The deduced amino acid sequence does not contain any identifiable N-terminal sequence for secretion. In addition, Fuc-α-1,3-GlcNAc remained nonhydrolyzed in the culture medium from the alfF mutant (BL371), suggesting that the EII complex internalized the disaccharide into the L. casei cells. Surprisingly, the mutant deficient in the PTS general component enzyme I (BL126) (43) presented a growth rate on Fuc-α-1,3-GlcNAc similar to that of the wild type, suggesting that its utilization is independent of enzyme I. A BLAST search using the deduced amino acid sequence of enzyme I against the genomic sequence of L. casei showed relevant homology (28% identity) only with the putative pyruvate phosphate dikinase, and it is unlikely that it acts as an enzyme I. Therefore, no other enzyme I paralogues are encoded in the BL23 genome. This observation, together with the fact that AlfB hydrolyzed nonphosphorylated FUSs (32), suggested that the specific EII complex encoded by alfEFG might transport Fuc-α-1,3-GlcNAc without phosphorylating it. The internalization of a sugar by the PTS in a phosphorylation-independent manner has previously been shown in lactobacilli. The mannose-specific EII complex of Lactobacillus pentosus, Lactobacillus plantarum, and L. casei can transport d-xylose by a mechanism of facilitated diffusion (7).
L. casei BL23 uses only the GlcNAc moiety of Fuc-α-1,3-GlcNAc for growth, which is in agreement with the existence of genes probably involved in its catabolism in the genome, such as LCABL_20280, coding for a GlcNAc-6-P deacetylase, or LCABL_31070, encoding a presumed glucosamine-6-phosphate deaminase. The l-fucose moiety was not utilized by L. casei, and it appeared quantitatively in the supernatants. In agreement with this, no l-fucose catabolic genes are found in the BL23 genome. In addition, no growth with l-fucose was observed for several Lactobacillus strains (37), and although L. rhamnosus GG was able to metabolize mucin, it was unable to utilize l-fucose (35). The l-fucose moiety in L. casei BL23 was excreted into the growth medium by a yet unknown mechanism that could be mediated by the specific EII complex encoded by alfEFG or some other uncharacterized permease(s). In analogy, most strains of Streptococcus thermophilus are unable to metabolize galactose, and it is expelled from the cells when grown in the presence of lactose. In this case, an exchange reaction mediated by the transport protein LacS, acting as an antiporter system, occurs between intracellular formed galactose and extracellular lactose (12, 42).
L. casei is a common resident of the gastrointestinal tracts of humans and animals (6), and the alfBR and alfEFG system could constitute a system for scavenging mucosa-derived carbohydrates. In order to exploit this carbon source, L. casei must rely on the glycosidic hydrolases of others members of the microbiota, which would cleave host glycans and release FUSs that can be transported and cleaved by its intracellular α-l-fucosidases. This is the case for other intestinal microorganisms which lack the required hydrolases for mucin degradation and harvest monosaccharides generated by the action of glycosylhydrolases from other bacteria (40). This is the first time that an α-l-fucosidase and an associated transport system from a probiotic bacterium have been linked to the utilization of a particular FUS. It exemplifies the versatility of L. casei in sugar utilization and probably represents an adaptation for development at the intestinal mucosal niche.
Supplementary Material
ACKNOWLEDGMENTS
This work was financed by funds of the Spanish Ministry for Science and Innovation (MICINN)/FEDER through projects AGL2010-18696 and Consolider Fun-c-Food CSD2007-00063 and of the Valencian Government through project GV/2011/079. J.R.-D. was supported by a JAE-doc contract from CSIC.
We thank Vicente Monedero for fruitful discussions and for critical reading of the manuscript.
Footnotes
Published ahead of print 27 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alcantara C, et al. 2008. Regulation of Lactobacillus casei sorbitol utilization genes requires DNA-binding transcriptional activator GutR and the conserved protein GutM. Appl. Environ. Microbiol. 74:5731–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Ashida H, et al. 2009. Two distinct alpha-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19:1010–1017 [DOI] [PubMed] [Google Scholar]
- 4. Azcarate-Peril MA, et al. 2008. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl. Environ. Microbiol. 74:4610–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Becker DJ, Lowe JB. 2003. Fucose: biosynthesis and biological function in mammals. Glycobiology 13:41R–53R doi:10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 6. Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL. 2009. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol. Evol. 1:239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaillou S, Pouwels PH, Postma PW. 1999. Transport of d-xylose in Lactobacillus pentosus, Lactobacillus casei, and Lactobacillus plantarum: evidence for a mechanism of facilitated diffusion via the phosphoenolpyruvate:mannose phosphotransferase system. J. Bacteriol. 181:4768–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cho YJ, et al. 2011. Genome sequence of Lactobacillus salivarius GJ-24, a probiotic strain isolated from healthy adult intestine. J. Bacteriol. 193:5021–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. 2009. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137:321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foligne B, et al. 2007. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J. Gastroenterol. 13:236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hutkins RW, Ponne C. 1991. lactose uptake driven by galactose efflux in Streptococcus thermophilus: evidence for a galactose-lactose antiporter. Appl. Environ. Microbiol. 57:941–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kankainen M, et al. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106:17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. King MJ. 1994. Blood group antigens on human erythrocytes—distribution, structure and possible functions. Biochim. Biophys. Acta 1197:15–44 [DOI] [PubMed] [Google Scholar]
- 15. Landete JM, et al. 2010. Requirement of the Lactobacillus casei MaeKR two-component system for l-malic acid utilization via a malic enzyme pathway. Appl. Environ. Microbiol. 76:84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leloup L, Ehrlich SD, Zagorec M, Morel-Deville F. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levander L, Gunnarsson P, Grenegard M, Ryden I, Pahlsson P. 2009. Effects of α1-acid glycoprotein fucosylation on its Ca2+ mobilizing capacity in neutrophils. Scand. J. Immunol. 69:412–420 [DOI] [PubMed] [Google Scholar]
- 18. Martin R, et al. 2003. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 143:754–758 [DOI] [PubMed] [Google Scholar]
- 19. Maze A, et al. 2010. Complete genome sequence of the probiotic Lactobacillus casei strain BL23. J. Bacteriol. 192:2647–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meng D, et al. 2007. Bacterial symbionts induce a FUT2-dependent fucosylated niche on colonic epithelium via ERK and JNK signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 293:G780–G787 [DOI] [PubMed] [Google Scholar]
- 21. Misonou Y, et al. 2009. Comprehensive clinico-glycomic study of 16 colorectal cancer specimens: elucidation of aberrant glycosylation and its mechanistic causes in colorectal cancer cells. J. Proteome Res. 8:2990–3005 [DOI] [PubMed] [Google Scholar]
- 22. Monedero V, Gosalbes MJ, Perez-Martinez G. 1997. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J. Bacteriol. 179:6657–6664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morrow AL, Ruiz-Palacios GM, Jiang X, Newburg DS. 2005. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J. Nutr. 135:1304–1307 [DOI] [PubMed] [Google Scholar]
- 24. Munoz-Provencio D, Monedero V. 2011. Shotgun phage display of Lactobacillus casei BL23 against collagen and fibronectin. J. Microbiol. Biotechnol. 21:197–203 [DOI] [PubMed] [Google Scholar]
- 25. Osumi D, et al. 2009. Core fucosylation of E-cadherin enhances cell-cell adhesion in human colon carcinoma WiDr cells. Cancer Sci. 100:888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36 doi:10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Posno M, et al. 1991. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 57:1822–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rajilic-Stojanovic M, Smidt H, de Vos WM. 2007. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9:2125–2136 [DOI] [PubMed] [Google Scholar]
- 30. Rico J, Yebra MJ, Perez-Martinez G, Deutscher J, Monedero V. 2008. Analysis of ldh genes in Lactobacillus casei BL23: role on lactic acid production. J. Ind. Microbiol. Biotechnol. 35:579–586 [DOI] [PubMed] [Google Scholar]
- 31. Rochat T, et al. 2007. Anti-inflammatory effects of Lactobacillus casei BL23 producing or not a manganese-dependant catalase on DSS-induced colitis in mice. Microb. Cell Fact. 6:22 doi:10.1186/1475-2859-6-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rodriguez-Diaz J, Monedero V, Yebra MJ. 2011. Utilization of natural fucosylated oligosaccharides by three novel alpha-l-fucosidases from a probiotic Lactobacillus casei strain. Appl. Environ. Microbiol. 77:703–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruas-Madiedo P, Gueimonde M, Fernandez-Garcia M, de los Reyes-Gavilan CG, Margolles A. 2008. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl. Environ. Microbiol. 74:1936–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 35. Sanchez B, Saad N, Schmitter JM, Bressollier P, Urdaci MC. 2010. Adhesive properties, extracellular protein production, and metabolism in the Lactobacillus rhamnosus GG strain when grown in the presence of mucin. J. Microbiol. Biotechnol. 20:978–984 [DOI] [PubMed] [Google Scholar]
- 36. Sanfelix-Haywood N, Coll-Marques JM, Yebra MJ. 2011. Role of alpha-phosphoglucomutase and phosphoglucose isomerase activities at the branching point between sugar catabolism and anabolism in Lactobacillus casei. J. Appl. Microbiol. 111:433–442 [DOI] [PubMed] [Google Scholar]
- 37. Schwab C, Ganzle M. 2011. Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides. FEMS Microbiol. Lett. 315:141–148 [DOI] [PubMed] [Google Scholar]
- 38. Sela DA. 2011. Bifidobacterial utilization of human milk oligosaccharides. Int. J. Food Microbiol. 149:58–64 [DOI] [PubMed] [Google Scholar]
- 39. Sela DA, et al. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 78:795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sonnenburg JL, Angenent LT, Gordon JI. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5:569–573 [DOI] [PubMed] [Google Scholar]
- 41. Titgemeyer F, Hillen W. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek 82:59–71 [PubMed] [Google Scholar]
- 42. Veenhoff LM, Poolman B. 1999. Substrate recognition at the cytoplasmic and extracellular binding site of the lactose transport protein of Streptococcus thermophilus. J. Biol. Chem. 274:33244–33250 [DOI] [PubMed] [Google Scholar]
- 43. Viana R, et al. 2000. Enzyme I and HPr from Lactobacillus casei: their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol. Microbiol. 36:570–584 [DOI] [PubMed] [Google Scholar]
- 44. Yebra MJ, Monedero V, Zuñiga M, Deutscher J, Perez-Martinez G. 2006. Molecular analysis of the glucose-specific phosphoenolpyruvate:sugar phosphotransferase system from Lactobacillus casei and its links with the control of sugar metabolism. Microbiology 152:95–104 [DOI] [PubMed] [Google Scholar]
- 45. Yebra MJ, Veyrat A, Santos MA, Perez-Martinez G. 2000. Genetics of l-sorbose transport and metabolism in Lactobacillus casei. J. Bacteriol. 182:155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.