Abstract
To gain more insight into the role of chromosomal instability (CIN), the cytogenetic hallmark of most solid tumors, we performed fluorescence in situ hybridization (FISH) on interphase nuclei of cytological specimens enabling the correct detection of chromosome copies in intact tumor cells of 18 well (G1), moderately (G2), or poorly (G3) differentiated hepatocellular carcinomas (HCCs). A close correlation between the morphological dedifferentiation and increasing copy numbers and variation of FISH signals was seen for chromosomes 1 and 8, respectively (P ≤ 0.0002). Four HCC G1 had constant chromosome patterns for chromosomes 1 and/or 8 with a mean of signals per nucleus ≤5.08 and ≤3 different signal combinations, indicating a low level of CIN, as confirmed by FISH using probes for centromeres of chromosomes 3, 7, and 17. In contrast to this, five HCC G2–3 revealed ≥8.46 signals per nucleus and 23–41 different signal combinations, indicating high levels of CIN. In the remaining cases, signal counts from 5.96–8.46 and 7–15 combinations were seen. Here, nuclei with constant aberration patterns and low copy numbers occurred alongside nuclei with inconstant patterns and high copy numbers. It is evident that in these cases a transition from well to moderately differentiated HCC developed in parallel to an increase in CIN, possibly induced by a major dysregulation of mitotic control mechanisms. In conclusion, CIN may induce a stepwise increase of aneuploidy in HCC that is mirrored by the morphological dedifferentiation of tumor cells.
Most solid tumors are characterized by complex chromosome rearrangements (1, 2). These aneuploidies are believed to develop because of an increased chromosomal instability (CIN). This form of instability has been defined as an increased rate of chromosome aberrations, in contrast to microsatellite instability, which induces alterations of DNA repeat sequences but no changes in chromosome number or structure (3, 4). CIN is associated with aggressive tumor behavior and poor prognosis (5). Although there are some clues that CIN may be caused by exposure to various carcinogens (6), the mechanisms leading to CIN and subsequent aneuploidy are still poorly understood (7).
Whether CIN is a primary or a secondary event in tumor development is currently a subject of controversial discussion (8, 9). Experiments to elucidate this question are based mainly on the evaluation of cell lines. However, interpretation of these results may be hampered by the fact that an increased CIN may be induced by prolonged culture passages in vitro.
To overcome these problems, we chose hepatocellular carcinoma (HCC) as a suitable model to investigate CIN. HCC is one of the most significant tumors worldwide, leading to more than 250,000 deaths per year (10), with hepatitis B and C and the ingestion of aflatoxins or ethanol as the main etiological factors (11). Recurrent chromosomal imbalances of HCC are gains of 1q, 8q, 17q, and 20q as well as losses of 4q, 8p, 13q, 16q, and 17p (12–15). In >90% of HCC, chromosomal aberrations can be detected by using centromere-specific probes for chromosomes 1 and 8, indicating a significant role of CIN in the development of HCC (15).
Because only limited information on CIN can be obtained by using techniques that pool DNA, such as comparative genomic hybridization, we performed molecular cytogenetic analysis with fluorescence in situ hybridization (FISH) on cytological specimens from primary HCC. The advantage of this methodological approach is that cells with preserved morphology can be studied, and therefore the exact copy number of defined chromosomes can be determined on the single cell level (15).
Using this technique, we demonstrated that (i) there was a close correlation between the morphological dedifferentiation and increase and variability in copy numbers of defined chromosomes, (ii) there was no correlation between this CIN and the diameter of the tumors, and (iii) within certain tumors, there was a heterogeneity of tumor cells regarding both the morphology and the degree of CIN.
Materials and Methods
Fine needle aspirates were taken from 18 patients with HCC treated at Hannover Medical School (clinical data given in Table 1). Aspirations were performed with needles of 0.7-mm diameter with ultrasound control (16). Morphological evaluation on May–Grünwald–Giemsa-stained smears according to Ishak et al. (17) revealed seven well, eight moderate, and three moderate-to-poorly differentiated HCCs.
Table 1. Clinical and cytomorphological data of the patients with HCC analyzed by FISH on cytological specimens.
| Case | Sex | Age, yr | Tumor diameter, mm | Single (s)/ multiple (m) tumors | Grading | Serology for hepatitis/ other etiologic factors |
|---|---|---|---|---|---|---|
| T5329-02 | M | 76 | 12 | m | G2 | Hepatitis B |
| T5850-02 | M | 64 | 18 | m | G1 | Hepatitis C |
| T6265-02 | M | 56 | 38 | m | G2-3 | Hepatitis B |
| T6660-02 | M | 48 | 24 | m | G2-3 | Hepatitis B and C |
| T6754-02 | M | 66 | 73 | m | G2 | Negative/ethanol |
| T7361-02 | M | 53 | 4 | m | G1 | Negative/ethanol |
| T7795-02 | M | 85 | 61 | m | G1 | Negative |
| T8542-02 | M | 74 | 47 | m | G1 | Negative/ethanol |
| T9230-02 | M | 61 | 72 | m | G1 | Negative/ethanol |
| T9532-02 | M | 75 | 58 | s | G1 | Negative |
| T10339-02 | M | 69 | 64 | m | G2 | Negative |
| T58-03 | M | 61 | 14 | s | G2 | Negative/ethanol* |
| T154-03 | F | 71 | l.n.† | m | G2 | Hepatitis C/coincidental CLL‡ |
| T408-03 | F | 63 | 75 | m | G1 | Negative |
| T411-03 | F | 75 | 42 | m | G2 | Hepatitis C |
| T698-03 | M | 69 | 55 | s | G3 | Hepatitis B |
| T1485-03 | M | 73 | 89 | s | G2 | Negative |
| T1600-03 | M | 53 | 36 | m | G2 | Negative |
Recurrent HCC in transplanted liver.
Aspiration from a metastatic lymph node.
Chronic lymphocytic leukemia.
FISH experiments were carried out simultaneously with probes for the centromeres of chromosomes 1 and 8 in all 18 patients and in a second experiment with a mixture of probes for centromeres of chromosomes 3, 7, and 17 in 6 patients from whom more than one smear was available (centromere probes 1, 3, 7, 8, and 17, Abbott, Wiesbaden, Germany). Pepsin digestion (99 ml of distilled water, 1 ml of 1 M HCl, and 5 mg of pepsin) for 3 min at room temperature was followed by washing for 1 min in distilled water and incubation for 10 min in paraformaldehyde (1.5%). After washing for 1 min in distilled water and drying the slides, 0.5 μl of chromosome 1 and 8 probes or of chromosome 3, 7, and 8 probes in 10 μl of hybridization buffer (Abbott) was pipetted onto the slide, placed under a glass coverslip, sealed with rubber cement, heated to 80°C for 10 min, and incubated overnight at 37°C in a humidified chamber. The coverslip was removed, and the slides were washed twice in 0.4× SSC, 0.3% Igepal at 75°C for 2 min. Counterstaining was done with 5 μl of 4′,6-diamidino-2-phenylindole (40 ng/ml) (Qiagen, Heidelberg, Germany).
Evaluation of signals was carried out by using an epifluorescence microscope (Axioskop, Zeiss) equipped with filters specific for fluorescein, Cy3, aqua blue, or 4′,6-diamidino-2-phenylindole and a 100-W mercury lamp. In each tumor, 50 nuclei were evaluated and documented with the isis software package (Metasystems, Altlussheim, Germany).
To determine the cut-off levels for the detection of numerical chromosomal aberrations by using centromere-specific probes for chromosomes 1, 3, 7, 8, and 17, 2,000 peripheral blood lymphocytes (i.e., 400 cells each from five healthy donors with normal karyotypes) and 2,000 normal hepatocytes from liver cell aspirates of five patients with regenerative nodules and/or fatty changes were analyzed. According to Ward et al. (18), the thresholds for gains and losses of the respective chromosomes were calculated as the mean + 3SD.
Results
Determination of Cut-Off Levels. Analysis of 2,000 cells from peripheral blood lymphocytes from healthy donors with centromere-specific probes for chromosomes 1, 3, 7, 8, and 17 showed one signal in 2.25–3.10% of the cells (SD 1.08–1.74%) and three or more signals in 0.35 and 1.20% of cells (SD 0.29–0.78%). Thus, the cut-off levels (mean + 3SD) were determined as 6.15–7.48% for losses and 1.21–3.14% for gains.
Analysis of 2,000 normal hepatocytes from five liver aspirates with the probes mentioned above showed one signal in 2.40–3.20% of the cells (SD 0.63–1.55%) and three or more signals in 1.3–1.65% of the cells (SD 0.29–0.57%). The cut-off levels (mean + 3SD) were determined as 4.28–7.84% for losses and 2.41–3.26% for gains. The detailed findings are given in Table 2. The percentage of tetrasomic cells, as indicated by four signals each for chromosomes 1 and 8 or by four signals each for chromosomes 3, 7, and 17, in the same cell was <1.75%.
Table 2. Control studies to determine the cut-off level for gains and losses of chromosomes 1, 3, 7, 8, and 17, used as markers for CIN.
| Signals/nucleus, %
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| One signal
|
Three and more signals
|
|||||||||
| Probe | cen1 | cen3 | cen7 | cen8 | cen17 | cen1 | cen3 | cen7 | cen8 | cen17 |
| Leukocytes | ||||||||||
| Mean | 2.90 | 2.25 | 2.90 | 3.10 | 2.85 | 0.35 | 1.20 | 1.05 | 0.30 | 0.80 |
| SD | 1.08 | 1.74 | 1.40 | 1.36 | 1.44 | 0.29 | 0.48 | 0.33 | 0.33 | 0.78 |
| Cut-off level* | 6.15 | 7.48 | 7.10 | 7.19 | 7.18 | 1.21 | 2.64 | 2.03 | 1.28 | 3.14 |
| Hepatocytes | ||||||||||
| Mean | 2.65 | 3.20 | 2.80 | 3.20 | 2.40 | 1.65 | 1.30 | 1.55 | 1.35 | 1.55 |
| SD | 0.86 | 1.22 | 1.36 | 1.55 | 0.63 | 0.29 | 0.37 | 0.33 | 0.42 | 0.57 |
| Cut-off level* | 5.23 | 6.85 | 6.89 | 7.84 | 4.28 | 2.51 | 2.41 | 2.53 | 2.60 | 3.26 |
Mean + 3SD.
HCC. FISH analysis on cytological smears by using centromere-specific probes for chromosomes 1 and 8 (Fig. 1a–c) showed that all tumors were aneuploid. The mean number of signals per nucleus for chromosome 1 varied between 2 and 7.76 (range: 2–21, SD 0–3.49) (Table 3). For chromosome 8, the values varied between 1.16 and 14.66 (range: 1–28, SD 0–5.34). The mean number of signals per nucleus for both chromosomes varied from 3.66 to 19.56 (range: 3–40, SD 0–6.68). In nine cases, more signals were seen for chromosome 1, and in the other nine cases more signals were present for chromosome 8.
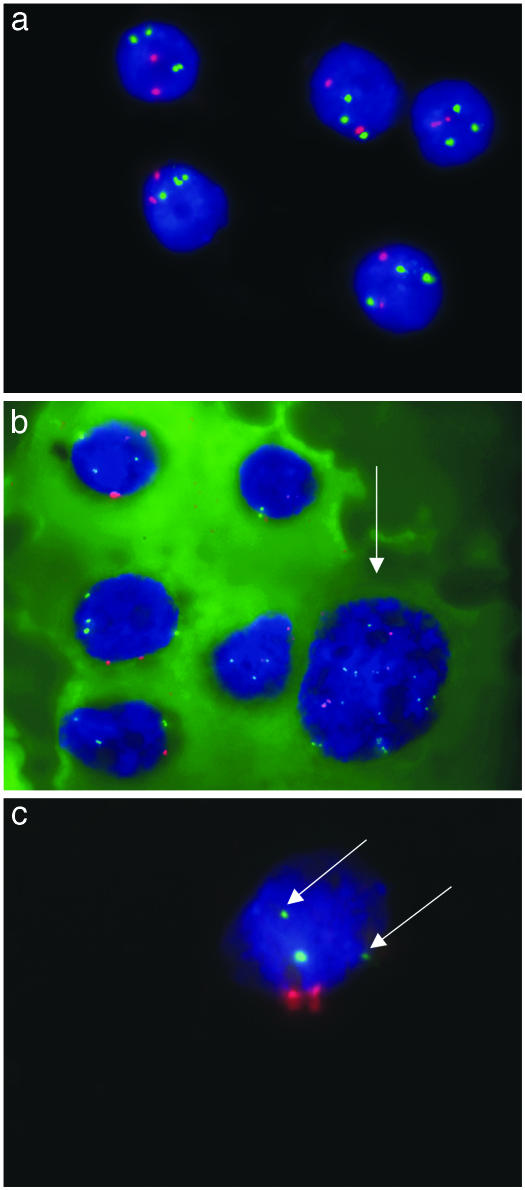
Fig. 1.
FISH analysis of cytological specimens by using centromere-specific probes for chromosomes 1 and 8 (red and green, respectively). (a) In HCC G1 (T9230-02), a constant pattern with two red signals for chromosome 1 and three green signals for chromosome 8 was found. (Microscopic magnification, 100 × 10.) (b) In contrast, the cytological specimen of T1600-03 (HCC G2) reveals six cohesive intact nuclei with a constant pattern of two red signals for chromosome 1 and three to four green signals for chromosome 8 in five nuclei. In the sixth nucleus (arrow), a highly aberrant pattern was found with 4 and 11 signals, respectively. (c) T698-03 (HCC G3) showed two red signals for chromosome 1 and three signals for chromosome 8. However, two of the signals were clearly smaller (arrows) than the third signal.
Table 3. Mean and SD for chromosomes 1 and 8 both singly and combined.
| Chromosome 1
|
Chromosome 8
|
Chromosomes 1 and 8
|
||||
|---|---|---|---|---|---|---|
| Case | Mean | SD | Mean | SD | Mean | SD |
| T408-03 G1* | 2.00 | 0.00 | 3.00 | 0.00 | 5 | 0.00 |
| T10339-02 G1 | 3.02 | 0.14 | 2.02 | 0.14 | 5.04 | 0.28 |
| T9230-02 G1* | 2.04 | 0.28 | 3.04 | 0.28 | 5.08 | 0.56 |
| T698-03 G3 | 2.06 | 0.31 | 3.04 | 0.28 | 5.10 | 0.58 |
| T7795-02 G1 | 2.50 | 0.51 | 1.16 | 0.37 | 3.66 | 0.75 |
| T8542-02 G1* | 3.30 | 0.68 | 2.70 | 0.65 | 6.00 | 1.01 |
| T7361-02 G1* | 2.68 | 0.77 | 3.18 | 1.21 | 5.86 | 1.28 |
| T5850-02 G2 | 4.22 | 1.34 | 3.72 | 0.57 | 7.94 | 1.49 |
| T154-03 G2* | 2.98 | 0.96 | 3.36 | 1.05 | 6.34 | 1.57 |
| T6754-02 G2 | 3.28 | 1.03 | 4.22 | 1.09 | 7.5 | 1.78 |
| T1485-03 G2 | 4.9 | 1.31 | 2.72 | 0.73 | 7.62 | 1.79 |
| T6660-02 G2-3 | 3.50 | 1.27 | 3.32 | 1.15 | 6.82 | 2.18 |
| T9532-02 G2 | 2.98 | 1.17 | 3.96 | 1.32 | 6.94 | 2.28 |
| T1600-03 G2 | 3.4 | 1.78 | 5.06 | 2.35 | 8.46 | 3.89 |
| T5329-02 G2 | 7.00 | 2.81 | 5.20 | 1.80 | 12.20 | 4.23 |
| T58-03 G2* | 5.22 | 2.15 | 9.46 | 3.42 | 14.68 | 5.10 |
| T411-03 G2 | 7.76 | 3.49 | 6.48 | 2.54 | 14.24 | 5.81 |
| T6265-02 G2-3 | 4.90 | 2.29 | 14.66 | 5.34 | 19.56 | 6.68 |
Cases are listed in order of increasing SD for chromosomes 1 and 8 combined. Comparing this SD with the morphological grading leads to the most significant correlation of all parameters analyzed (P < 0.0002, Mann—Whitney test). Three groups of cases with low and high levels of CIN and an intermediate group according to the criteria defined in the text are separated by a space.
Cases also studied by using centromere-specific probes for chromosomes 3, 7, and 17.
The maximum range of signals within a single case was seen for chromosome 1 in case T411-03 G2 with 3–21 signals. For chromosome 8, the maximum range was detected in case T6265-02 G2–3 with 3–25 signals. Regarding both chromosomes, the maximum range with 7–40 signals was seen in the same sample.
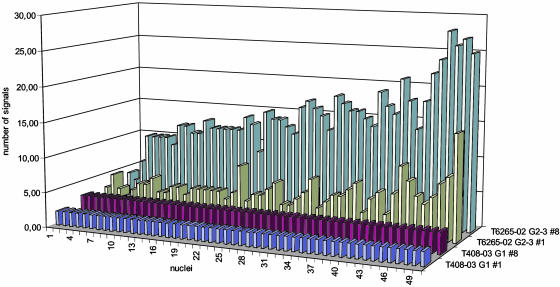
The modal number of chromosome copies for chromosome 1 varied from 2 to 7 and for chromosome 8 from 1 to 12 (Table 4). The percentage of nuclei not displaying the modal number, defined as CIN index by Nakamura et al. (19), ranged from 0% to 78% for chromosome 1 and from 0% to 84% for chromosome 8. The number of different chromosome combinations had the lowest value of 1 in case T408-03 G1 with all nuclei displaying two signals for chromosome 1 and three signals for chromosome 8. The highest value occurred in case T6265-02 G2–3 with 41 different combinations of signal copies (Fig. 2).
Table 4. Modal number of chromosome copies, CIN index, and number of different chromosome combinations for the cases analyzed.
| Chromosome 1
|
Chromosome 8
|
No. of different chromosome combinations
|
|||
|---|---|---|---|---|---|
| Case | Modal | CIN index | Modal | CIN index | |
| T408-03 G1* | 2 | 0 | 3 | 0 | 1 |
| T10339-02 G1 | 3 | 2 | 2 | 2 | 2 |
| T9230-02 G1* | 2 | 2 | 3 | 2 | 2 |
| T698-03 G3 | 2 | 4 | 3 | 2 | 3 |
| T7795-02 G1 | 3 | 0 | 1 | 16 | 3 |
| T8542-02 G1* | 3 | 42 | 3 | 50 | 7 |
| T7361-02 G1* | 2 | 50 | 3 | 62 | 10 |
| T5850-02 G1 | 4 | 74 | 4 | 72 | 11 |
| T154-03 G2* | 3 | 56 | 4 | 58 | 12 |
| T6754-02 G2 | 4 | 62 | 5 | 56 | 13 |
| T1485-03 G2 | 5 | 62 | 3 | 54 | 14 |
| T6660-02 G2-3 | 3 | 40 | 3 | 64 | 13 |
| T9532-02 G2 | 3 | 56 | 3 | 50 | 15 |
| T1600-03 G2 | 2 | 58 | 3 | 78 | 23 |
| T5329-02 G2 | 6 | 78 | 4 | 66 | 25 |
| T58-03 G2* | 4 | 66 | 9 | 74 | 26 |
| T411-03 G2 | 7 | 78 | 7 | 76 | 31 |
| T6265-02 G2-3 | 4 | 64 | 12 | 84 | 41 |
CIN index was defined as the percentage of cells not displaying the modal number of chromosomes (18), whereas the number of different chromosome combinations indicates the number of subclones detectable in the tumor cell populations. The cases are listed in the same order as in Table 2.
Cases also studied by using centromere-specific probes for chromosomes 3, 7, and 17.
Fig. 2.
As an example, case T408-03 G1 with the lowest and case T6265-02 G2–3 with the highest variability of FISH signals for chromosome 1 and 8 are shown. In case T408-03 G1, all nuclei revealed two signals for chromosome 1 and 3 signals for chromosome 8. In contrast, in case T6265-02 G2–3, 2–15 signals for chromosome 1 and 3–25 signals for chromosome 8 occurred in 41 different combinations, indicating a high CIN.
Grouping HCCs with regard to their cytogenetic patterns showed that five samples had a constant chromosome pattern with a mean of signals per cell not exceeding 5.10 and an SD below 0.75, a modal signal number of a maximum of 3, a CIN index of 0–16, and a number of different chromosome combinations from 1 to 3. Four of these samples represented well differentiated HCC (T408-03, T10339-02, T9230-02, and T7795-02). Case T698-03 was an exception in that it was a high-grade G3 tumor. However, two of the signals for chromosome 8 were clearly smaller than the third signal, pointing to a structural alteration involving the centromeric region of chromosome 8 (Fig. 1c). Up to 20 of these signals per nucleus were seen in cases T58-03 and T6265-03. In contrast, cases T1600-03, T5329-02, T58-03, T411-03, and T6265-02 had mean signal counts higher than 8.46 signals per cell and an SD of >3.89. High variability of signals in these cases was also indicated by modal numbers of 2–12, a CIN index of at least 58, and a number of different chromosome combinations from 23 to 41. In the remaining eight HCCs, the mean number of chromosomes ranged from 5.86 to 7.94 with an SD from 1.01 to 2.28, a modal number of 2–5, a CIN index from 42 to 74, and variant chromosome combinations from 7 to 15. These HCCs demonstrated nuclei with constant aberration patterns occurring alongside nuclei with inconstant aberrations even in the same pseudoacini (Fig. 1b).
In six of these patients, more than one smear was available to perform additional hybridization experiments. To rule out the possibility that using a different probe set may have changed the CIN index, these six HCCs were also analyzed with centromere-specific probes for chromosomes 3, 7, and 17. These six HCCs comprised two well differentiated HCCs (T408-03 G1 and T9230-02 G1) of the group with a low CIN, three moderately differentiated HCCs with an intermediate CIN (T8542-03 G1, and T7361-03 G1, T154-03 G2), and one moderately differentiated HCC (T58-03 G2) with a high CIN, as determined by analysis of chromosomes 1 and 8. In the experiments analyzing chromosomes 3, 7, and 17, the mean number of signals per nucleus varied between 2.00 and 5.88 (SD 0.00–1.95), the maximum ranged between 3 and 11 signals per nucleus, the modal number of signals was between 2 and 6, the CIN index varied from 0 to 70, and the number of different signal combinations per cell ranged between 1 and 38. Detailed findings are given in Table 5.
Table 5. Mean, SD (singly and combined), modal number, CIN, and number of different chromosome combinations for chromosomes 3, 7, and 17.
| Chromosome 3
|
Chromosome 7
|
Chromosome 17
|
Chromosomes 3 + 7 + 17
|
Chromosome 3
|
Chromosome 7
|
Chromosome 17
|
No. of different chromosome combinations
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Modal | CIN | Modal | CIN | Modal | CIN | |
| T408-03 G1 | 2.26 | 0.44 | 2.22 | 0.42 | 2.38 | 0.49 | 6.86 | 0.83 | 2 | 22 | 2 | 26 | 2 | 36 | 9 |
| T9230-02 G1 | 2.00 | 0.00 | 2.00 | 0.00 | 2.00 | 0.00 | 6.00 | 0.00 | 2 | 0 | 2 | 0 | 2 | 0 | 1 |
| T8542-02 G1 | 2.66 | 0.92 | 4.02 | 1.17 | 3.10 | 1.25 | 9.78 | 2.85 | 2 | 46 | 3 | 58 | 2 | 64 | 24 |
| T7361-02 G1 | 3.54 | 0.91 | 3.46 | 0.93 | 3.34 | 0.87 | 10.34 | 2.27 | 4 | 60 | 4 | 44 | 4 | 52 | 19 |
| T154-03 G2 | 4.70 | 1.58 | 5.34 | 1.95 | 4.50 | 1.79 | 14.54 | 4.70 | 4 | 58 | 5 | 70 | 3 | 68 | 36 |
| T58-03 G2 | 4.64 | 1.31 | 5.88 | 1.52 | 4.12 | 1.35 | 14.64 | 3.53 | 4 | 68 | 6 | 70 | 4 | 66 | 38 |
Comparing the results for chromosomes 3, 7, and 17 with those obtained for chromosomes 1 and 8 demonstrated that the two well differentiated HCCs G1 (T408-03 G1 and T9230-02 G1), grouped into HCCs with low levels of CIN according to the FISH studies using probes for chromosomes 1 and 8, showed the lowest mean number of signals for chromosomes 3, 7, and 17, the lowest number of cells with aberrant signal constellations, and the lowest number of signal variations. Accordingly, the moderately differentiated HCC T58-03 G2, grouped into HCCs with high levels of CIN according to the FISH studies using probes for chromosomes 1 and 8, showed the highest mean number of signals for chromosomes 3, 7, and 17, the highest number of cells with aberrant signal constellations, and the highest number of signal variations. The values of the three HCCs (T8542-02 G1, T7361-02 G1, and T1541-03 G2) that grouped into HCCs with intermediate levels of CIN when probes for chromosomes 1 and 8 were used also showed intermediate levels of CIN when probes for chromosomes 3, 7, and 17 were used. Thus, the finding that dedifferentiation of HCC is paralleled by increasing levels of CIN, obtained by FISH using probes for chromosomes 1 and 8, was totally confirmed by using probes for additional chromosomes.
Moreover, in the HCC with intermediate levels of CIN we again observed cells with constant signal constellations alongside cells with variable signal numbers.
Statistical Analysis. Comparing the added mean number of signals for chromosomes 1 and 8 of the group of HCC G1 to those of the group of HCC G2–3 revealed a statistically significant difference (P < 0.0014, Mann–Whitney test). This significance was even higher when comparing the SD for the added values of chromosomes 1 and 8 with the morphology (P < 0.0002). A lower or no significance was seen when comparing HCC G1 with HCC G2–3 with the modal number of chromosomes 1 and 8 (P < 0.0374 and 0.0129), the CIN index (P < 0.091 for chromosome 1, P < 0.0164 for chromosome 8), and the number of cells displaying variant chromosome combinations (P < 0.006).
Statistical analysis also confirmed significant correlations between the results obtained for chromosomes 1 and 8 and the findings for chromosomes 3, 7, and 17. Here, the Spearman rank correlation test revealed a significant correlation between the number of signals (P < 0.019), the SD of the signals (P < 0.042), and the number of different signal combinations (P < 0.019).
No correlations were seen between any of these values and the clinical data, including sex, age, serological status, and, in particular, the tumor size.
Discussion
Based on the advantages of FISH analysis of cytological specimens of primary HCC, three main observations were made in this study.
First, there was a close correlation between the morphological dedifferentiation and increase in copy numbers as well as variation of FISH signals for chromosomes 1 and 8, as confirmed by results of additional hybridization for chromosomes 3, 7, and 17. Thus, dedifferentiation in HCC is accompanied by an increasing CIN, as described in other malignant neoplasias such as follicular lymphoma (20) or chronic lymphocytic leukemia (21). The term CIN is used, although the FISH approach did not directly measure an increased rate of aberrations but evaluated the status of chromosomal imbalances at a given point of time only. However, the chromosomal heterogeneity observed in the cells may serve as a good indicator for CIN.
Second, there was no correlation between CIN and the diameter of the tumors. Well differentiated HCC with constant chromosome patterns can build up small as well as large tumor nodes with a diameter of up to 160 mm. Vice versa, moderate-to-low differentiated HCC can be found in large as well as in small tumors of 14 mm. Thus, well differentiated HCC may be able to grow for longer times, conserving distinct patterns of chromosomal abnormalities. An increased CIN may also occur within moderate or low differentiated HCC at an early stage of tumor development.
Third, within certain tumors, there was a heterogeneity of tumor cells regarding both the morphology and the degree of CIN. In these specimens, nuclei with constant aberration patterns occurred next to nuclei with inconstant aberrations even in the same pseudoacini. It is evident that in these cases a transition from well to moderately differentiated HCC developed in parallel to a cytogenetic change from constant to inconstant aberration pattern. Remarkably, the progression from well to less differentiated HCC is seen in the majority of cases in our study. Similar findings have been reported in histological specimens with dedifferentiation of well differentiated HCC in ≈50% of cases (22) and in 75% of patients within 7–34 months being reported in a follow-up study (23).
However, this progression does not seem to be an obligatory event. As has been shown, well differentiated HCC can grow to tumors of a large size under preserved cytogenetic stability. This observation argues against an increasing aneuploidy as a kind of autocatalytic process as described in colon carcinoma cell lines (24), at least in tumors with low-level CIN. Nevertheless, once the tumor cells gain a higher level of CIN, aneuploidy seems to increase more and more, probably indicating a growth advantage in a Darwinian sense (25). This stepwise increase of CIN seems to be more concordant with alterations of distinct proteins involved in mitotic mechanisms described exemplarily in colon carcinoma by Lengauer and coworkers (3, 4, 26).
Candidate proteins important in these mechanisms are speculative at this point. As examples, securin (pituitary tumor transforming gene), hBUB1, and MAD2 involved in chromatid attachment and segregation (27–29), Hec1 acting with Mad1/Mad2 at the kinetochore (30), and CDK2 involved in duplication of centrosomes (31) may be discussed. Integrity of chromosomes is also influenced by genes such as ATM (32) and CHK2 (33), closely controlling DNA integrity in the interphase, as also done by the p53 DNA damage checkpoint playing a key role at the end of the G2 phase (34).
In conclusion, CIN leads to a stepwise increase of aneuploidy in HCC reflected in particular by morphological dedifferentiation of tumor cells. The progression from well differentiated HCC to moderately or low differentiated HCC seems to be induced by a major dysregulation of mitotic control, leading to increasing aneuploidy.
Acknowledgments
We thank Willi Arndt and Markus Meyer for excellent technical assistance, and Gill Teicke for support in preparing this manuscript. This work was funded by grants from the Deutsche Forschungsgemeinschaft (KFO 119) and Deutsche Krebshilfe (Project 10-992).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CIN, chromosomal instability; FISH, fluorescence in situ hybridization; HCC, hepatocellular carcinoma.
References
- 1.Mitelman, F. (1994) Catalog of Chromosome Aberrations in Cancer (Wiley–Liss, New York).
- 2.Sandberg, A. A. (1990) The Chromosomes in Human Cancer and Leukemia (Elsevier, New York).
- 3.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1997) Nature 386, 623–627. [DOI] [PubMed] [Google Scholar]
- 4.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1998) Nature 396, 643–649. [DOI] [PubMed] [Google Scholar]
- 5.DeVita, V., Hellmann, S. & Rosenberg, S., eds. (1997) Cancer: Principles and Practice (Lipincott–Raven, New York).
- 6.Bardelli, A., Cahill, D. P., Lederer, G., Speicher, M. R., Kinzler, K. W., Vogelstein, B. & Lengauer, C. (2001) Proc. Natl. Acad. Sci. USA 98, 5770–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nowak, M. A., Komarova, N. L., Sengupta, A., Jallepalli, P. V., Shih, L. M., Vogelstein, B. & Lengauer, C. (2002) Proc. Natl. Acad. Sci. USA 99, 16226–16231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, R., Sonik, A., Stindl, R., Rasnick, D. & Duesberg, P. (2000) Proc. Natl. Acad. Sci. USA 97, 3236–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duesberg, P. (2002) Cancer Res. 62, 6345–6349. [PubMed] [Google Scholar]
- 10.Bosch, F. & Munoz, N. (1991) Adv. Appl. Biotechnol. 13, 55–56. [Google Scholar]
- 11.Schirmacher, P. & Dienes, H. P. (1999) in Molecular Biology in Cancer Medicine, eds. Kurzrock, R. & Talpaz, M. (Dunitz, London), pp. 355–366.
- 12.Marchio, A., Meddeb, M., Pineau, P., Danglot, G., Tiollais, P., Bernheim, A. & Dejean, A. (1997) Genes Chromosomes Cancer 18, 59–65. [PubMed] [Google Scholar]
- 13.Wong, N., Lai, P., Lee, S. W., Fan, S., Pang, E., Liew, C. T., Sheng, Z., Lau, J. W. & Johnson, P. J. (1999) Am. J. Pathol. 154, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kusano, N., Shiraishi, K., Kubo, K., Oga, A., Okita, K. & Sasaki, K. (1999) Hepatology 29, 1858–1862. [DOI] [PubMed] [Google Scholar]
- 15.Wilkens, L., Bredt, M., Flemming, P., Schwarze, Y., Becker, T., Mengel, M., von Wasielewski, R., Klempnauer, J. & Kreipe, H. (2001) J. Mol. Diagn. 3, 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caselitz, M., Masche, N., Bleck, J. S., Gebel, M., Atay, Z., Stern, C., Manns, M. P. & Kubicka, S. (2003) Z. Gastroenterol. (Verh.) 41, 1–6. [DOI] [PubMed] [Google Scholar]
- 17.Ishak, K. G., Goodman, Z. D. & Stocker, J. T. (2001) Tumors of the Liver and Intrahepatic Bile Ducts (Armed Forces Institute of Pathology, Washington, DC).
- 18.Ward, B. E., Gersen, S. L., Carelli, M. P., McGuire, N. M., Dackowski, W. R., Weinstein, M., Sandlin, C., Warren, R. & Klinger, K. W. (1993) Am. J. Hum. Genet. 52, 854–865. [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura, H., Saji, H., Idiris, A., Kawasaki, N., Hosaka, M., Ogata, A., Saijo, T. & Kato, H. (2003) Clin. Cancer Res. 9, 2294–2299. [PubMed] [Google Scholar]
- 20.Martinez-Climent, J. A., Alizadeh, A. A., Segraves, R., Blesa, D., Rubio-Moscardo, F., Albertson, D. G., Garcia-Conde, J., Dyer, M. J., Levy, R., Pinkel, D., et al. (2003) Blood 101, 3109–3117. [DOI] [PubMed] [Google Scholar]
- 21.Bea, S., Lopez-Guillermo, A., Ribas, M., Puig, X., Pinyol, M., Carrio, A., Zamora, L., Soler, F., Bosch, F., Stilgenbauer, S., et al. (2002) Am. J. Pathol. 161, 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenmochi, K., Sugihara, S. & Kojiro, M. (1987) Liver 7, 18–26. [DOI] [PubMed] [Google Scholar]
- 23.Sugihara, S., Nakashima, O., Kojiro, M., Majima, Y., Tanaka, M. & Tanikawa, K. (1992) Cancer 70, 1488–1492. [DOI] [PubMed] [Google Scholar]
- 24.Duesberg, P., Rausch, C., Rasnick, D. & Hehlmann, R. (1998) Proc. Natl. Acad. Sci. USA 95, 13692–13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cahill, D. P., Kinzler, K. W., Vogelstein, B. & Lengauer, C. (1999) Trends Cell Biol. 9, M57–M60. [PubMed] [Google Scholar]
- 26.Jallepalli, P. & Lengauer, C. (2001) Nat. Rev. Cancer 1, 109–117. [DOI] [PubMed] [Google Scholar]
- 27.Jallepalli, P. V., Waizenegger, I. C., Bunz, F., Langer, S., Speicher, M. R., Peters, J. M., Kinzler, K. W., Vogelstein, B. & Lengauer, C. (2001) Cell 105, 445–457. [DOI] [PubMed] [Google Scholar]
- 28.Michel, L. S., Liberal, V., Chatterjee, A., Kirchwegger, R., Pasche, B., Gerald, W., Dobles, M., Sorger, P. K., Murty, V. V. & Benezra, R. (2001) Nature 409, 355–359. [DOI] [PubMed] [Google Scholar]
- 29.Cahill, D. P., Lengauer, C., Yu, J., Riggins, G. J., Willson, J. K., Markowitz, S. D., Kinzler, K. W. & Vogelstein, B. (1998) Nature 392, 300–303. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Lluesma, S., Stucke, V. M. & Nigg, E. A. (2002) Science 297, 2267–2270. [DOI] [PubMed] [Google Scholar]
- 31.Hinchcliffe, E. H., Li, C., Thompson, E. A., Maller, J. L. & Sluder, G. (1999) Science 283, 851–854. [DOI] [PubMed] [Google Scholar]
- 32.Boultwood, J. (2001) J. Clin. Pathol. 54, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bell, D. W., Varley, J. M., Szydlo, T. E., Kang, D. H., Wahrer, D. C., Shannon, K. E., Lubratovich, M., Verselis, S. J., Isselbacher, K. J., Fraumeni, J. F., et al. (1999) Science 286, 2528–2531. [DOI] [PubMed] [Google Scholar]
- 34.Vogelstein, B., Lane, D. & Levine, A. J. (2002) Nature 408, 307–310. [DOI] [PubMed] [Google Scholar]