Abstract
Shiga toxigenic Escherichia coli O157 is the leading cause of hemolytic uremic syndrome (HUS) worldwide. The frequencies of stx genotypes and the incidences of O157-related illness and HUS vary significantly between Argentina and Australia. Locus-specific polymorphism analysis revealed that lineage I/II (LI/II) E. coli O157 isolates were most prevalent in Argentina (90%) and Australia (88%). Argentinean LI/II isolates were shown to belong to clades 4 (28%) and 8 (72%), while Australian LI/II isolates were identified as clades 6 (15%), 7 (83%), and 8 (2%). Clade 8 was significantly associated with Shiga toxin bacteriophage insertion (SBI) type stx2 (locus of insertion, argW) in Argentinean isolates (P < 0.0001). In Argentinean LI/II strains, stx2 is carried by a prophage inserted at argW, whereas in Australian LI/II strains the argW locus is occupied by the novel stx1 prophage. In both Argentinean and Australian LI/II strains, stx2c is almost exclusively carried by a prophage inserted at sbcB. However, alternative q933- or q21-related alleles were identified in the Australian stx2c prophage. Argentinean LI/II isolates were also distinguished from Australian isolates by the presence of the putative virulence determinant ECSP_3286 and the predominance of motile O157:H7 strains. Characteristics common to both Argentinean and Australian LI/II O157 strains included the presence of putative virulence determinants (ECSP_3620, ECSP_0242, ECSP_2687, ECSP_2870, and ECSP_2872) and the predominance of the tir255T allele. These data support further understanding of O157 phylogeny and may foster greater insight into the differential virulence of O157 lineages.
INTRODUCTION
Escherichia coli O157 is a food-borne pathogen of global significance. Human infection can result in progressive sequelae extending from bloody diarrhea to hemolytic uremic syndrome (HUS), giving rise to the designation of this pathogen as enterohemorrhagic E. coli (EHEC) (5, 33). The predominant source of O157 is cattle (12) and undercooked beef products (2); however, secondary sources, including leafy green vegetables, apple cider, and dairy products which have been contaminated with manure, are also vehicles for food-borne infection (6).
Molecular typing and microbial genomics have facilitated the characterization and comparison of O157 strains isolated from human and animal sources. These studies have been directed to the identification of O157 factors influencing successful human infection. Such analyses have included characterization of Shiga toxin genotypes (32), locus-specific polymorphism assays (LSPA) (40, 44), clade typing (24, 34), allelic variation of virulence genes (4), and Shiga toxin bacteriophage insertion (SBI) site analysis (3, 36, 38).
For some time, it has been known that O157 strains carrying stx2 predominate in human infection, causing more severe disease symptoms than stx2c strains (10, 28). Evidence for a hypervirulent clade of O157 was first demonstrated by Manning et al. (24) in their analysis of O157 outbreak isolates associated with raw spinach consumption in the United States in 2006. This evidence was further supported by genotype comparisons demonstrating that hypervirulent clade 8 isolates can also be designated LSPA-6 lineage I/II (LI/II) with unique SBI genotypes (19). LI/II clade 8 strains are characterized by insertion of the stx2 prophage in argW (18), frequently also possess the stx2c prophage inserted in sbcB, and do not possess stx1 prophage (24).
While the studies described above have been informative, they have focused largely on isolates from geographic regions in the Northern Hemisphere. In those studies, which assessed the genotypes of isolates from diverse geographic regions, some limited indications of strain divergence have been observed. For example, O157:H− strains carrying stx2c (either alone or in association with stx1) dominate from both clinical and cattle sources in Australia (8). Human O157 isolates in Australia also possess SBI genotypes that are different from O157 isolates of human origin in the United States (39), suggesting the possibility of different stx prophage configurations in human isolates in these separate countries. More recent striking data indicate that O157 strains with the stx2 stx2c genotype are predominant in both Argentinean cattle (56%) and clinical cases, where such strains are implicated in >90% of postenteric HUS (22). However, O157 strains with the stx2 stx2c genotype are very rare in Australia (8). Notable differences in Argentinean and Australian O157 epidemiologies are also reflected in the incidence of human cases. A total of 0.24 cases per 100,000 population in Australia (27) and 13.9 cases per 100,000 children younger than 5 years in Argentina (35) are caused mainly by E. coli O157.
To further examine the relatedness of Southern Hemisphere E. coli O157 isolates, we have now compared human and bovine O157 isolates from Argentina and Australia. Strain motility, stx genotyping, q allele genotyping, LSPA-6 genotyping, single nucleotide polymorphism (SNP) clade typing, stx prophage analysis, and putative virulence factor genotyping have been applied to characterize properties of O157 isolates from both countries.
MATERIALS AND METHODS
Bacterial strains.
E. coli O157 strains from Argentina (human, n = 30; cattle, n = 30) and Australia (human, n = 30; cattle, n = 30) comprising isolates that vary in source, pulsed-field gel electrophoresis (PFGE) type, stx genotype, and geographical and temporal isolation were included in this study. Bacterial strains were recovered from −80°C protect preservers (Oxoid, Basingstoke, United Kingdom) in Luria-Bertani (LB) broth at 37°C overnight and stored on tryptone soya agar (TSA; Oxoid) for the duration of the study. Serotype O157 and motility were determined as described by Fegan and Desmarchelier (8). Escherichia coli K-12 Q358 Smr (15) lysogens were created from Argentinean and Australian isolates carrying stx1-, stx2-, and stx2c-encoding prophages. E. coli O157 strains Sakai, EDL933, EC623 (16), EC1812 (16); K-12 strains MG1655 and Q358 Smr; and Salmonella Braenderup H9812 (ATCC BAA-664) were used as controls when appropriate.
Analysis of stx genes.
Isolates were investigated for the presence of stx1 and stx2 using previously published multiplex primers and PCR conditions (30). Restriction fragment length polymorphism was performed on isolates that tested positive for stx2 to discriminate between stx2 and stx2c as previously described (24). Isolates that tested positive for the presence of stx2 and/or stx2c were subsequently tested for stx2-specific q933 and stx2c-specific q21 alleles using primer pairs Q-stx2-F (5′-AAAGCGGAGGGGATTGTTGAAGGC-3′)/stx2_rev (5′-CCGGGAATAGGATACCGAAG-3′) and Qc-stx2-F (5′-GAACAGCATGAGTGGCTGAA-3′)/stx2_rev, respectively, with the following cycling conditions; 96°C for 10 s; 60°C for 30 s; 72°C for 45 s. A representative set of stx2 (987-bp) and stx2c (1,177-bp) amplicons were gel purified and used as templates in capillary sequence reactions (Australian Genome Research Facility, St. Lucia, Australia) to confirm the sequence of q933 and q21 regions upstream of the stx2 and stx2c genes, respectively.
Determination of Shiga toxin bacteriophage insertion (SBI) loci and stx lysogen formation.
Primer sets targeting SBI site boundary sequences were used to determine the stx prophage occupancy of E. coli O157 loci yehV (mlrA) and wrbA using the method of Shaikh and Tarr (36). Additional primers were designed to determine the occupancy of argW, sbcB, prfC, and yecE (Table 1). The SBI sites of E. coli K-12 stx lysogens were determined in the same manner. Mitomycin C (0.5 μg ml−1) was used to induce stx prophage from wild-type O157 strains (1). E. coli K-12 stx lysogen candidates were selected as survivors following stx phage infection (1). Survivors were selected as colonies growing within zones of stx phage lysis in semisolid layered agar plates or as colonies streaked from stx phage infection broth cultures. stx lysogen candidates were confirmed using primer pairs specific for stx1, stx2, or stx2c as appropriate (24, 30); the absence of O157 eae and ehxA genes in lysogens was also confirmed by PCR (30).
Table 1.
Bacteriophage insertion site primers
| Target | Primer | Sequence | Cycle conditions |
|---|---|---|---|
| argW left junction | int4045F | 5′-ACCATCGAGTAGGCGGTATG-3′ | 96°C for 10 s, 60°C for 30 s, 72°C for 40 s |
| int1810R | 5′-ATTTCAGCAGGGCCAGAGTA-3′ | ||
| argW right junction | yfdCF | 5′-ACTGGAGCGATTTCATCTGG-3′ | 96°C for 10 s, 60°C for 30 s, 72°C for 40 s |
| phi1810stx2F | 5′-GGTTGAGCGGGATATGAAAA-3′ | ||
| sbcB left junction | sbcBF | 5′-ATTGTCGCGCTAAAGCTGAT-3′ | 96°C for 10 s, 60°C for 30 s, 72°C for 45 s |
| stx2cphiB | 5′-CAACGATGCTCGTTATGGTG-3′ | ||
| sbcB right junction | stx2cphiA | 5′-GGACAACAGCGCACAGTAAA-3′ | 96°C for 10 s, 60°C for 30 s, 72°C for 45 s |
| sbcBR1 | 5′-CGGGCTTGCAGTAAAAGACT-3′ | ||
| prfC | prfCF | 5′-CGATGCCGCTTACTCAAGA-3′ | 96°C for 10 s, 60°C for 30 s, 72°C for 45 s |
| prfCR | 5′-GAACAGCAGCACCTTCTCG-3′ | ||
| yecE | yecEF | 5′-ATTGCCGAAGATGCCTGTAG-3′ | 96°C for 10 s, 60°C for 30 s, 72°C for 45 s |
| yecER | 5′-CATACAGCGCGCTTACCATA-3′ |
LSPA-6 typing.
Isolates were further characterized using the LSPA-6 method as previously described (40). Briefly, fluorescent-labeled amplicons were diluted 1/20 and separated by capillary electrophoresis using an Applied Biosystems 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA), DS-33 matrix, and GeneScan600 LIZ size standard. Amplicon size was determined using Peakscanner software (version 1.0; Applied Biosystems). Three isolates (EC623, EC1812, and MG1655) with known LSPA-6 patterns were included in each run as positive controls.
O157 clade and tir255T/A allele genotyping.
O157 virulence clades were identified by targeted SNP typing using SYBR green-based real-time PCR with hairpin primers. Clades 1 to 3 and 8 were identified using the SNPs described by Riordan et al. (34). The remaining O157 clades (4 to 7 and 9) were identified using SNPs or combinations of SNPs previously shown to be specific for individual clades (24). Polymorphisms of tir were detected using a probe-based real-time PCR method (38).
PCR genotyping of O157 virulence factors.
The gene encoding the H7 flagellum antigen, fliCH7, was detected using the method of Gannon et al. (11). Isolates were screened for the locus tags corresponding to putative virulence determinants ECSP_0242, ECSP_1773, ECSP_2687, ECSP_2870, ECSP_2872, ECSP_3286, and ECSP_3620 as previously described (18).
Statistical analysis.
Statistical analyses were performed using a 2-by-2 contingency table and Fisher's exact test (Minitab15; Minitab Inc., Minneapolis, MN). P values were two-tailed, and groups were considered significantly different if P values were <0.05. When multiple comparisons were performed, Bonferroni P value correction was incorporated.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database (accession numbers HQ993494 to HQ993501).
RESULTS
Motility.
All Argentinean strains (60/60) were E. coli O157:H7 (motile), while 14/60 Australian strains were E. coli O157:H7 and the remainder were E. coli O157:H− (nonmotile). Argentinean O157 strains were significantly more often motile than Australian O157 strains (P = 0.0001).
stx genotyping.
Argentinean and Australian O157 isolates in this study comprised seven and five different stx genotypes, respectively (Table 2). Among Argentinean isolates, stx2 stx2c represented the largest group (26/60) followed by stx2 alone (9/60) and stx2c alone (8/60). The predominant stx genotypes of Australian isolates were identified as stx1 stx2c (37/60) and stx2c alone (17/60). A significant difference was observed between the predominant genotypes from Argentina (stx2 stx2c) and Australia (stx1 stx2c) (P < 0.0001).
Table 2.
Distribution of stx genotypes in Argentinean and Australian E. coli O157
| stx genotypea | No. of isolates |
|||||
|---|---|---|---|---|---|---|
| Argentina |
Australia |
|||||
| Human (n = 30) | Cattle (n = 30) | Total (n = 60) | Human (n = 30) | Cattle (n = 30) | Total (n = 60) | |
| stx1 | 1 | 2 | 3 | 3 | 3 | |
| stx1 stx2 | 2 | 2 | 1 | 1 | 2 | |
| stx1 stx2 stx2c | 3 | 3 | 6 | |||
| stx1 stx2c | 2 | 4 | 6 | 17 | 20 | 37 |
| stx2 | 6 | 3 | 9 | |||
| stx2 stx2c | 16 | 10 | 26 | 1 | 1 | |
| stx2c | 2 | 6 | 8 | 9 | 8 | 17 |
stx genotype refers to the Shiga toxin genotype.
LSPA-6 typing.
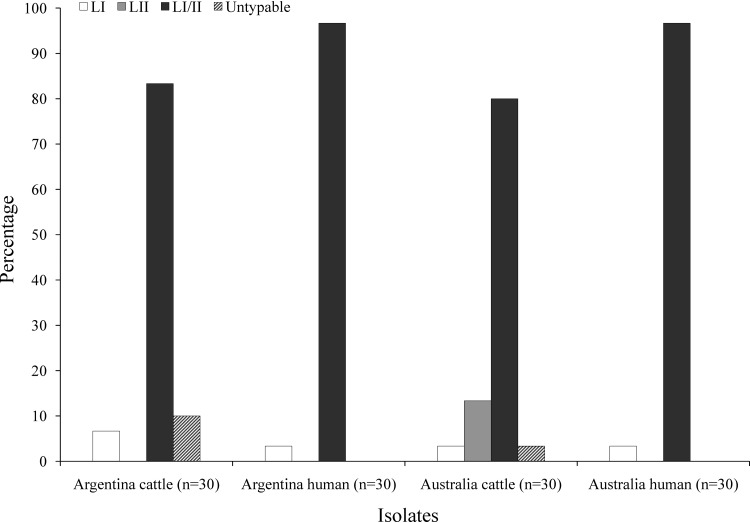
LSPA-6 lineage I/II dominated in isolates from both Argentina (90%) and Australia (88%) (Fig. 1). There was no significant difference in the prevalence of LI/II isolates from both countries. LI isolates were present in both countries, accounting for 4% of the 120 isolates tested. All stx2-positive LI isolates from both Argentina and Australia carried the stx2 (wrbA) prophage. In contrast, LII isolates were present only in Australia, with 7% (4/60) of isolates shown to possess this genotype. No lineage designation could be assigned to three Argentinean and one Australian isolate due to the absence of Z5935 allele amplicons from these isolates.
Fig 1.
Distribution of LSPA-6 genotypes among E. coli O157 isolates from Argentina and Australia. No lineage designation could be assigned to three Argentinean isolates and one Australian isolate due to the absence of the Z5935 allele amplicons. These isolates have been referred to as untypeable.
Virulence clade determination.
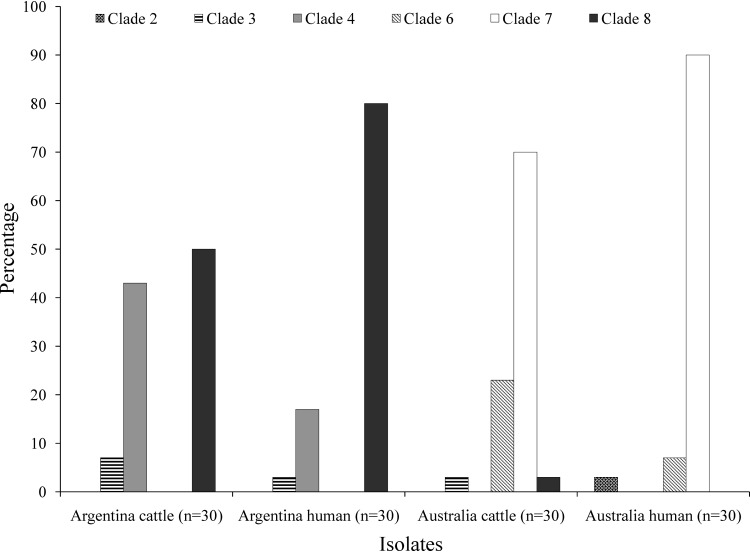
Argentinean LI/II isolates (n = 54) were shown to belong to clades 4 (28%) and 8 (72%). Australian LI/II isolates (n = 53) were identified as clades 6 (15%), 7 (83%), and 8 (2%). No clade 4 isolates were characterized among Australian O157 and, conversely, no clade 6 or 7 isolates were observed among Argentinean O157. Clinical isolates from Argentina and Australia were predominately clade 8 (80%) and clade 7 (90%), respectively (Fig. 2). However, among cattle isolates, clade 4 (43%) and clade 8 (50%) were prevalent in Argentina, while clade 6 (23%) and clade 7 (70%) predominated in Australia. In Argentina, clade 8 isolates dominated in both cattle (50%) and humans (80%) but were present in significantly more human isolates than cattle isolates (P = 0.0292). A similar pattern of clade dominance was observed for Australian isolates, where clade 7 dominated in both cattle (70%) and humans (90%). However, the different distribution of clade 7 in human and cattle isolates was not statistically significant.
Fig 2.
Distribution of E. coli O157 virulence clades in isolates from Argentina and Australia. Isolates are grouped according to country and source, and the distribution of clades within each group is displayed as a percentage of total isolates within the group.
Shiga toxin bacteriophage insertion site characterization.
The insertion of Shiga toxin prophage in previously described O157 chromosomal loci yehV, wrbA, sbcB, argW, yecE, and prfC was investigated (Table 3). In Argentinean O157, stx2- and stx2c-associated prophages were inserted in the argW (37/43 isolates) and sbcB (46/46 isolates) loci, respectively. The stx2 prophage of four Argentinean O157 strains was not inserted in any of the loci studied. Australian O157 strains also showed association of stx2c with prophage insertion in the sbcB locus (54/55 isolates). In a single stx2c-positive Australian strain, the presumptive stx2c prophage was not inserted in any of the loci studied. A separate single Australian isolate showed stx2-associated prophage insertion in the argW locus. In stx1-positive LI/II Argentinean strains (n = 14), the stx1 prophage was not inserted in any of the loci examined. However, in these strains (and in all other Argentinean LI/II and lineage untypeable strains [n = 43]), the yehV locus showed evidence that it was occupied by a non-stx prophage. LI Argentinean (n = 2) and Australian (n = 2) E. coli O157 isolates carrying stx1 stx2 showed association with prophage insertion in the yehV and wrbA loci, respectively. In contrast, all other Australian O157 strains carrying stx1 (n = 40) showed stx1 association with prophage insertion in the argW locus, suggesting the presence of a novel O157 stx1 bacteriophage in these strains.
Table 3.
Characterization of E. coli O157 isolates by O157 clade type and stx bacteriophage insertion site
| Country | Clade | No. of isolates | SBIa |
||
|---|---|---|---|---|---|
| stx1 | stx2 | stx2c | |||
| Argentina | 3 | 1 | yehV | ||
| 2 | yehV | wrbA | |||
| 4 | 2 | Unk | |||
| 3 | Unk | ||||
| 6 | Unk | sbcB | |||
| 5 | sbcB | ||||
| 1 | Unk | sbcB | |||
| 1 | argW | sbcB | |||
| 8 | 3 | sbcB | |||
| 6 | Unk | argW | sbcB | ||
| 6 | argW | ||||
| 24 | argW | sbcB | |||
| Australia | 2 | 1 | yehV | wrbA | |
| 3 | 1 | yehV | wrbA | ||
| 6 | 1 | sbcB | |||
| 8 | argW | sbcB | |||
| 7 | 3 | argW | |||
| 1 | argW | Unk | |||
| 28 | argW | sbcB | |||
| 16 | sbcB | ||||
| 8 | 1 | argW | sbcB | ||
SBI, Shiga toxin bacteriophage insertion locus; Unk, unknown. The SBI for some stx prophages of O157 isolates remain unknown. In such cases, stx lysogens were not isolated and SBI mapping did not correlate with stx genotype data for any of the five Shiga toxin prophage loci examined.
The occurrence of SBI type stx2 (argW)/stx2c (sbcB) in Argentinean isolates was significantly different from the occurrence in Australian isolates (P < 0.0001) (Table 3). Similarly, the occurrence of SBI type stx1 (argW)/stx2c (sbcB) in Australian isolates was significantly different to the occurrence in Argentinean isolates (P < 0.0001). In addition, clade 8 was significantly associated with SBI type stx2 (argW) in Argentinean isolates (P < 0.0001).
Induced stx1, stx2, and stx2c phages insert at phage-specific loci to form E. coli K-12 lysogens.
The stx prophage of Argentinean and Australian O157 strains was further investigated by prophage induction and lysogeny of an E. coli K-12 host strain in order to demonstrate unambiguous linkage of stx genotypes with particular phages (Table 4). Representative O157 strains were induced with mitomycin C followed by harvest of resultant phage lysates and infection of E. coli strain Q358 Smr. Putative stx1, stx2, and stx2c lysogens were selected as Q358 Smr survivors following phage infection and demonstration of lysogen resistance to streptomycin. All lysogen strains were confirmed by stx-specific PCR to carry only the predicted stx genes introduced by the specific infecting stx phage. The absence of PCR amplification of O157 virulence genes eae and ehxA was confirmed for all stx lysogens. Despite repeated efforts, K-12 lysogens from Argentinean O157 stx1 or stx2c prophage were not obtained. SBI analysis then demonstrated that the stx2 (argW) phage induced from Argentinean O157 strains inserted in the argW locus of E. coli Q358 Smr. SBI analysis also demonstrated that stx2 (wrbA) phage induced from Argentinean O157 strains inserted in the wrbA locus of E. coli Q358 Smr. Similarly, stx2c phage induced from Australian O157 demonstrated insertion specificity for the sbcB gene in E. coli Q358 Smr lysogens. Lysogenic insertion of stx1 phage from Australian O157 LI/II isolates, into the E. coli K-12 argW locus, was also demonstrated. Mitomycin C induction of all stx1, stx2, and stx2c lysogens demonstrated that de novo stx1, stx2, and stx2c phages, able to form infectious plaques on E. coli Q358 Smr indicator lawns, were produced from the newly created lysogens. Confirmation of stx1 linkage to prophage specifically integrating at the argW SBI locus of their lysogen host demonstrated the novelty of the stx1 (argW) phage from Australian O157 strains.
Table 4.
Genotypes of E. coli K12 Q358 Smr lysogen strains carrying Argentinean and Australian O157 stx1, stx2, or stx2c prophages
| O157 donor strain | Lysogen genotypea |
Integration site | q allele | |||||
|---|---|---|---|---|---|---|---|---|
| stx1 | stx2 | stx2c | eae | ehxA | Smr | |||
| I-004 | − | + | − | − | − | + | wrbA | q933 |
| I-003 | − | + | − | − | − | + | wrbA | q933 |
| 313/99 | − | + | − | − | − | + | argW | q933 |
| 129/01 | − | + | − | − | − | + | argW | q933 |
| 210/03 | − | + | − | − | − | + | argW | q933 |
| 326/03 | − | + | − | − | − | + | argW | q933 |
| 755/05 | − | + | − | − | − | + | argW | q933 |
| 112/07 | − | + | − | − | − | + | argW | q933 |
| 145/98 | − | + | − | − | − | + | argW | q933 |
| FP-196 | − | + | − | − | − | + | argW | q933 |
| 353/00 | − | + | − | − | − | + | argW | q933 |
| EC3185 | + | − | − | − | − | + | argW | NAb |
| EC3206 | + | − | − | − | − | + | argW | NA |
| EC2441 | − | − | + | − | − | + | sbcB | q21 |
| EC3197 | − | − | + | − | − | + | sbcB | q21 |
| EC3204 | − | − | + | − | − | + | sbcB | q933 |
+, present; −, absent.
NA, not applicable for stx1 prophage.
q gene allelic variation upstream of stx2c in Argentinean and Australian O157.
The prevalences of q933 and q21 alleles in Australian and Argentinean O157 isolates were determined with q allele-specific PCR. Argentinean strains carrying stx2 (n = 43) tested positive for q933, while those carrying stx2c (n = 46) tested positive for q21. Unexpectedly, Australian strains carrying stx2c (n = 55) showed evidence for q933 and q21 gene allelic variation upstream of stx2c. Eighteen Australian O157 strains carrying stx2c possessed q933 (33%) and 36 possessed q21 (65%). A single stx2c-carrying Australian isolate showed evidence for deletion disruption of the q-stx2cA region which prevented q allele designation in this isolate. To confirm the unusual presence of the q933 allele upstream of stx2c in 18 Australian isolates and verify that designated q933 and q21 amplicons matched the appropriate gene sequences, the DNA sequences of 7 representative q allele amplicons were compared. The sequences of q933 and q21 gene regions from representative Argentinean stx2 and Australian stx2c (including both q933 and q21 allelic forms) O157 strains showed greater than 99% identity to stx2 q933 and stx2c q21 nucleotide regions from strain TW14359, respectively. Additional confirmation of stx2c q933 and stx2cq21 gene allelic variation was shown in stx2c lysogen strains. Representative E. coli K-12 lysogens transduced with Australian stx2c prophage were shown to possess the q21- or q933-related allelic forms (Table 4). Using these approaches, the presence of novel stx2cq933 in some strains of Australian O157 was verified.
Identification of putative virulence determinant genes and allelic variants.
Following our designation of the most prevalent O157 strains from both Argentina and Australia as LI/II, we investigated if putative TW14359 virulence factor genes were also present (Table 5). The presence of the putative heme binding protein gene (ECSP_3286) carried by the TW14359 stx2 (argW) prophage in 97% (36/37) of Argentinean O157 isolates was significantly different (P < 0.0001) from the presence in a single Australian O157 strain also carrying stx2 (argW) prophage. ospB (ECSP_2687), carried by the TW14359 stx2c (sbcB) prophage, was present in 43/46 Argentinean and 54/55 Australian O157 strains also carrying the stx2c (sbcB) prophage. Three Argentinean isolates shown to carry stx2 but not stx2c tested positive for ECSP_2687. These isolates also contained an occupied sbcB locus, suggesting that they may carry stx2c-negative sbcB prophage. Of the additional potential virulence determinants associated with the TW14359 genome, ECSP_1773 was present in 16/60 Argentinean strains and 14/60 Australian strains. ECSP_2870 and ECSP_2872 were present together in a higher proportion of Argentinean strains (38/60) than Australian strains (24/60); however, this was not statistically significant (P > 0.007). In the Australian strains, ECSP_2870 and ECSP_2872 were present together in significantly more human isolates (17/30) than animal isolates (7/30) (P < 0.05). Ankyrin repeat elements (ECSP_0242) were present in 56/60 Argentinean and the majority of Australian strains (55/60). Similarly, the nitric oxide reductase gene norV (ECSP_3620) was present in 57/60 Argentinean and the majority of Australian strains (58/60). All isolates carrying norV were non-LI and also belonged to either clade 4, 6, 7, or 8. All Argentinean (n = 3) and Australian (n = 2) O157 isolates that carried a ΔnorV allele (204-bp deletion homologous to EDL933 and Sakai strains) were shown to belong to LI. Therefore, significant associations of ankyrin repeat presence with LI/II (P < 0.05) and norV presence with LI/II (P < 0.05) were evident. Additional analysis of the translocated intimin receptor gene tir alleles, previously used to discriminate clinical O157 strains, revealed that with the exception of four Australian isolates possessing tir255A, all other Argentinean (60/60) and Australian O157 isolates (56/60) possessed the tir255T allele.
Table 5.
Escherichia coli O157 isolates positive for putative virulence determinants
| Determinanta | No. (%) of isolates |
|
|---|---|---|
| Argentina (n = 60) | Australia (n = 60) | |
| tir255T | 60 (100) | 56 (93) |
| ECSP_0242 | 56 (93) | 55 (92) |
| ECSP_1773 | 16 (27) | 14 (23) |
| ECSP_2687 | 46 (77) | 54 (90) |
| ECSP_2870/2872 | 38 (63) | 24 (40) |
| ECSP_3286 | 36 (60)b | 1 (2)b |
| ECSP_3620 | 57 (95) | 58 (97) |
ECSP locus tags indicate putative virulence determinants identified in the E. coli O157 TW14359 genome (18).
Considered to be significantly different (P < 0.0007).
DISCUSSION
The prevalences of particular stx genotypes in E. coli O157 isolates from different geographic origins have previously been reported (8, 22, 25). Fegan and Desmarchelier (8) tested 102 Australian E. coli O157 isolates from human and animal sources for the presence of stx1, stx2, and stx2c. Their findings demonstrated the following stx genotypes: stx1 stx2c (74%), stx2c (16%), stx1 stx2 (5%), stx2 stx2c (3%), and stx1 (3%). Studies from Argentina have also demonstrated predominance of the particular genotype stx2 stx2c (22, 25). Isolates in the current study represented multiple genotypes in proportions similar to the diversity previously demonstrated in Argentina and Australia. Consistent with this, we observed a significant difference between the dominant stx2 stx2c (Argentina) and stx1 stx2c (Australia) genotypes in this study.
The high prevalence of dominant but contrasting stx genotypes in Argentina and Australia provoked our investigation of Shiga-toxin bacteriophage insertion (SBI) sites in diverse sets of epidemiologically representative O157 isolates from each country. Through this we have demonstrated that stx1-positive Australian isolates dominantly carry novel stx1 prophage in the argW locus, which had previously been described only as a locus for stx2 prophage insertion (18, 29). Additionally, we have demonstrated that, in common with the U.S. spinach-associated outbreak strain TW14359 (18), Argentinean stx2 isolates most commonly carry stx2 prophage in argW. These data suggest that stx1-positive Australian isolates can be distinguished from Argentinean isolates on the basis of unique SBI profiles. Importantly, the novel combination of stx1 linkage with argW prophage insertion was verified by transduction of representative stx1 (argW) prophage to form E. coli K-12 lysogens. While both stx1 (argW) and stx2 (argW) prophages and their argW integration site were identified with the same primer pairs (specific to the 5′ and 3′ prophage/chromosome junctions, implying phage genome sequence similarity), the absence of the gene for putative heme binding protein (ECSP_3286) in Australian stx1 (argW) prophage genomes (K. S. Gobius, unpublished data), in addition to the alternative stx alleles, suggests further prophage sequence divergence.
In the context of our discovery of the novel stx1 (argW) prophage in Australian O157, we also prioritized attempts to characterize the stx1 prophage in 14 non-LI Argentinean O157 strains. Since none of the loci, yehV, wrbA, sbcB, argW, yecE, or prfC, which have been previously noted as SBI sites in enterohemorrhagic E. coli (29), appeared to be occupied with stx1 prophage, we attempted prophage induction, followed by lysogen formation in E. coli K-12 to identify the integration site(s). Despite numerous attempts at lysogeny, this approach to SBI characterization was also unsuccessful. As each of the 14 strains showed evidence consistent with prophage insertion in the yehV locus, it is tempting to speculate that the stx1 regulon may be carried by a prophage at this site. However, since all other Argentinean LI/II and lineage untypeable strains also appear to possess non-stx prophages inserted at yehV, it is unlikely that stx1 prophage inserts at this locus. As a consequence of this, the stx1 prophage integration site for Argentinean isolates carrying stx1 phage remains undetermined.
Consistent with previous studies (8, 25), we also observed a significant difference in the motility of O157 from Argentina (predominantly motile strains) and Australia (predominantly nonmotile strains), further supporting potential differentiation of the O157 populations in each country.
LSPA-6 genotyping data revealed that the predominant O157 isolates of both Argentinean and Australian origins share a similar genetic backbone and are designated LI/II genotype 211111. Overall, the current data contrast with previous studies that have determined lineage heterogeneity of O157 strains based on host and/or pathogenicity. In North America, LI and LI/II strains are dominant in humans infected with O157, and LII strains dominate in the cattle reservoir (38, 43, 44). In addition, Yokoyama et al. (42) recently established that in Chiba Prefecture, Japan, LI strains (52.5%), LI/II strains (31.5%), and LII strains (16%) are associated with human patients and asymptomatic carriers. Franz et al. (9) have also demonstrated in the Netherlands that LII isolates are most dominant in cattle, while LI/II strains, followed by LI strains, predominate among human isolates. Therefore, it is a notable contrast that in both Argentina and Australia, the single LSPA-6 phylogenetic lineage LI/II shows dominance in both cattle reservoirs and clinical disease manifestation. While we have shown that O157 strains with LSPA-6 LI and LII genotypes are also present in cattle and clinical isolates from Argentina and Australia, it appears that these alternative strains comprise a minor proportion of the O157 populations in both countries.
Following LSPA-6 genotyping, we examined the virulence clade types (24, 34) of Argentinean and Australian O157 isolates. Clade typing revealed additional phylogenetic insights, with clade 4, 6, 7, and 8 isolates observed to contribute to the composition of the LI/II genotype. Clade 8 isolates were first described by Manning et al. (24) in the United States and have subsequently been characterized in other countries, including Japan (41), Sweden (7), and Norway (13). The presence of clade 8 O157 strains in Argentina and Australia indicates that clade 8 strains are geographically widespread across several continents. Clade 6 and 7 isolates have been previously noted among U.S. O157 isolates which were designated LI/II (23); however, to the best of our knowledge, our study provides the first confirmation of virulence clade 4 association with LI/II. Furthermore, alternative country-specific and host-specific clade bias was observed between the two countries. In Argentina, clade 8 isolates dominated in both cattle and humans but were most prevalent in human cases. A similar pattern of clade dominance was observed for Australian isolates, where clade 7 dominated in both cattle and humans but was most prevalent in human cases.
Due to the severity of the U.S. spinach-associated O157 outbreak in 2006, a representative LI/II clade 8 outbreak strain, TW14359, has been characterized by SNP and genome analysis (18, 24). Seven putative virulence factors, not present in previously characterized LI and LII strains, were identified and considered with respect to their association with the alternative LSPA-6 lineages. Only one (ECSP_3286) of the seven putative virulence factors tested was shown to be more frequent in Argentinean isolates. The increased frequency of ECSP_3286, carried by the TW14359 stx2 (argW), prophage, in Argentinean strains can be attributed to the different rate of stx2 (argW) carriage in Argentinean and Australian isolates. On the basis of observed stx1 prophage presence and ΔnorV correlation, Kulasekara et al. (18) suggested that the presence of stx1 prophage is selective for the ΔnorV allele. In contrast, our data from Australian strains suggest that the ΔnorV allele is not predisposed by the presence of stx1. Alternatively, our data indicate that since Australian O157 isolates carrying stx1 generally also carry an undeleted norV allele, rather than stx1 selecting for the ΔnorV allele, it is likely the prophage type (i.e., stx1 [yehV] prophage) is associated with genomes possessing the ΔnorV allele. Extending this reasoning, it is also possible that complete norV genes are a feature of LI/II genomes, whereas ΔnorV alleles may be a feature of at least LI genomes. Previously, it has also been suggested that the tir255T→A polymorphism may act as a marker for virulence in E. coli O157 (4); however, a more specific association of tir255T with LI/II strains was recently observed by Laing et al. (19). With the exception of four Australian isolates, all other O157 isolates (93%) examined in this study carried tir255T. Our data confirm the association of tir255T with LI/II but demonstrate its association with multiple clades (4, 6, 7, and 8) within this lineage, suggesting, in contrast to previous studies (4, 9), that this SNP may not be a specific predictor for strains likely to cause human disease.
We have shown that stx2c prophage is common in LI/II O157 isolates (including Argentinean clade 4 and 8 and Australian clade 6 and 7 isolates). The presence of stx2c prophage in such isolates is consistent with the nonrandom concentration of stx2c in clades 4, 6, 7, and 8 in U.S. clinical O157 isolates observed by Manning et al. (24); however, we have noted q gene region heterogeneity in the Australian stx2c prophage. Allelic variation in the q gene region upstream of stx2 and stx2c was first reported by LeJeune et al. (20), who demonstrated that the q21 allele is characteristic for the stx2c prophage. For Australian O157 strains in this study, we have described the presence of the stx2c prophage carrying heterogenic q933 or q21 alleles immediately 5′ of the stx2c genes. The variable q933- and q21-related alleles were associated with stx2c genes in both LI/II stx1 stx2c or LI/II stx2c Australian O157 strains. To date, all previously described O157 stx2c genes have possessed q21 alleles encoding an apparent variant Q antiterminator protein (17, 20, 26, 37, 43); however, to our knowledge, the functionality of the Q21 protein has not been demonstrated. Thus, it could be anticipated that the variable q933- and q21-related alleles might regulate different levels of Stx2c, though further work is required to demonstrate such a relationship.
The presence of E. coli O157 isolates with related LI/II phylogeny, and which show evidence as the most prevalent strains in both Argentina and Australia, raises the possibility that these strains possess an ancient relationship. Leopold et al. (21) recently used high-throughput pyrosequencing to determine the nucleotide sequence of several O157 strains and developed a set of SNPs to examine probable ancestral phylogenetic linkages of O157 strains. Their data indicate that strain TW14359 is a representative of EHEC 1 clade, subgroup 3, cluster 1 strains (alternatively defined as LI/II clade 8 [19, 24]) which evolved at an earlier time than the more recent EHEC 1 clade, subgroup 3, cluster 3 strains EDL933 (31) and Sakai (14). Leopold et al. also suggest the likelihood that radiating evolution, with subsequent genetic bottlenecking, followed the emergence of the cluster 1/LI/II founder organism, to result in the extinction of particular radiating branches and the current limited pool of cluster 1 (LSPA-6 LI/II) strains. Since the current study supports the cluster 1 (LSPA-6 LI/II) designation of the dominant O157 strains from both Argentina and Australia, further phylogenomic investigation may determine the precise radial branch location of these strains.
In conclusion, we have demonstrated that diverse sets of E. coli O157 isolated in Argentina and Australia are differentiated by separate dominant phylogenetic clades. In Argentina, LI/II clade 8 strains with stx2 prophage integrated in the argW chromosomal insertion site were most prevalent, whereas in Australia, LI/II clade 7 strains with stx1 prophage integrated in the argW chromosomal insertion site were most prevalent. This O157 genotype differentiation was present in isolates causing human illness, as well as isolates from the animal reservoirs of each respective country. It is enticing to speculate that the correlation of separate country-specific genotypes in clinical and animal O157 isolates may provide the basis for observed differences in both the prevalence and severity of human disease caused by O157 in Australia (low) and Argentina (high). These data now provide evidence for the geographical segregation of E. coli O157 strains in which distinctive clade and Shiga toxin prophage combinations may provide genetic markers for E. coli O157 strains with various pathogenic potential. Examination of more extensive collections from each country, coupled with animal model comparisons of the virulence of different Argentinean and Australian O157 clades, is now recommended to provide further confirmation of these data.
ACKNOWLEDGMENTS
We thank John Bates and Helen Smith (Reference Microbiology and Molecular Epidemiology Laboratories, Queensland Health, Australia) for the kind provision of human O157 isolates. Fruitful discussion with Narelle Fegan, expert statistical advice from Richard Wilson, and helpful comments from the reviewers are gratefully acknowledged.
This study was supported by strategic research funds from CSIRO and Department of Primary Industries Victoria.
Footnotes
Published ahead of print 27 April 2012
REFERENCES
- 1. Allison HE, et al. 2003. Immunity profiles of wild-type and recombinant Shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect. Immun. 71:3409–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anonymous. 1993. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—western United States, 1992–1993. MMWR Morb. Mortal. Wkly. Rep. 42:258–263 [PubMed] [Google Scholar]
- 3. Besser TE, et al. 2007. Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl. Environ. Microbiol. 73:671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bono JL, et al. 2007. Association of Escherichia coli O157:H7 tir polymorphisms with human infection. BMC Infect. Dis. 7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caprioli A, Morabito S, Brugere H, Oswald E. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289–311 [DOI] [PubMed] [Google Scholar]
- 6. Erickson MC, Doyle MP. 2007. Food as a vehicle for transmission of Shiga toxin-producing Escherichia coli. J. Food Prot. 70:2426–2449 [DOI] [PubMed] [Google Scholar]
- 7. Eriksson E, Soderlund R, Boqvist S, Aspan A. 2011. Genotypic characterization to identify markers associated with putative hypervirulence in Swedish Escherichia coli O157:H7 cattle strains. J. Appl. Microbiol. 110:323–332 [DOI] [PubMed] [Google Scholar]
- 8. Fegan N, Desmarchelier P. 2002. Comparison between human and animal isolates of Shiga toxin-producing Escherichia coli O157 from Australia. Epidemiol. Infect. 128:357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Franz E, et al. 2012. Genetic features differentiating bovine, food, and human isolates of Shiga toxin-producing Escherichia coli O157 in The Netherlands. J. Clin. Microbiol. 50:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Friedrich AW, et al. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84 [DOI] [PubMed] [Google Scholar]
- 11. Gannon VP, et al. 1997. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J. Clin. Microbiol. 35:656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hancock D, Besser T, Lejeune J, Davis M, Rice D. 2001. The control of VTEC in the animal reservoir. Int. J. Food Microbiol. 66:71–78 [DOI] [PubMed] [Google Scholar]
- 13. Haugum K, Brandal LT, Lobersli I, Kapperud G, Lindstedt BA. 2011. Detection of virulent Escherichia coli O157 strains using multiplex PCR and single base sequencing for SNP characterization. J. Appl. Microbiol. 110:1592–1600 [DOI] [PubMed] [Google Scholar]
- 14. Hayashi T, et al. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11–22 [DOI] [PubMed] [Google Scholar]
- 15. Karn J, Brenner S, Barnett L, Cesareni G. 1980. Novel bacteriophage lambda cloning vector. Proc. Natl. Acad. Sci. U. S. A. 77:5172–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J, et al. 2001. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, beta-glucuronidase-negative enterohemorrhagic Escherichia coli O157. J. Bacteriol. 183:6885–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koitabashi T, et al. 2006. Genetic characterization of Escherichia coli O157: H7/- strains carrying the stx2 gene but not producing Shiga toxin 2. Microbiol. Immunol. 50:135–148 [DOI] [PubMed] [Google Scholar]
- 18. Kulasekara BR, et al. 2009. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 77:3713–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laing CR, et al. 2009. In silico genomic analyses reveal three distinct lineages of Escherichia coli O157:H7, one of which is associated with hyper-virulence. BMC Genomics 10:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. LeJeune JT, Abedon ST, Takemura K, Christie NP, Sreevatsan S. 2004. Human Escherichia coli O157:H7 genetic marker in isolates of bovine origin. Emerg. Infect. Dis. 10:1482–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leopold SR, et al. 2009. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc. Natl. Acad. Sci. U. S. A. 106:8713–8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leotta GA, et al. 2008. Characterisation of Shiga toxin-producing Escherichia coli O157 strains isolated from humans in Argentina, Australia and New Zealand. BMC Microbiol. 8:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu K, Knabel SJ, Dudley EG. 2009. rhs genes are potential markers for multilocus sequence typing of Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 75:5853–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manning SD, et al. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Masana MO, et al. 2010. Prevalence, characterization, and genotypic analysis of Escherichia coli O157:H7/NM from selected beef exporting abattoirs of Argentina. J. Food Prot. 73:649–656 [DOI] [PubMed] [Google Scholar]
- 26. Matsumoto M, et al. 2008. Identification and epidemiological description of enterohemorrhagic Escherichia coli O157 strains producing small amounts of Shiga toxin 2 in Aichi Prefecture, Japan. Jpn. J. Infect. Dis. 61:442–445 [PubMed] [Google Scholar]
- 27. McPherson M, et al. 2009. Serogroup-specific risk factors for Shiga toxin-producing Escherichia coli infection in Australia. Clin. Infect. Dis. 49:249–256 [DOI] [PubMed] [Google Scholar]
- 28. Nishikawa Y, et al. 2000. Relationship of genetic type of Shiga toxin to manifestation of bloody diarrhea due to enterohemorrhagic Escherichia coli serogroup O157 isolates in Osaka City, Japan. J. Clin. Microbiol. 38:2440–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ogura Y, et al. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:17939–17944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paton AW, Paton JC. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perna NT, et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 32. Persson S, Olsen KE, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 11:603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Riordan JT, Viswanath SB, Manning SD, Whittam TS. 2008. Genetic differentiation of Escherichia coli O157:H7 clades associated with human disease by real-time PCR. J. Clin. Microbiol. 46:2070–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rivas M, Miliwebsky ES, Chinen I, Deza N, Leotta GA. 2006. Epidemiology of the hemolytic uremic syndrome in Argentina. Diagnosis of the etiological agent, reservoirs and routes of transmission. Medicina (Buenos Aires) 66:27–32 [PubMed] [Google Scholar]
- 36. Shaikh N, Tarr PI. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strauch E, et al. 2008. Bacteriophage 2851 is a prototype phage for dissemination of the Shiga toxin variant gene 2c in Escherichia coli O157:H7. Infect. Immun. 76:5466–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Whitworth J, et al. 2010. Diverse genetic markers concordantly identify bovine origin Escherichia coli O157 genotypes underrepresented in human disease. Appl. Environ. Microbiol. 76:361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whitworth JH, et al. 2008. International comparison of clinical, bovine, and environmental Escherichia coli O157 isolates on the basis of Shiga toxin-encoding bacteriophage insertion site genotypes. Appl. Environ. Microbiol. 74:7447–7450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang Z, et al. 2004. Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70:6846–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yokoyama E, et al. 2011. Emergence of enterohemorrhagic Escherichia coli serovar O157 strains in clade 8 with highly similar pulsed-field gel electrophoresis patterns. J. Food Prot. 74:1324–1327 [DOI] [PubMed] [Google Scholar]
- 42. Yokoyama E, et al. 2011. Biased distribution of IS629 among strains in different lineages of enterohemorrhagic Escherichia coli serovar O157. Infect. Genet. Evol. 11:78–82 [DOI] [PubMed] [Google Scholar]
- 43. Zhang Y, et al. 2010. Lineage and host source are both correlated with levels of Shiga toxin 2 production by Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 76:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ziebell K, et al. 2008. Genotypic characterization and prevalence of virulence factors among Canadian Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 74:4314–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]