Abstract
A novel rutin-α-l-rhamnosidase hydrolyzing α-l-rhamnoside of rutin, naringin, and hesperidin was purified and characterized from Aspergillus niger DLFCC-90, and the gene encoding this enzyme, which is highly homologous to the α-amylase gene, was cloned and expressed in Pichia pastoris GS115. The novel enzyme was classified in glycoside-hydrolase (GH) family 13.
TEXT
α-l-Rhamnosidase (EC 3.2.1.40) cleaves terminal α-l-rhamnose from a large number of natural products, such as flavonoids, saponins, and many other natural glycosides, and this enzyme has been used in the food industry for eliminating the naringin bitterness of citrus juices (4, 20, 25, 26), removing hesperidin crystals from orange juices and releasing 7-O-β-d-glucoside-hesperetin, an important precursor in sweetener production (17, 24), and enhancing wine aromas (7, 6, 22). In addition, the deglycosylation of flavonoids in vitro by α-l-rhamnosidase has ushered in a new era of drug development (5, 8, 19, 27). Hitherto, α-l-rhamnosidases from fungi, bacteria, and animals have been purified and characterized (29), and several genes encoding α-l-rhamnosidases have also been cloned (1, 2, 3, 9, 16). Thus far, the α-l-rhamnosidases are categorized into four glycoside-hydrolase (GH) families, 28, 78, 106, and NC (nonclassified), in the CAZy (carbohydrate-active enzymes) database based on amino acid sequence similarities (10, 11).
In this paper, a novel rutin-α-l-rhamnosidase was purified and characterized from Aspergillus niger DLFCC-90, and the gene (rhaR) encoding this enzyme was also cloned and expressed in Pichia pastoris GS115.
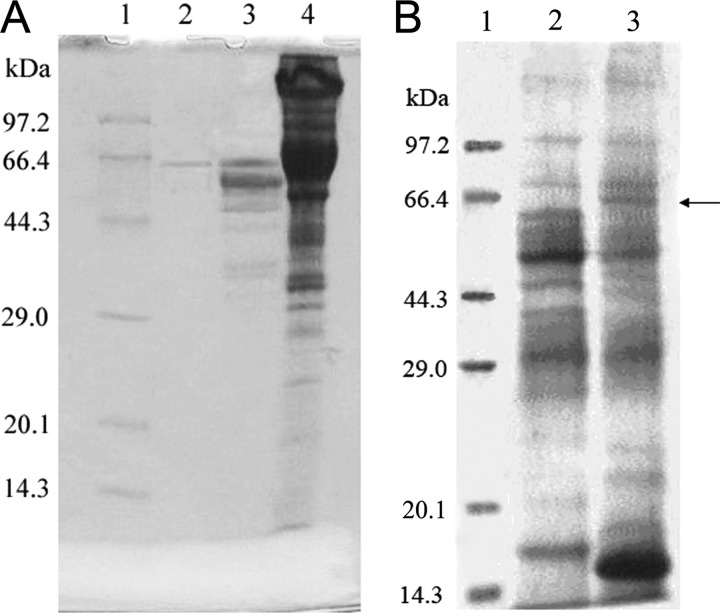
The strain A. niger DLFCC-90, obtained from the Culture Collection of Biotechnology Engineering of Dalian Polytechnic University (Dalian, China), was cultured with shaking at 28 to 30°C for 72 to 96 h in a wort medium of 5.0 Baume degrees containing 2% extract from flowers of Sophora japonica (Huai Hua in Chinese). To obtain pure enzyme, the cell-free culture was treated by a 3-step method, i.e., ammonium sulfate precipitation (75% saturation), a Sephadex G-75 column (Amersham Pharmacia) eluted with 0.02 M acetate buffer (pH 5.0), and a DEAE 52-cellulose column (Amersham Pharmacia) eluted with a linear gradient of KCl (0.0 to 0.5 M) in 0.02 M acetate buffer (pH 5.0). The determination of the protein concentration was performed as described in reference 13. The enzymatic activity was measured using 2.0 mg/ml rutin (Sigma) in 0.02 M acetate buffer (pH 5.0) as a substrate after a reaction at 50°C for 18 h, and products of rhamnose and isoquercitrin from the enzyme reaction were detected to calculate the enzyme activity (18, 28). After the above-described 3 steps of purification, the enzyme's specific activity was increased 5.3-fold. The purified enzyme migrated as a single band on the 12% SDS–PAGE gel (12) with an apparent molecular mass of about 66 kDa (Fig. 1A), which was similar to those for the previously reported α-l-rhamnosidase from Curvularia lunata (6) and naringinase from Aspergillus sojae (5). The 3-step-purified enzyme gave one major peak by high-performance liquid chromatography (HPLC) analysis (Tosoh TSKgel G2000SW; Φ, 7.8 mm by 300 mm), which indicates that it was almost pure protein. The purified enzyme can convert rutin to isoquercitrin from the qualitative analysis of the enzymatic product by the methods of HPLC (Knauer C-18; Φ, 3 mm by 300 mm) and nuclear magnetic resonance (NMR) (Bruker DR-400; Germany) (30).
Fig 1.
SDS-PAGE of the rutin-α-l-rhamnosidase. (A) SDS-PAGE of the enzyme from A. niger DLFCC-90. Lane 2, DEAE-purified enzyme; lane 3, Sephadex-purified enzyme; lane 4, ammonium sulfate precipitation of the culture supernatant proteins. (B) SDS-PAGE of rutin-α-l-rhamnosidase expressed in P. pastoris GS115. Lane 2, expression of the enzyme without methanol; lane 3, expression of the enzyme with methanol as an inducer. The arrow marks the predicted enzyme expressed in P. pastoris GS115. (A and B) Lane 1 in both panels, molecular mass markers (TaKaRa, Dalian, China).
The optimal pH and temperature of the purified enzyme (with rutin as a substrate) were 5.0 and 50°C, respectively. The enzyme had over 75% activity in the pH interval from 2.0 to 6.5, had over 80% activity below 60°C after 1 h of incubation, and still retained 40% activity at 70°C, but the activity was inactivated at 80°C after 1 h of incubation. The Km and Vmax of this enzyme for rutin were approximately 11.7 mM and 56.1 mM · h−1, respectively.
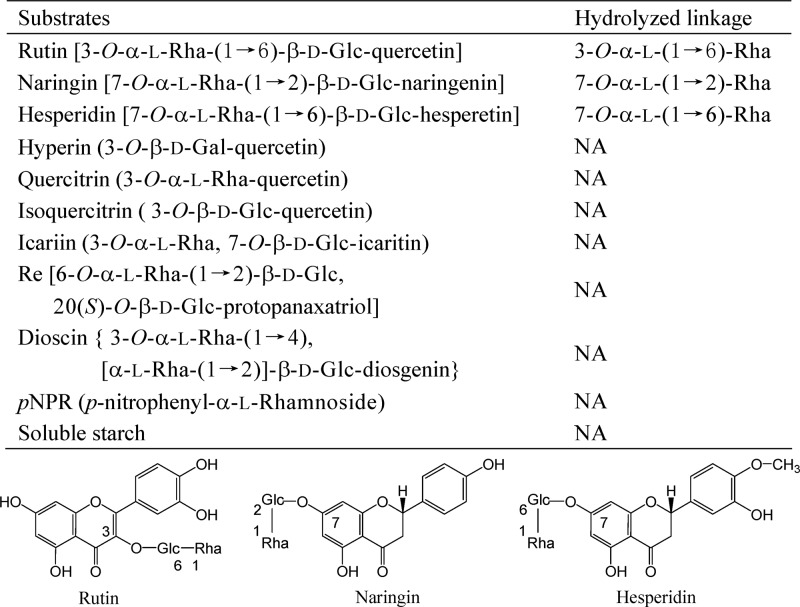
The substrate specificity of rutin-α-l-rhamnosidase was analyzed with flavonoids (28), saponins (6), p-nitrophenyl-α-l-rhamnoside (pNPR) (6), and soluble starch (7) as described previously (all substrates from Sigma). The purified enzyme can hydrolyze 3-O-α-(1→6)-l-rhamnoside of rutin, 7-O-α-(1→6)-l-rhamnoside of hesperidin, and 7-O-α-(1→2)-l-rhamnoside of naringin (Fig. 2) but cannot hydrolyze pNPR, α-l-rhamnosides of saponins, such as ginsenoside Re and dioscin, or other glycosides of flavonoids, such as 3-O-β-d-galactoside of hyperin, 3-O-α-l-rhamnoside of quercitrin, 3-O-α-l-rhamnoside or 7-O-β-d-glucoside of icariin, or 3-O-β-d-glucoside of isoquercitrin (Fig. 2).
Fig 2.
Substrate specificity of the rutin-α-l-rhamnosidase. Rha, rhamnose; Glc, glucose; Gal, galactose; NA, no activity.
The N-terminal amino acid sequence of the new enzyme was identified as LSAAEWRTQSIYFLLT… using a liquid sequenator from Applied Biosystems (model 491) (23). Based on the homology blasted from NCBI, the upstream primer (5′-TGAACTCCAACTTCGGCACT-3′) and downstream primer (5′-ATGTAGCTGAGGACGTTTTTSG-3′) were designed. The total RNA was isolated from A. niger DLFCC-90 with the Catrimox-14 RNA isolation kit (TaKaRa, Dalian, China). Using isolated total RNA as the template, first-strand cDNA was synthesized by a reverse transcription (RT)-PCR method with the TaKaRa 3′-Full rapid amplification of cDNA ends (RACE) core set, version 2.0, and then a partial cDNA of 640 bp was subsequently amplified using the two specific primers mentioned above. Using primers based on the known sequence described above, the 5′ and 3′ cDNA ends of rhaR were amplified as 612 bp and 745 bp long with the TaKaRa 5′-Full RACE kit and the TaKaRa 3′-Full RACE kit, respectively. The complete sequence of rhaR was identified to be 1,865 bp long from the three overlapping PCR amplification products mentioned above. The open reading frame (ORF) of rhaR encodes 505 amino acids, with a signal peptide of 21 amino acids. The molecular mass of this enzyme, calculated from the amino acid sequences, is 53 kDa, which is much smaller than the 66 kDa estimated from the SDS-PAGE gel (Fig. 1A), indicating that the enzyme secreted from A. niger DLFCC-90 is possibly glycosylated (14, 15, 16).
The new enzyme gene (rhaR) exhibits high similarity with the α-amylase gene (EC 3.2.1.1) from aspergilli. The ORF of rhaR is completely identical to those of two predicted α-amylase genes from aspergilli (GenBank accession numbers XM_001394298 and HQ186299).
The rhaR cDNA was subcloned into the vector pPIC9K (Invitrogen) and then transformed into P. pastoris GS115 (Invitrogen), and the recombinant Pichia strain was grown in BMGY medium using methanol as an inducer (21). There appeared to be a methanol-inducible (0.5%, vol/vol) protein with an apparent molecular mass of 66 kDa, which is in agreement with the molecular mass of the enzyme from A. niger DLFCC-90, but there was no specific band in noninduced cells (Fig. 1B). The crude cell extracts from P. pastoris GS115 which harbored expression vector showed derhamnosylation activity for rutin, naringin, and hesperidin (data not shown).
The amino acid sequence of rutin-α-l-rhamnosidase was compared with that of an α-amylase (Protein Data Bank [PDB] accession number 2AAA). Both enzymes consist of 484 amino acids, and similarity of above 95% was found between these two enzymes (Fig. 3). The new rutin-α-l-rhamnosidase contains amino acids (Asp206, Glu230, and Asp297) relating to α-amylase catalysis, but the new enzyme hydrolyzes α-l-rhamnoside of flavonoid and cannot hydrolyze soluble starch (Fig. 2), unlike α-amylase. The new rutin-α-l-rhamnosidase is therefore obviously a novel type of flavonoid glycoside-hydrolase. Based on amino acid sequence similarities, the novel enzyme was classified as GH family 13.
Fig 3.
Sequence alignment of rutin-α-l-rhamnosidase and 2AAA. The identical residues in the individual columns are indicated with a black background. The conserved amino acid residues (Asp206, Glu230, and Asp297) of 2AAA are indicated by black rectangles.
Nucleotide sequence accession number.
The nucleotide sequence for rhaR has been submitted to the GenBank database (accession number EU200666).
Protein structure accession number.
The predicted amino acid sequence of the new rutin-α-l-rhamnosidase has been submitted to the NCBI Protein Database (accession number ABW81206).
ACKNOWLEDGMENTS
This work was supported by the Program for Liaoning Innovative Research Team in University (LNIRT; 2009T007) and the National Science Foundation of the People's Republic of China (NSFC).
Footnotes
Published ahead of print 27 April 2012
REFERENCES
- 1. Avila M, et al. 2009. Physiological and biochemical characterization of the two α-l-rhamnosidases of Lactobacillus plantarum NCC245. Microbiology 155:2739–2749 [DOI] [PubMed] [Google Scholar]
- 2. Beekwilder J, et al. 2009. Characterization of rhamnosidases from Lactobacillus plantarum and Lactobacillus acidophilus. Appl. Environ. Microbiol. 75:3447–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Birgisson H, et al. 2004. Two new thermostable α-l-rhamnosidases from a novel thermophilic bacterium. Enzyme Microb. Technol. 34:561–571 [Google Scholar]
- 4. Busto MD, Meza V, Ortega N, Perez-Mateos M. 2007. Immobilization of naringinase from Aspergillus niger CECT 2088 in poly (vinyl alcohol) cryogels for the debittering of juices. Food Chem. 104:1177–1182 [Google Scholar]
- 5. Chang HY, et al. 2011. Purification and characterisation of Aspergillus sojae naringinase: the production of prunin exhibiting markedly enhanced solubility with in vitro inhibition of HMG-CoA reductase. Food Chem. 124:234–241 [Google Scholar]
- 6. Feng B, et al. 2007. Purification, characterization, and substrate specificity of a glucoamylase with steroidal saponin-rhamnosidase activity from Curvularia lunata. Appl. Microbiol. Biotechnol. 76:1329–1338 [DOI] [PubMed] [Google Scholar]
- 7. Gallego MV, Piñaga F, Ramón D, Vallés S. 2001. Purification and characterization of an α-l-rhamnosidase from Aspergillus terreus of interest in winemaking. J. Food Sci. 66:204–209 [Google Scholar]
- 8. González-Barrio R, et al. 2004. Production of bioavailable flavonoid glucosides in fruit juices and green tea by use of fungal α-l-rhamnosidases. J. Agric. Food Chem. 52:6136–6142 [DOI] [PubMed] [Google Scholar]
- 9. Hashimoto W, Miyake O, Nankai H, Murata K. 2003. Molecular identification of an α-l-rhamnosidase from Bacillus sp. strain GL1 as an enzyme involved in complete metabolism of gellan. Arch. Biochem. Biophys. 415:235–244 [DOI] [PubMed] [Google Scholar]
- 10. Henrissat B. 1991. A classification of glycosyl hydrolases based on amino-acid sequence similarities. Biochem. J. 280:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henrissat B, Davies GJ. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7:637–644 [DOI] [PubMed] [Google Scholar]
- 12. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 13. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 14. Manzanares P, De Graaff LH, Visser J. 1997. Purification and characterization of an α-l-rhamnosidase from Aspergillus niger. FEMS Microbiol. Lett. 157:279–283 [DOI] [PubMed] [Google Scholar]
- 15. Manzanares P, Orejas M, Ibanez E, Vallés S, Ramon D. 2000. Purification and characterization of an α-l-rhamnosidase from Aspergillus nidulans. Lett. Appl. Microbiol. 31:198–202 [DOI] [PubMed] [Google Scholar]
- 16. Manzanares P, Van Den Broeck HC, De Graaff LH, Visser J. 2001. Purification and characterization of two different α-l-rhamnosidases, RhaA and RhaB, from Aspergillus aculeatus. Appl. Environ. Microbiol. 67:2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mazzaferro L, Pinuel L, Minig M, Breccia J. 2010. Extracellular monoenzyme deglycosylation system of 7-O-linked flavonoid β-rutinosides and its disaccharide transglycosylation activity from Stilbella fimetaria. Arch. Microbiol. 192:383–393 [DOI] [PubMed] [Google Scholar]
- 18. Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426–428 [Google Scholar]
- 19. Monti D, Pisvejcová A, Kren V, Lama M, Riva S. 2004. Generation of an α-l-rhamnosidase library and its application for the selective derhamnosylation of natural products. Biotechnol. Bioeng. 87:763–771 [DOI] [PubMed] [Google Scholar]
- 20. Prakash S, Singhal RS, Kulkarni PR. 2002. Enzymic debittering of Indian grapefruit (Citrus paradisi) juice. J. Sci. Food Agric. 82:394–397 [Google Scholar]
- 21. Racape J, et al. 2005. Ca2+-dependent lipid binding and membrane integration of PopA, a harpin-like elicitor of the hypersensitive response in tobacco. Mol. Microbiol. 58:1406–1420 [DOI] [PubMed] [Google Scholar]
- 22. Spagna G, Barbagallo RN, Martino A, Pifferi PG. 2000. A simple method for purifying glycosidases: α-l-rhamnopyranosidase from Aspergillus niger to increase the aroma of Moscato wine. Enzyme Microb. Technol. 27:522–530 [DOI] [PubMed] [Google Scholar]
- 23. Tarentino AL, Quinones G, Hauer CR, Changchien LM, Plummer TH. 1995. Molecular cloning and sequence analysis of Flavobacterium meningosepticum glycosylasparaginase: a single gene encodes the α and β subunits. Arch. Biochem. Biophys. 316:399–406 [DOI] [PubMed] [Google Scholar]
- 24. Terada Y, Kometani T, Nishimura T, Takii H, Okada S. 1995. Prevention of hesperidin crystal formation in canned mandarin orange syrup and clarified orange juice by hesperidin glycosides. Food Sci. Technol. Int. 1:29–33 [Google Scholar]
- 25. Tsen HY, Tsai SY, Yu GK. 1989. Fiber entrapment of naringinase from Penicillium sp. and application to fruit juice debittering. J. Ferment. Bioeng. 67:186–189 [Google Scholar]
- 26. Tsen HY, Yu GK. 1991. Limonin and naringin removal from grapefruit juice with naringinase entrapped in cellulose triacetate fibers. J. Food Sci. 56:31–34 [Google Scholar]
- 27. Vila-Real H, Alfaia AJ, Rosa ME, Calado AR, Ribeiro MHL. 2010. An innovative sol-gel naringinase bioencapsulation process for glycosides hydrolysis. Process Biochem. 45:841–850 [Google Scholar]
- 28. Wei SH, Yu HS, Jin FX. 2003. Separating and purification of flavonoid active compounds. J. Dalian Inst. Light Ind. 22:114–116 [Google Scholar]
- 29. Yadav V, Yadav PK, Yadav S, Yadav KDS. 2010. α-l-Rhamnosidase: a review. Process Biochem. 45:1226–1235 [Google Scholar]
- 30. Yang Y, et al. 2009. Separation and purification of three flavonoids from Helichrysum arenarium (L.) Moench by HSCCC. Chromatographia 69:963–967 [Google Scholar]