Abstract
We have developed and validated two new fluorescence-based PCR assays to detect the Wolbachia wMel strain in Aedes aegypti and the wRi and wAu strains in Drosophila simulans. The new assays are accurate, informative, and cost-efficient for large-scale Wolbachia screening.
TEXT
The intracellular bacterium Wolbachia pipientis parasitizes and spreads in many arthropod hosts (13, 14). One classic example is the rapid sweep of the Wolbachia Riverside strain (wRi) across populations of Drosophila simulans in California (11). This evolutionarily optimized mechanism has inspired the use of Wolbachia as a driver to alter insect population structure (10). The recent establishment of wMel-infected, dengue virus-suppressing Aedes aegypti populations in Australia paves the way for similar programs in other countries (5, 12). In future release operations, rapid monitoring of wMel in Ae. aegypti will remain an ongoing requirement.
Current molecular methods to detect Wolbachia in Ae. aegypti are based on PCR followed by electrophoresis (2, 3, 5, 7–9, 15). These assays might be adequate for routine applications, but they are not ideal for large-scale field experiments. Under field conditions, mosquito specimens often comprise a mixture of Wolbachia-infected and uninfected individuals. One potential problem is the amplification of trace amounts of exogenous Wolbachia DNA from Wolbachia-negative samples. It is therefore desirable to develop a robust screening assay that can simultaneously detect wMel infection and quantify wMel density.
Three sets of primers were developed for the Ae. aegypti assay: (i) Aedes universal primer pair mRpS6_F (5′-AGTTGAACGTATCGTTTCCCGCTAC) and mRpS6_R (5′-GAAGTGACGCAGCTTGTGGTCGTCC), which target the conserved region of the RpS6 gene, to detect the presence of Aedes DNA (Fig. 1a); (ii) Ae. aegypti primers aRpS6_F (5′-ATCAAGAAGCGCCGTGTCG) and aRpS6_R (5′-CAGGTGCAGGATCTTCATGTATTCG), which target the Ae. aegypti-specific polymorphisms within the variable region of RpS6, to distinguish Ae. aegypti from non-Ae. aegypti specimens (Fig. 1a); (iii) Wolbachia-specific primers w1_F, (5′-AAAATCTTTGTGAAGAGGTGATCTGC) and w1_R (5′-GCACTGGGATGACAGGAAAAGG), to detect the presence of Wolbachia DNA (Fig. 1b).
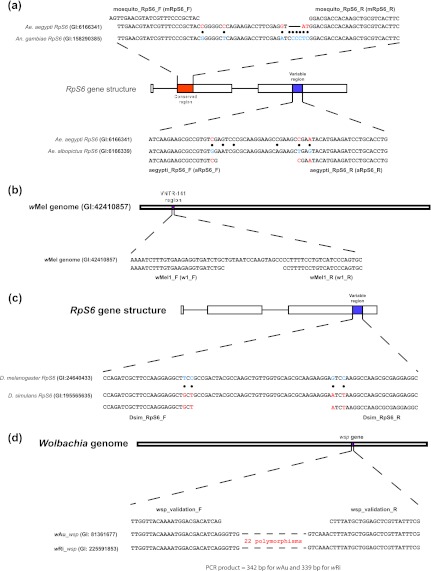
Fig 1.
Development of gene markers to detect and quantify Wolbachia infection in Ae. aegypti and D. simulans. (a) Coding sequences of the RpS6 gene from Ae. aegypti and Anopheles (An.) gambiae were aligned; a conserved region was selected to place the universal Aedes primers mRpS6_F and mRpS6_R. Nucleotide alignments of the coding sequences from Ae. aegypti and Ae. albopictus RpS6 were used to identify a variable region in which to position a pair of Ae. aegypti-specific primers (aRpS6_F and aRpS6_R), with 1 to 2 diagnostic nucleotides at the 3′ end of each primer. (b) The complete genome sequence of Wolbachia wMel was used to design a Wolbachia-specific marker at the VNTR-141 locus (see reference 9). The GenBank identifiers (GI) of the source sequences are given. Solid circles indicate polymorphic sites. (c) The variable region of the coding sequences in the RpS6 gene between Drosophila melanogaster and D. simulans is shown. Primers were designed to amplify D. simulans but not D. melanogaster. Solid circles indicate polymorphic sites. (d) Universal primers were placed at the conserved regions between the wRi and the wAu strains. These primers flank an ∼290-bp highly variable region that contains 22 polymorphic sites. The GI numbers of the source sequences are given. Note: reverse primers (mRpS6_R, aRpS6_R w1_R, Dsim_RpS6_R, and w1_R) are illustrated in the sense direction; their 5′-to-3′ sequences are described in the text.
PCR was carried out using the Roche LightCycler 480 system in a 384-well format (see the supplemental material). Aedes aegypti mosquitoes infected with Wolbachia (wMel) produced robust amplification for all three markers (Fig. 2a). Ae. aegypti mosquitoes that were Wolbachia negative supported amplification of mRpS6 and aRpS6, but not w1 (Fig. 2b). Aedes notoscriptus mosquitoes showed strong amplification for the generic mosquito marker (mRpS6) but failed to support amplification of the Ae. aegypti-specific marker (aRpS6) and the Wolbachia marker (w1) (Fig. 2c). These results indicate that the assay is able to simultaneously distinguish (i) between Ae. aegypti and Ae. notoscriptus and (ii) between Wolbachia-infected and uninfected Ae. aegypti mosquitoes. We have termed this new genotyping method the RT/HRM (real-time PCR/high-resolution melt) assay. The RT/HRM assay results were consistent with the two traditional PCR/electrophoresis-based assays, namely, the Braig assay (2) and the Caragata assay (3).
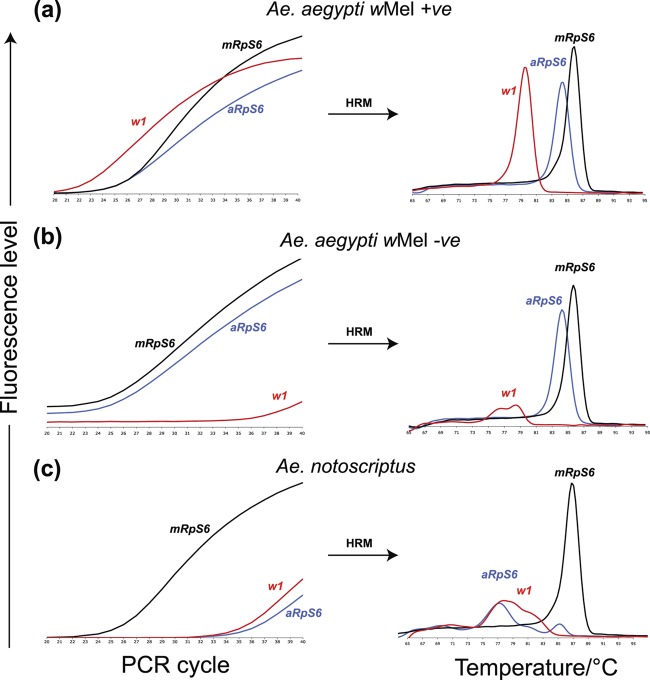
Fig 2.
Performance and expected outcomes of the Aedes RT/HRM assay. (Left graphs) PCR amplification profile; (right graphs) amplicon-specific melting peaks (i.e., Tm). (a) Ae. aegypti that carries wMel supports robust amplification of all three markers, with average Cp values (means ± 95% confidence intervals) of 27.86 ± 0.49 for mRpS6, 27.54 ± 0.49 for aRpS6, and 24.97 ± 0.83 for w1. The amplicons of these three markers had distinct Tm values: 85.65 ± 0.03°C for mRpS6, 84.33 ± 0.02°C for aRpS6, and 79.47 ± 0.02°C for w1. (b) Ae. aegypti that does not carry wMel supports robust amplification of the mRpS6 and aRpS6 markers from mosquito host DNA but not of the w1 primer from the Wolbachia DNA. (c) An Ae. notoscriptus mosquito could support amplification of only the universal Aedes marker (mRpS6), and not the Ae. aegypti-specific (aRpS6) or the wMel-specific (w1) marker.
The ability to quantify wMel in Ae. aegypti is important for preventing detection of false positives in field samples. Primer efficiencies were not significantly different from 100% based on standard curve analysis of four wMel+ genomic DNA dilutions (0.1×, 0.05×, 0.025×, and 0.0125×). We used the crossing point (Cp) difference between the aRpS6 and w1 markers to estimate Wolbachia load. The average density, estimated as 2[(Cp of aRpS6) − (Cp of w1)], was ∼6 copies of wMel per copy of RpS6 of the host genome. We also subjected the same DNA dilutions to the traditional electrophoresis-based PCR method. All dilutions produced a single expected PCR product of similar intensity (see Fig. S3 in the supplemental material). This indicates that the RT/HRM method is able to detect and quantify wMel at low concentrations, whereas the traditional assay is less quantitative.
Unlike in Ae. aegypti, in which Wolbachia has been artificially introduced, some populations of D. simulans in Australia are naturally infected with Wolbachia. One such strain is wAu, which does not induce host cytoplasmic incompatibility (4). The distribution of wAu in Australia has been documented, and the infection is generally found at low frequencies in populations of D. simulans on the east coast of Australia (4). Recently, sequencing of the Wolbachia wsp gene from D. simulans isofemale lines collected at Coffs Harbour in 2008 suggested that the wRi strain (11) might be present in this population (A. R. Weeks, unpublished data).
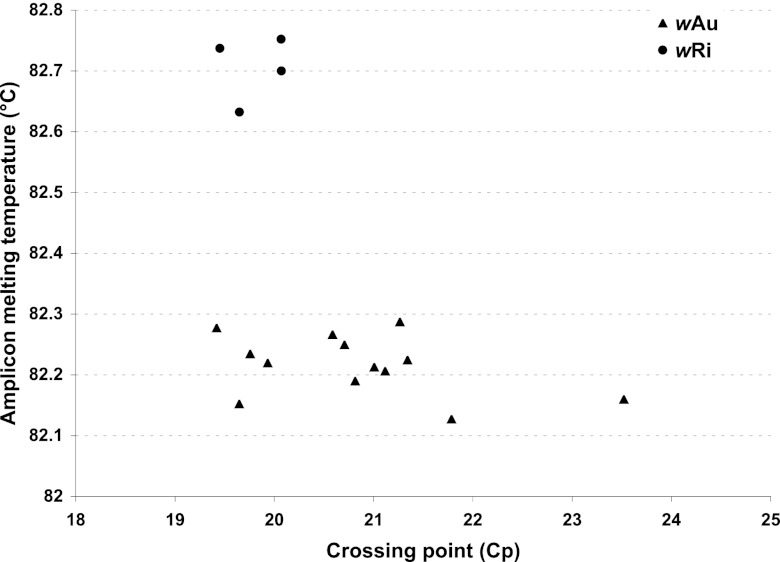
To confirm the presence of Wolbachia wRi and wAu strains in Drosophila simulans, we developed a new assay (Fig. 1c and d). We designed a pair of RpS6 primers (Dsim_RpS6_F, 5′-CCAGATCGCTTCCAAGGAGGCTGCT-3′; Dsim_RpS6_R, 5′-GCCTCCTCGCGCTTGGCCTTAGAT-3′) to check for successful DNA isolation (Fig. 1c). To detect and differentiate Wolbachia wRi and wAu infection in D. simulans, we designed a set of Wolbachia-specific primers (wsp_validation_F, 5′-TTGGTTACAAAATGGACGACATCAG-3′; wsp_validation_R, 5′-CGAAATAACGAGCTCCAGCATAAAG-3′). The priming sites of the Wolbachia primers are located at conserved sequences flanking a variable region (22 polymorphisms) of the wsp gene between the wAu and the wRi sequences (Fig. 1d). Among the 28 D. simulans flies from Coffs Harbor successfully genotyped, 17 were Wolbachia positive. A closer inspection of the melting temperatures (Tm) of the wsp products revealed two distinct Tm clusters that differed by ∼0.5°C (Fig. 3). Sequencing of amplicons confirmed that the high-Tm cluster (∼82.7°C) was the wRi allele and the lower-Tm cluster (∼82.2°C) was wAu. Since amplicon Tm is condition dependent, we believe that the 0.5°C Tm difference between wRi and wAu is a more useful diagnostic than their respective Tm's. Based on this Tm-based genotyping method, there were 4 occurrences of wRi (14.3%) 13 of wAu (46.4%), and 11 flies uninfected (39.3%) in Coffs Harbor, Australia.
Fig 3.
Classification of Wolbachia wRi and wAu infection status based on amplicon melting temperature differences. The graph shows a plot of melting temperature against the crossing point (Cp) of the wsp validation PCR amplicon in 17 Wolbachia-positive individuals. The proposed Wolbachia genotypic clusters are indicated by triangles (wAu) and circles (wRi).
The wRi strain of Wolbachia has not previously been detected in Australian D. simulans populations (1, 4) and likely represents a new infection. The origin of the infection is unclear, but given the strong cytoplasmic incompatibility associated with this strain (6) and its incompatibility with the endemic wAu strain (4), it is likely that the distribution of the Australian Wolbachia infections in D. simulans populations will change over time.
While the traditional assays are sufficient for small-scale Wolbachia screening, the new RT/HRM assays provide high-throughput options to detect and quantify Wolbachia infection in Ae. aegypti and D. simulans at all life stages. We have successfully reduced the unit cost of genotyping such that large-scale field monitoring can be more feasible. Although the reagent costs have been minimized, the RT/HRM assay does require an initial capital investment in (or access to) an RT-PCR instrument capable of performing HRM analysis. In addition, the current specificity of the RT/HRM assays means that further adjustments will be needed if additional Wolbachia strains (e.g., wMelpop) are introduced into Ae. aegypti or D. simulans populations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Heng Lin Yeap and Jason Axford for providing the mosquito samples and also Michael Turelli for helpful discussions.
The study was funded by a grant from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative as well as the CSIRO Cluster Collaboration Fund “Urbanism, Climate Change and Health” and fellowships from the Australian Research Council.
Footnotes
Published ahead of print 20 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ballard JW. 2004. Sequential evolution of a symbiont inferred from the host: Wolbachia and Drosophila simulans. Mol. Biol. Evol. 21:428–442 [DOI] [PubMed] [Google Scholar]
- 2. Braig HR, Zhou W, Dobson SL, O'Neill SL. 1998. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J. Bacteriol. 180:2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caragata EP, et al. 2011. Improved accuracy of the transcriptional profiling method of age grading in Aedes aegypti mosquitoes under laboratory and semi-field cage conditions and in the presence of Wolbachia infection. Insect Mol. Biol. 20:215–224 [DOI] [PubMed] [Google Scholar]
- 4. Hoffmann AA, Clancy D, Duncan J. 1996. Naturally-occurring Wolbachia infection in Drosophila simulans that does not cause cytoplasmic incompatibility. Heredity 76:1–8 [DOI] [PubMed] [Google Scholar]
- 5. Hoffmann AA, et al. 2011. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476:454–457 [DOI] [PubMed] [Google Scholar]
- 6. Hoffmann AA, Turelli M, Simmons GM. 1986. Unidirectional incompatibility between populations of Drosophila simulans. Evolution 40:692–701 [DOI] [PubMed] [Google Scholar]
- 7. Holden PR, Brookfield JF, Jones P. 1993. Cloning and characterization of an ftsZ homologue from a bacterial symbiont of Drosophila melanogaster. Mol. Gen. Genet. 240:213–220 [DOI] [PubMed] [Google Scholar]
- 8. O'Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM. 1992. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. U. S. A. 89:2699–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Riegler M, Sidhu M, Miller WJ, O'Neill SL. 2005. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr. Biol. 15:1428–1433 [DOI] [PubMed] [Google Scholar]
- 10. Turelli M, Hoffmann AA. 1999. Microbe-induced cytoplasmic incompatibility as a mechanism for introducing transgenes into arthropod populations. Insect Mol. Biol. 8:243. [DOI] [PubMed] [Google Scholar]
- 11. Turelli M, Hoffmann AA. 1991. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353:440–442 [DOI] [PubMed] [Google Scholar]
- 12. Walker T, et al. 2011. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476:450–453 [DOI] [PubMed] [Google Scholar]
- 13. Werren JH, Windsor DM. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. Biol. Sci. 267:1277–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Werren JH, Zhang W, Guo LR. 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. Biol. Sci. 261:55–63 [DOI] [PubMed] [Google Scholar]
- 15. Zhou W, Rousset F, O'Neill S. 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265:509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.