Abstract
Members of the genus Wolbachia are intracellular bacteria that are widespread in arthropods and establish diverse symbiotic associations with their hosts, ranging from mutualism to parasitism. Here we present the first detailed analyses of Wolbachia in butterflies from India with screening of 56 species. Twenty-nine species (52%) representing five families were positive for Wolbachia. This is the first report of Wolbachia infection in 27 of the 29 species; the other two were reported previously. This study also provides the first evidence of infection in the family Papilionidae. A striking diversity was observed among Wolbachia strains in butterfly hosts based on five multilocus sequence typing (MLST) genes, with 15 different sequence types (STs). Thirteen STs are new to the MLST database, whereas ST41 and ST125 were reported earlier. Some of the same host species from this study carried distinctly different Wolbachia strains, whereas the same or different butterfly hosts also harbored closely related Wolbachia strains. Butterfly-associated STs in the Indian sample originated by recombination and point mutation, further supporting the role of both processes in generating Wolbachia diversity. Recombination was detected only among the STs in this study and not in those from the MLST database. Most of the strains were remarkably similar in their wsp genotype, despite divergence in MLST. Only two wsp alleles were found among 25 individuals with complete hypervariable region (HVR) peptide profiles. Although both wsp and MLST show variability, MLST gives better separation between the strains. Completely different STs were characterized for the individuals sharing the same wsp alleles.
INTRODUCTION
Wolbachia species are intracellular obligatory symbionts belonging to the family Anaplasmataceae that infect a large variety of arthropods and filarial nematodes (6, 50). These bacteria establish diverse symbiotic associations with their hosts ranging from mutualism to parasitism (51). They are known to manipulate biology of their host by inducing male killing, feminization, parthenogenesis, cytoplasmic incompatibility, and speciation through reproductive isolation (46, 50, 51). Their main strategy of transfer is vertical cytoplasmic inheritance; however, horizontal transfer across different hosts also occurs (5, 21) and accounts for the widespread distribution of these bacteria, which infect around 16% to 66% of insect species (14, 17). A remarkable genetic diversity exists in Wolbachia, and gene phylogenies show the existence of 11 supergroups (A to K) (7, 11, 30, 38, 39). Studies of Wolbachia using multilocus sequence typing (MLST) have demonstrated discriminatory power of these approaches in accurately characterizing and identifying various Wolbachia strains (2, 3, 36, 41, 43, 54, 55). Wolbachia infections have been reported in various Lepidoptera families such as Lycaenidae, Pieridae, Nymphalidae, Hesperiidae, Pyralidae, Noctuidae, and Lasiocampidae (14, 18, 23, 41, 48).
Butterflies are mainly day-flying insects of the order Lepidoptera, comprising the true butterflies (superfamily Papilionoidea), the skippers (superfamily Hesperioidea), and the moth-butterflies (superfamily Hedyloidea). They exhibit genetic polymorphisms, mimicry, and aposematism. Some butterflies have evolved symbiotic and parasitic relationships with social insects, such as ants (15, 37). Butterflies serve as important plant pollinators and help to pollinate more than 50 economically important crop plants (8). Some species in their larval stages are pests and damage domestic crops or trees (10, 13).
The considerable ecological, biological, and behavioral diversity of butterflies suggests the need for further characterization of Wolbachia to understand the impact of infection on their reproduction, evolution, and speciation. Data on the molecular biology and phenotypic effects of Wolbachia from some butterfly species show the presence of supergroup A and B Wolbachia strains (12, 14, 18, 23, 41, 48). Wolbachia strains in butterflies have been implicated in basic biological processes such as sex ratio distortion, sex determination, sperm-egg compatibility, and speciation (33, 42, 47). However, the distribution of Wolbachia strains among butterfly species is largely unknown.
India's diverse fauna includes a rich variety of butterflies, comprising 1,501 species, which accounts for one-fifth of the known butterfly species in the world (16, 26). Western Ghats harbor 330 known species (16) belonging to 166 genera and 5 families (Lycaenidae, Pieridae, Nymphalidae, Papilionidae, and Hesperiidae [27]) and including 37 endemic species and another 23 shared only with Sri Lanka (16). Curiously, this tropical group from India and particularly Western Ghats, which is a biodiversity hot spot, has not yet been explored for Wolbachia infection. In the present report we show (i) the presence of Wolbachia among a sample of butterflies from Western Ghats belonging to five families; (ii) the diversity of Wolbachia strains within these butterflies, determined by using MLST and wsp genes; (iii) the phylogenetic relatedness of butterfly Wolbachia strains; and (iv) the role of recombination and point mutation in generating new sequence types (STs) in Wolbachia.
MATERIALS AND METHODS
Insects and DNA extraction.
Butterflies used in this study were collected during 2006 to 2008 from different regions of Western Ghats, India (Table 1). Legs of the butterflies were removed and preserved in absolute ethanol at −20°C until DNA extraction. Legs were used for screening, and the rest of the specimen was preserved for identification. Use of legs in screening for Wolbachia is a common practice (19, 24, 29, 31, 33). DNA was extracted from tissue using a QIAamp DNA minikit (Qiagen) following the manufacturer's instructions. In cases where specimens were small, abdomens were used. The specimens of all the butterflies under study were morphologically identified at the specimen collection and preservation center of the Department of Zoology, Modern College, Pune, India.
Table 1.
Screening of Wolbachia in butterflies from different families
| Family | No. of specimens | Butterfly | Collection locationa | No. of specimens |
Body part used for DNA isolation | |
|---|---|---|---|---|---|---|
| Total | Positive | |||||
| Papilionidae | 8 | Graphium agamemnon | F | 2 | 0 | Leg |
| Papilio demoleusb | D, F | 2 | 1 | Leg | ||
| Papilio polytes | F | 4 | 0 | Leg | ||
| Nymphalidae | 53 | Junonia arithya | F | 1 | 0 | Leg |
| Acraea violae | F | 2 | 0 | Leg | ||
| Ariadne merioneb | B, F, G | 3 | 1 | Leg | ||
| Byblia ilithyia | B | 1 | 0 | Leg | ||
| Charaxes dolon | F | 1 | 0 | Leg | ||
| Danaus chrysippusb | E, F | 3 | 1 | Leg | ||
| Danaus genutia | F | 1 | 0 | Leg | ||
| Euploea core | F, H | 2 | 0 | Leg | ||
| Euthalia lubentina | F | 1 | 0 | Leg | ||
| Euthalia nais | F | 1 | 0 | Leg | ||
| Hypolimnas bolina | C, F | 7 | 3 | Leg | ||
| Hypolimnas misippus | F | 4 | 0 | Leg | ||
| Junonia almana | F | 1 | 0 | Leg | ||
| Junonia hierta | F | 1 | 0 | Leg | ||
| Junonia lemoniasb | C, F | 3 | 1 | Leg | ||
| Lethe europa | F | 1 | 0 | Leg | ||
| Melanitis ledab | D, F | 2 | 1 | Leg | ||
| Moduza procris | F | 1 | 0 | Leg | ||
| Neptis hylasb | F | 2 | 1 | Leg | ||
| Parantica agleab | E, H | 2 | 1 | Leg | ||
| Polyura athamas | F | 1 | 0 | Leg | ||
| Precis iphitab | E, H | 2 | 1 | Leg | ||
| Tirumala limniaceb | F | 2 | 1 | Leg | ||
| Tirumala septentrionis | H | 1 | 0 | Leg | ||
| Vanessa cardui | F | 1 | 0 | Leg | ||
| Ypthima asteropeb | C, E | 5 | 1 | Abdomen | ||
| Ypthima sp. | C | 1 | 0 | Abdomen | ||
| Pieridae | 36 | Anaphaeis aurota | F | 1 | 0 | Abdomen |
| Catopsilia pomonab | E, F | 10 | 2 | Leg | ||
| Cepora nerissab | H | 1 | 1 | Leg | ||
| Colotis amatab | A | 2 | 2 | Abdomen | ||
| Delias eucharisb | F | 1 | 1 | Leg | ||
| Eurema brigitta | F | 2 | 0 | Abdomen | ||
| Eurema hecabe | C, D, E, F, G, H | 11 | 5 | Abdomen | ||
| Eurema laetab | C, F | 2 | 1 | Abdomen | ||
| Ixias marianne | F | 2 | 0 | Leg | ||
| Ixias pyreneb | F | 1 | 1 | Leg | ||
| Leptosia ninab | H | 1 | 1 | Abdomen | ||
| Pareronia valeriab | F, H | 2 | 2 | Leg | ||
| Lycaaenidae | 16 | Caleta caletab | E | 1 | 1 | Abdomen |
| Castalius rosimonb | C, F | 2 | 2 | Abdomen | ||
| Curetis thetis | F | 1 | 0 | Abdomen | ||
| Jalmenus evagorasb | E | 2 | 2 | Abdomen | ||
| Jamides bochus | F | 1 | 0 | Abdomen | ||
| Pseudozizeeria mahab | F | 2 | 2 | Abdomen | ||
| Talicada nyseusb | F | 3 | 3 | Abdomen | ||
| Tarucus narab | C | 1 | 1 | Abdomen | ||
| Zizeeria knysnab | F | 2 | 2 | Abdomen | ||
| Euchrysops cnejus | C | 1 | 0 | Abdomen | ||
| Hesperiidae | 5 | Telicota ancilla | H | 1 | 0 | Abdomen |
| Borbo cinnara | B | 1 | 0 | Abdomen | ||
| Taractrocera ceramusb | E | 2 | 1 | Abdomen | ||
| Udaspes folusb | H | 1 | 1 | Abdomen | ||
A, Ahemadnagar; B, Alandi; C, Junnar; D, Khanapur; E, Mulshi; F, Pune city; G, Satara; and H, Thane.
Butterfly found to harbor Wolbachia for the first time.
Wolbachia DNA amplification and sequencing.
The quality of DNA extracted from samples was checked by PCR targeting butterfly DNA using arthropod-specific 28S primers, amplified as described by Werren et al. (53), and samples with weak or no amplification were extracted again. All the specimens were screened initially for Wolbachia infection by PCR for the wsp (9) and ftsZ (5) genes using primers and previously described protocols. Primer details and PCR protocols for amplification of the five reported Wolbachia MLST genes (ftsZ, coxA, fbpA, hcpA, and gatB) and wsp genes are described elsewhere (2). The sequence data were analyzed against the Wolbachia MLST database (http://pubmlst.org/wolbachia/). All PCR products were purified using the polyethylene glycol (PEG)-NaCl method (44). The successfully amplified products of the five MLST genes and the wsp gene were sequenced bidirectionally with the respective primers using a BigDye terminator cycle sequencing kit, version 3.1 (Applied Biosystems). Sequences were obtained using an automatic DNA sequencer (3730 DNA analyzer; ABI).
At least one HVR for wsp gene was sequenced for 38 individuals. wsp typing assigned the wsp allele to 25 individuals with complete profiles for four HVRs. Alleles for 13 individuals could not be assigned due to incomplete HVR profiles. However, HVR peptide numbers were assigned to these individuals. Four strains could not be amplified with the wsp gene.
Wolbachia genetic diversity.
Estimates of genetic diversity (Pi), number of variable sites (VI), and ratios of synonymous substitutions per synonymous site to nonsynonymous substitutions per nonsynonymous site (Ka/Ks) were performed by DNAsp, version 4.10.2 (40).
Recombination and pairwise genetic distance analysis.
MaxChi (45) and GENECONV (35) programs in the RDP3 package (32) were used to perform the recombination analysis of the concatenated MLST gene alignment from butterfly STs. A Bonferroni correction was applied, and 100 permutations were generated. The highest acceptable P value cutoff was set at 0.05. The pairwise genetic distance of different Wolbachia strains was tested by using the Kimura 2-parameter method in MEGA4 (25, 49). Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS) program, version 17.0. Comparisons between the supergroups were performed using the Mann-Whitney U test for independent pairwise comparisons. A percent similarity matrix was calculated in Phylip 3.69. A concatenated data set for all MLST genes was considered for pairwise comparisons. The percent differences in pairwise comparisons between supergroups were evaluated by a Wilcoxon signed-rank test. A P value of less than 0.05 was considered statistically significant.
Phylogenetic analysis.
We retrieved STs and wsp alleles currently available in the MLST database for all members of supergroup B and Lepidoptera representatives of supergroup A. Wolbachia gene sequences (MLST and wsp) generated in this study were aligned with homologous sequences deposited in the Wolbachia MLST database using ClustalX, version 2.0.9 (28). All sequences were manually edited using MEGA4 (49). Unrooted phylogenetic trees were constructed using Bayesian inference and the neighbor-joining method for a concatenated data set for the five MLST genes and a separate data set for the wsp gene. For Bayesian inference of phylogeny, the program MrBayes 3.1.2 was used (20). The analysis for each gene consisted of 3,000,000 generations with sampling every 100 generations. The first 12,000 trees (40%) were discarded as “burn-in.” Before the probabilistic phylogenetic analyses were carried out, appropriate models of sequence evolution for each data set were chosen via the Akaike information criterion (AIC) using the program MrModeltest 2.2 (34). The selected model of nucleotide substitution was GTR+I+G for concatenated MLST gene sequences and the wsp gene. The final alignments consisted of 2,079 bp for concatenated MLST gene sequences and 495 bp for wsp gene fragments. Only the strains with full STs (complete five MLST alleles) were selected to construct the phylogenetic tree for the concatenated data set. The strains with incomplete allelic profiles were therefore omitted from the concatenated analysis. Similarly, strains with at least three complete HVRs were selected to construct the wsp phylogenetic tree. Three independent runs were performed for each data set. In phylogenetic trees, levels of confidence for each node are shown in the form of Bayesian posterior probabilities (BPP). BPP below 0.50 are not shown. NJ trees were constructed using MEGA 4.1 with 1,000 bootstrap replicates and the Kimura 2-parameter method as a model of nucleotide substitution.
Nucleotide sequence accession numbers.
Sequences obtained in this study were deposited in the Wolbachia MLST and GenBank databases with the alleles and accession numbers, respectively, noted in Table 2.
Table 2.
Complete and partial MLST and WSP profiles of Wolbachia isolates
| ID | Collection reference no. | Host |
Collection locationa | Strain | wsp allele (accession no.) |
wsp profile |
Allele (accession no.) |
ST | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Family | HVR1 | HVR2 | HVR3 | HVR4 | gatB | coxA | hcpA | ftsZ | fbpA | ||||||
| 208 | B4 | Ariadne merione | Nymphalidae | F | Amer_B_wMer | 64 (JN236173) | 35 | 35 | 38 | 44 | 96 (JN236109) | 14 (JN236015) | 40 (JN236146) | 73 (JN236075) | 4 (JN236043) | 153 |
| 209 | B21 | Colotis amata | Pieridae | A | Cama_B_wAma1 | (JN236208) | 8 | 10 | 4 (JN236103) | 14 (JN236010) | 40 (JN236141) | 7 (JN236070) | 4 (JN236038) | 147 | ||
| 210 | B22 | Colotis amata | Pieridae | A | Cama_B_wAma2 | 10 (JN236195) | 10 | 8 | 10 | 8 | 16 (JN236104) | 14 (JN236011) | 40 (JN236142) | 36 (JN236071) | 4 (JN236039) | 150 |
| 211 | F37 | Caleta caleta | Lycaenidae | E | Ccal_B_wCal | (JN236206) | 10 | 8 | 10 | 9 (JN236128) | 40 (JN236152) | |||||
| 212 | F48 | Cepora nerissa | Pieridae | H | Cner_B_wNer | 10 (JN236181) | 10 | 8 | 10 | 8 | 4 (JN236127) | 14 (JN236021) | 3 (JN236154) | 36 (JN236092) | 4 (JN236058) | 145 |
| 213 | F22 | Catopsilia pomona | Pieridae | F | Cpom_B_wPom | 10 (JN236202) | 10 | 8 | 10 | 8 | 39 (JN236116) | 14 (JN236026) | 36 (JN236085) | |||
| 214 | F3 | Catopsilia pomona | Pieridae | F | Cpom_B_wPom | (JN236178) | 8 | 10 | 8 | 9 (JN236118) | ||||||
| 215 | F14 | Castalius rosimon | Lycaenidae | C | Cros_B_wRos | 10 (JN236182) | 10 | 8 | 10 | 8 | 9 (JN236112) | 40 (JN236150) | ||||
| 216 | B5 | Castalius rosimon | Lycaenidae | F | Cros_B_wRos | 4 (JN236044) | ||||||||||
| 217 | F36 | Danaus chysippus | Nymphalidae | E | Dchr_B_wChr | (JN236174) | 2 | 142 | 131 | 39 (JN236120) | 11 (JN236028) | 101 (JN236163) | 36 (JN236081) | 4 (JN236052) | 151 | |
| 218 | B28 | Delias eucharis | Pieridae | F | Deuc_B_wEuc | 10 (JN236197) | 10 | 8 | 10 | 8 | 39 (JN236107) | 14 (JN236014) | 40 (JN236145) | 36 (JN236074) | 4 (JN236042) | 41 |
| 219 | F78 | Eurema hecabe | Pieridae | F | Ehec_B_wHec3 | 10 (JN236184) | 10 | 8 | 10 | 8 | 102 (JN236137) | 14 (JN236034) | 29 (JN236170) | 36 (JN236099) | 42 (JN236065) | 157 |
| 220 | B7 | Eurema hecabe | Pieridae | F | Ehec_B_wHec1 | 10 (JN236199) | 10 | 8 | 10 | 8 | 39 (JN236108) | 14 (JN236016) | 40 (JN236147) | 36 (JN236078) | 4 (JN236046) | 41 |
| 221 | F47 | Eurema hecabe | Pieridae | E | Ehec_B_wHec1 | 10 (JN236188) | 10 | 8 | 10 | 8 | 39 (JN236126) | 14 (JN236020) | 40 (JN236153) | 36 (JN236091) | 4 (JN236057) | 41 |
| 222 | F53 | Eurema hecabe | Pieridae | H | Ehec_B_wHec1 | 10 (JN236189) | 10 | 8 | 10 | 8 | 39 (JN236132) | 14 (JN236030) | 40 (JN236164) | 36 (JN236095) | 4 (JN236061) | 41 |
| 223 | F7 | Eurema hecabe | Pieridae | D | Ehec_B_wHec2 | (JN236171) | 19 | 17 | 24 | 102 (JN236136) | 14 (JN236033) | 100 (JN236167) | 36 (JN236098) | 4 (JN236064) | 156 | |
| 224 | F9 | Eurema laeta | Pieridae | C | Elae_B_wLae | (JN236172) | 19 | 17 | 24 | 9 (JN236138) | 80 (JN236035) | 100 (JN236169) | 8 (JN236100) | 156 (JN236066) | 149 | |
| 225 | B11 | Hypolimnas bolina | Nymphalidae | F | Hbol_B_wBol | 10 (JN236200) | 10 | 8 | 10 | 8 | 9 (JN236102) | 14 (JN236008) | 40 (JN236140) | 73 (JN236068) | 4 (JN236037) | 148 |
| 226 | F15 | Hypolimnas bolina | Nymphalidae | C | Hbol_B_wBol | (JN236191) | 8 | 4 (JN236113) | 40 (JN236151) | 36 (JN236079) | ||||||
| 227 | B14 | Hypolimnas bolina | Nymphalidae | F | Hbol_B_wBol | 10 (JN236180) | 10 | 8 | 10 | 8 | 14 (JN236009) | 36 (JN236069) | ||||
| 228 | F38 | Junonia iphita | Nymphalidae | E | Jiph_B_wIph | 10 (JN236183) | 10 | 8 | 10 | 8 | 9 (JN236121) | 100 (JN236158) | 73 (JN236080) | 4 (JN236053) | ||
| 229 | F20 | Junonia lemonias | Nymphalidae | C | Jlem_B_wLem | 10 (JN236198) | 10 | 8 | 10 | 8 | 4 (JN236114) | 14 (JN236024) | 40 (JN236161) | 36 (JN236088) | 4 (JN236050) | 146 |
| 230 | F21 | Ixias pyrene | Pieridae | F | Ipyr_B_wPyr | 10 (JN236203) | 10 | 8 | 10 | 8 | 39 (JN236115) | 14 (JN236025) | 40 (JN236159) | 36 (JN236084) | 4 (JN236048) | 41 |
| 231 | F45 | Jalmenus evagoras | Lycaenidae | E | Jeva_B_wEva1 | (JN236176) | 18 | 16 | 23 | 101 (JN236124) | 79 (JN236029) | 99 (JN236156) | 73 (JN236089) | 155 (JN236055) | 155 | |
| 232 | F46 | Jalmenus evagoras | Lycaenidae | E | Jeva_B_wEva2 | (JN236175) | 18 | 16 | 101 (JN236125) | 79 (JN236019) | 40 (JN236155) | 73 (JN236090) | 4 (JN236056) | 154 | ||
| 233 | F49 | Udaspes folus | Hesperidae | H | Ufol_B_wFol | 10 (JN236179) | 10 | 8 | 10 | 8 | 39 (JN236129) | 36 (JN236093) | 4 (JN236059) | |||
| 234 | F65 | Zizeeria knysna | Lycaenidae | F | Zkny_B_wKny | 10 (JN236194) | 10 | 8 | 10 | 8 | 39 (JN236135) | 14 (JN236032) | 40 (JN236166) | 36 (JN236097) | 4 (JN236063) | 41 |
| 235 | F51 | Leptosia nina | Pieridae | H | Lnin_B_wNin | 10 (JN236177) | 10 | 8 | 10 | 8 | 39 (JN236131) | 14 (JN236023) | 40 (JN236162) | 7 (JN236094) | 4 (JN236062) | 152 |
| 236 | F5 | Melanitis leda | Nymphalidae | D | Melanitis leda | 39 (JN236130) | 9 (JN236022) | |||||||||
| 237 | B26 | Neptis hylas | Nymphalidae | F | Nhyl_B_wHyl | (JN236186) | 10 | 8 | 39 (JN236106) | 14 (JN236013) | 40 (JN236144) | 36 (JN236073) | 4 (JN236041) | 41 | ||
| 238 | F39 | Parantica aglea | Nymphalidae | E | Pagl_B_wAgl | 39 (JN236122) | 4 (JN236054) | |||||||||
| 239 | F6 | Papilio demoleus | Papilionidae | D | Pdem_B_wDem | 10 (JN236193) | 10 | 8 | 10 | 8 | 39 (JN236134) | |||||
| 240 | B8 | Pseudozizeeria maha | Lycaenidae | F | Pmah_B_wMah | 10 (JN236201) | 10 | 8 | 10 | 8 | 39 (JN236110) | 14 (JN236017) | 40 (JN236148) | 36 (JN236077) | 4 (JN236047) | 41 |
| 241 | F29 | Pseudozizeeria maha | Lycaenidae | F | Pmah_B_wMah | 10 (JN236205) | 10 | 8 | 10 | 8 | 39 (JN236117) | 14 (JN236027) | 40 (JN236160) | 36 (JN236083) | 4 (JN236051) | 41 |
| 242 | B25 | Pareronia valeria | Pieridae | F | Pval_B_wVal | (JN236185) | 10 | 8 | 10 | 39 (JN236105) | 14 (JN236012) | 40 (JN236143) | 36 (JN236072) | 4 (JN236040) | 41 | |
| 243 | F54 | Pareronia valeria | Pieridae | H | Pval_B_wVal | (JN236207) | 10 | 8 | 10 | 39 (JN236133) | 14 (JN236031) | 40 (JN236165) | 36 (JN236096) | 4 (JN236060) | 41 | |
| 244 | F34 | Teractrocera ceramus | Hesperidae | E | Tcer_B_wCer | (JN236187) | 10 | 8 | 36 (JN236082) | |||||||
| 245 | F16 | Tarucus nara | Lycaenidae | C | Tnar_B_wNar | 36 (JN236087) | 4 (JN236049) | |||||||||
| 246 | RP3 | Telicada nyseus | Lycaenidae | F | Tnys_B_wNys1 | 10 (JN236190) | 10 | 8 | 10 | 8 | 4 (JN236139) | 14 (JN236036) | 40 (JN236168) | 36 (JN236101) | 4 (JN236067) | 146 |
| 247 | B9 | Telicada nyseus | Lycaenidae | F | Tnys_B_wNys2 | 10 (JN236196) | 10 | 8 | 10 | 8 | 4 (JN236111) | 14 (JN236018) | 40 (JN236149) | 73 (JN236076) | 4 (JN236045) | 125 |
| 248 | F4 | Telicada nyseus | Lycaenidae | F | Tnys_B_wNys | 10 (JN236204) | 10 | 8 | 10 | 8 | 4 (JN236123) | 40 (JN236157) | ||||
| 249 | F35 | Ypthima asterope | Nymphalidae | E | Yast_B_wAst | 10 (JN236192) | 10 | 8 | 10 | 8 | 39 (JN236119) | 36 (JN236086) | ||||
A, Ahemadnagar; B, Alandi; C, Junnar; D, Khanapur; E, Mulshi; F, Pune city; G, Satara; and H, Thane.
RESULTS
A total of 118 individuals representing 56 species belonging to five families of Lepidoptera were screened for Wolbachia by PCR assay using Wolbachia-specific wsp and ftsZ gene primers. The infection status of each species and the number of individuals screened are listed in Table 1. Twenty-nine species representing all five families were positive for Wolbachia (Table 1). In total, 44 of the 118 individuals were positive.
At least one MLST gene was amplified and sequenced for all 44 individuals. Two individuals exhibited the presence of multiple Wolbachia infections, with double peaks in the chromatograms. These strains were removed from the analysis. Complete MLST profiles were generated for 26 Wolbachia strains, whereas repeated failures to PCR amplify particular Wolbachia genes (MLST and wsp) resulted in 16 incomplete profiles (Table 2).
Sequence typing was performed on the 26 complete MLST strains using the Wolbachia MLST database (http://pubmlst.org/wolbachia/) (Table 2). Characterization of allelic profiles indicated the presence of 15 STs in butterflies from this study. Of these, the allelic profiles and STs for 13 strains were new to the MLST database, whereas two STs (ST41 and ST125) were previously known (Table 2).
Divergence among the 26 STs accounted for 97 variable sites (VI) out of 2,073 sites (4.679%) with concatenated alignment of all five Wolbachia MLST genes (Table 3). The gene coxA showed the highest nucleotide divergence, with 36 variable sites out of 402 (8.955%), followed by gatB, with 28 variable sites out of 369 (7.588%) (Table 3). The average Ka/Ks per gene was found to be <1 (average Ka/Ks across genes is 0.16877), which indicates strain evolution mainly by synonymous substitutions. This is in line with a scenario of strong purifying selection.
Table 3.
Genetic features at five MLST and wsp loci
| Locus | Pis | Pia | Ka/Ks | % VI |
|---|---|---|---|---|
| gatB | 0.04352 | 0.00494 | 0.113511 | 7.588 |
| coaX | 0.06432 | 0.00534 | 0.083022 | 8.955 |
| hcpA | 0.02250 | 0.00510 | 0.226666 | 5.630 |
| fbpA | 0.00523 | 0.00220 | 0.420650 | 3.782 |
| ftsZ | 0.01270 | 0.0000 | 0.00000 | 1.379 |
| MLST concatenated | 0.04038 | 0.00575 | 0.142397 | 4.679 |
| wsp | 0.06589 | 0.03153 | 0.478524 | 29.580 |
Occurrence of recombination and point mutations.
The recombination analysis based on the MLST concatenated data indicated that recombination events occurred in three STs, ST151 (beginning breakpoint; 1234, ending breakpoint, 9) and ST154 and ST155 (beginning breakpoint; 773, ending breakpoint, 9). The events revealed that this localized divergence was the result of recombination involving the major parent ST149 for all three STs, the minor parent ST145 for ST151, and the minor parent ST153 for ST154 and ST155 (Table 4). Even if ST151, ST154, and ST155 are recombinant, the phylogeny inference based on MLST placed these three STs in supergroup B.
Table 4.
Occurrence of recombination in the concatenated data set
| Recombinant sequence | Major parent | Minor parent | Average P value by: |
|
|---|---|---|---|---|
| MaxChi | GENECONV | |||
| ST151 | ST149 | ST145 | 2.720 × 10−2 | 1.201 × 10−5 |
| ST154 | ST149 | ST153 | 2.298 × 10−3 | 1.729 × 10−4 |
| ST155 | ST149 | ST153 | 2.298 × 10−3 | 1.729 × 10−4 |
ST154 and ST155 were observed in Jalmenus evagoras specimens which were collected from the same location (Mulshi, Pune, India). These STs are results of the point mutations in hcpA (base 1004) and fbpA (base 1890). (All the base positions given here are based on the concatenated MLST gene data set.) ST145 (Cepora nerissa, Pieridae) and ST146 (Junonia lemonias, Nymphalidae, and Talicada nyseus, Lycaenidae) are very closely related and arose as a result of the point mutation at base 1181 in hcpA. Surprisingly, these two STs were found in distinctly different hosts from three localities and belonging to three different families. Hypolimnas bolina from the family Nymphalidae (this study) and Colias erate poliographus from the family Pieridae (from Japan) were found to harbor very closely related Wolbachia (ST148 and ST141, respectively). These STs were also formed as a result of point mutations in gene gatB at bases 315 and 359. ST125 (Hypolimnas bolina and Talicada nyseus) and ST147 were formed as a result of point mutations at bases 1503 (ftsZ) and 1709 (fbpA). ST41 was observed among a wide range of butterfly hosts, and it showed point mutations in gatB at positions 120 and 359 relative to ST150, which was found in Colotis amata. These results suggest that along with the recombination, point mutation events also play a crucial role in the genesis of new STs in Wolbachia.
Genetic distance among STs.
To detect whether the genetic variation (percent similarity) was statistically significant, a nonparametric Mann-Whitney U test was independently conducted between Wolbachia representatives of supergroup A from the MLST database, all supergroup B strains, and the samples from this study. Three different tests were conducted for the three data sets described above (Table 5). The data reveal that the genetic difference among Wolbachia strains in this study and the genetic difference among the rest of the supergroup B Wolbachia strains are not significantly different (U = 4565.5, P = 0.00). The genetic difference between supergroup A and supergroup B Wolbachia strains is significantly higher (U = 2086.5, P = 0.038) than that between supergroup A strains and butterfly symbiont strains in this study (U = 360.0, P = 0.001). This suggests that butterfly Wolbachia isolates from this study are very closely related to the rest of the supergroup B Wolbachia isolates in the MLST database.
Table 5.
Comparison of genetic variation (% similarity) in Wolbachia isolates
| Groups | Mann-Whitney U test value | P value |
|---|---|---|
| Supergroups A and B | 2,086.500 | 0.038 |
| Supergroup B and this study | 4,565.500 | 0.00 |
| Supergroup A and this study | 360.000 | 0.001 |
Diversity of Wolbachia strains in butterflies.
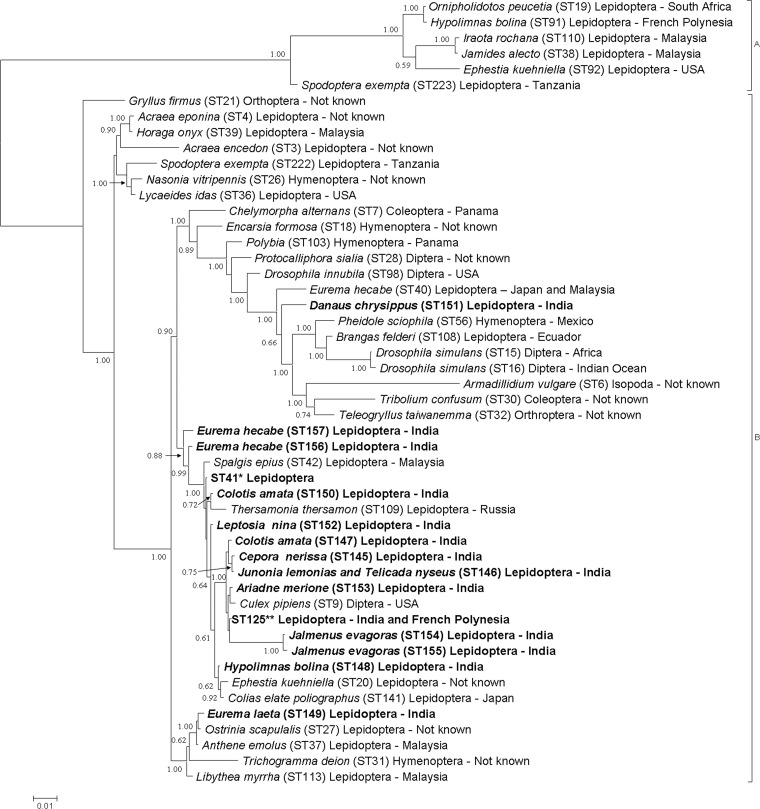
Phylogenetic reconstructions for all genes by Bayesian inference and neighbor-joining methods showed similar results. Phylogenetic reconstructions based on concatenated alignment of hcpA, gatB, coxA, ftsZ, and fbpA indicated a strong clustering of all butterfly Wolbachia isolates from this study within supergroup B; none of the strains belonged to supergroup A (Fig. 1). Wolbachia strains from Western Ghats Lepidoptera were classified in three major clusters. A large cluster with strong support (0.88) includes 11 STs that are newly described in this study, two STs found in this study but previously known to occur in other Lepidoptera, four STs previously found in other Lepidoptera, and one ST from Culex mosquitoes. A second clade contains three lepidopteran Wolbachia isolates from other regions of the world and ST149 from Western Ghats along with one Wolbachia isolate from the Hymenoptera. A third clade contains Wolbachia isolates from diverse insect taxa, as well as isolates identified from our sample in Danaus chrysippus (ST151).
Fig 1.
Unrooted phylogenetic relationships between Wolbachia strains from butterflies (bold) and those infecting other organisms, representing two supergroups (50 Wolbachia isolates), based on concatenated alignment of MLST loci (2,079 bp). Wolbachia supergroups are shown to the right side of the host species names. The bar shows substitutions per site. *, ST shared by samples from Pareronia valeria (B25 and F54), Neptis hylas (B26), Delias eucharis (B28), Eurema hecabe (B7, F47, and F53), Pseudozizeeria maha (B8 and F29), Ixias pyrene (F21), Zizeeria knysna (F65), Nacaduba angusta (Malaysia), Azanus mirza (Ghana), Celastrina argiolus (United States), Eurema mandarina (Japan) and Eurema hecabe (Japan) from MLST database. **, ST shared by Talicada nyseus (B9) from our study and Hypolimnas bolina (French Polynesia) and Spodoptera exempta (Tanzania) from the MLST database; STs and allele numbers are shown after each species name in parentheses.
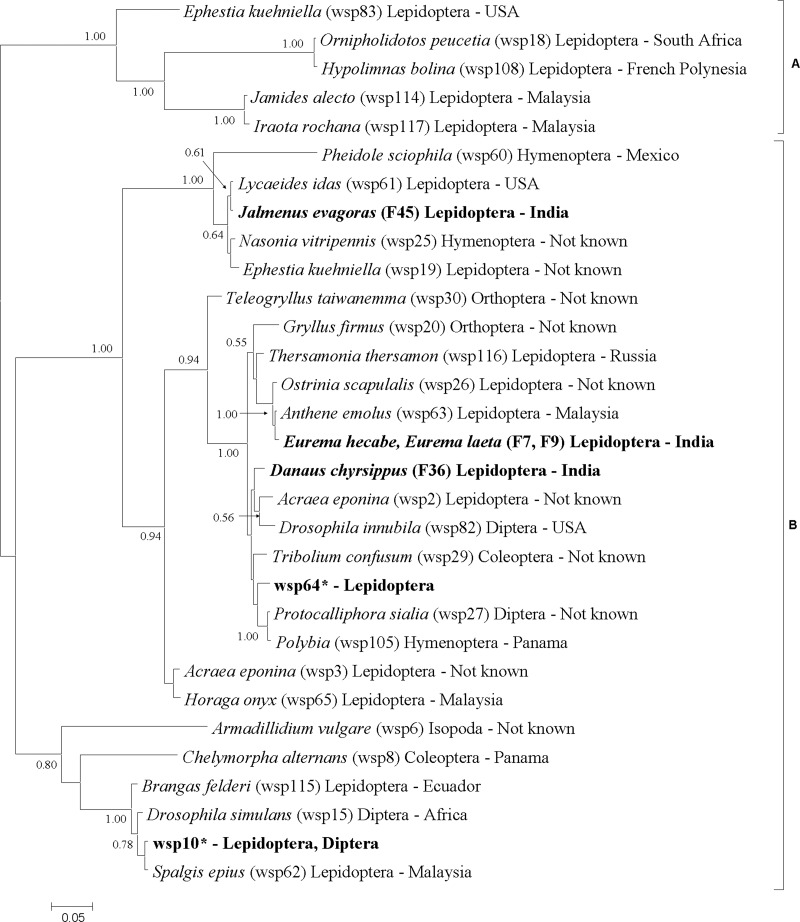
Most of the strains were remarkably akin in terms of their wsp genotypes. HVR1, HVR2, and HVR3 showed same variability, with the presence of five alleles in the sequenced data set. HVR4 showed the presence of two alleles, though this region was not sequenced for most of the individuals. Only two wsp alleles were found among 25 individuals with complete HVR peptide profiles. However, unique partial HVR peptide profiles indicated the possibility of the presence of three additional wsp alleles for (i) Danaus chrysippus (collection reference number F36), (ii) Eurema hecabe (F7) and Eurema laeta (F9), and (iii) Jalmenus evagoras (F45 and F46). Analysis of the relationships in phylogeny among butterfly strains supports five main clusters (Fig. 2). The most prevalent allele, wsp-10, was shared by 16 butterfly species from our study and six insect species representing Lepidoptera (French Polynesia, Ghana, Japan, Malaysia, and Taiwan) and Diptera (United States) from the MLST database (Fig. 2). The strains from Ariadne merione (B4) harbored wsp-64, which was shared with Colias erate poliographus (Japan), Eurema hecabe (Japan), and Surendra vivarna (Malaysia) (Fig. 2).
Fig 2.
Unrooted phylogenetic relationships between Wolbachia from butterflies (bold) and those infecting other organisms representing two supergroups (31 Wolbachia isolates), based on wsp loci (495 bp). The wsp alleles are shown after each species name in parentheses. Wolbachia supergroups are shown to the right side of the host species names. The bar shows substitutions per site. *, wsp-10 shared by Hypolimnas bolina (French Polynesia and India), Azanus mirza (Ghana), Castalius rosimon (India), Catopsilia pomona (India), Celastrina argiolus (United States), Cepora nerissa (India), Colotis amata (India), Culex pipiens (United States), Delias eucharis (India), Eurema hecabe (India), Eurema mandarina (Japan and Taiwan), Ixias pyrene (India), Junonia iphita (India), Junonia lemonias (India), Leptosia nina (India), Nacaduba angusta (Malaysia), Papilio demoleus (India), Pseudozizeeria maha (India), Talicada nyseus (India), Ypthima asterope (India), Zizeeria knysna (India), and Udaspes folus (India). **, wsp-64 shared by Ariadne merione (India), Colias erate poliographus (Japan), Eurema hecabe (Japan), and Surendra vivarna (Malaysia).
DISCUSSION
This is the first detailed analysis of Wolbachia in butterflies from India. For 27 butterfly species, this was also the first detection of infection by Wolbachia, though its occurrence in Hypolimnas bolina and Eurema hecabe was reported previously (14, 48). The butterfly Wolbachia strains belonged to supergroup B. Good-quality chromatograms were obtained for 42 Wolbachia strains, suggesting amplification of a single Wolbachia strain during the reaction. Mixed signals were observed in the chromatograms of samples from Eurema hecabe (F43) and Zizeeria knysna (B6), indicating the presence of more than one Wolbachia strain. Hence, these sequences were omitted from further analyses. This indicated the utility of MLST primers to detect multiple infections, as investigated by Baldo et al. (2).
It is interesting to note the existence of a number of new STs present in Indian butterflies that had not previously been found in butterflies or other arthropods. The phylogenetic tree constructed using the shared region of wsp nucleotides also demonstrated the affiliation of butterfly Wolbachia from this study with supergroup B. As has been observed for other Wolbachia strains (2, 4), the wsp phylogeny is not concordant with the MLST gene phylogeny. These results are consistent with the high rates of recombination between wsp and other MLST genes and within the wsp locus (2, 4). In our study, legs were used to screen for Wolbachia. It is possible that some infection types were missed by this method, although it likely also facilitates our analysis by reducing the complications of multiple infections in evaluations MLST types. Legs are commonly used in some Wolbachia screens (19, 24, 29, 31, 33), in part because the remainder of the specimen is then preserved for identification.
The MLST system works better than wsp typing in butterflies.
Both wsp typing (% VI, 29.580; Ka/Ks, 0.478524) and MLST (% VI, 4.679; Ka/Ks, 0.142397) were found to give variability among the strains. All the strains from this study consistently belonged to supergroup B according to both the typing methods. Some strains sharing common STs also shared wsp alleles. ST41 and wsp-10 were shared among five Western Ghats strains and five butterfly species from other geographic regions in the MLST database.
In contrast, many species differed in their STs and wsp alleles. Eight completely different STs were characterized for the individuals sharing same wsp allele 10 (Table 2). Two individual Talicada nyseus specimens (B9 and RP3) shared a wsp allele 10 but harbored two distinctly different STs (ST125 and ST146). Four different Eurema hecabe individuals shared a wsp allele (wsp-10), but MLST typing characterized two different strains in these four samples, with ST157 in one (F78) and ST41 in the other three (B7, F47, and F53). Strains grouped closely on the basis of wsp were found to differ in their phylogenetic affiliation as determined by MLST (Fig. 1 and 2). Phylogenetic reconstruction using wsp showed the same affiliation for Eurema hecabe (F7) and Eurema laeta symbionts (F9) (Fig. 1), while they had two distinctly different STs (ST156 and ST149) and were positioned at different places in MLST phylogeny (Fig. 1). The Ariadne merione symbiont (ST153) shared a wsp allele (wsp-64) with isolates from Colias erate poliographus (ST141), Eurema hecabe (ST40), and Surendra vivarna (ST40) but formed a separate clade in MLST phylogeny (Fig. 1 and 2). The Ariadne merione isolate (ST153) clustered with Culex pipiens (ST9) but had a distinctly different wsp allele (wsp-64 and wsp-10, respectively). The isolate from Jalmenus evagoras (ST155) was positioned at different places in both phylogenies (Fig. 1 and 2). In the wsp phylogeny, it formed a clade with Lycaeides idas (wsp-61), while in the MLST phylogeny, it formed a clade with a cluster including Drosophila simulans (ST15) and Teleogryllus taiwanemma (ST32) (Fig. 1 and 2). Danaus chrysippus (F36) harbored a unique strain that formed separate clades within supergroup B Wolbachia in both the phylogenetic reconstructions (Fig. 1 and 2). The MLST tree was found to show better separation among the strains (Fig. 2).
Extensive recombination in wsp (4) and throughout the genome at large was observed (5, 22, 52). The MLST system developed by Baldo and colleagues provides a standardized and rigorous framework for studies of Wolbachia strains (2). Combined with extensive sampling from related hosts, this MLST approach has successfully been applied to Wolbachia strains from the spider genus Agelenopsis (1), the spider Hylyphantes graminicola (54), the scorpion genus Opistophthalmus (3), the termite genera Odontotermes and Coptotermes (43), and the lone star tick, Amblyomma americanum (55). This study provides another example of Wolbachia strain diversity, specifically, that in a community of butterfly hosts.
Role of point mutation and recombination in forming new STs in Wolbachia.
The recombination was detected only within the butterflies in this study and not in existing STs from the MLST database (Table 4). Within supergroup B, three distinct clades, one comprising lepidopteran species with the exception of the Hymenoptera (Trichogramma), the second comprising lepidoptera with the exception of the Diptera (Culex), and the third including insects from the Hymenoptera, Coleoptera, Orthoptera, Diptera, and Lepidoptera, were observed. ST151, ST154, and ST155, which were revealed to be recombinant, fall within the two different clusters. Though all the strains were not monophyletic, all populations were identified as strains belonging to supergroup B (Fig. 1).
Point mutations were detected not only in Wolbachia isolates in closely related hosts but also in Wolbachia isolates found in butterflies from different families and localities. The isolates from Cepora nerissa (ST145), Junonia lemonias (ST146), and Talicada nyseus (ST146) show only one base change. These hosts belong to three different families and were collected from three different locations (Table 2). Similarly, Hypolimnas bolina (ST148 from India, in this study) and Colias erate poliographus (ST141 from Japan) were found to harbor very closely related Wolbachia isolates. These STs were also formed as a result of point mutations in gatB at bases 315 and 359. Two new STs (ST154 and ST155) were observed in Jalmenus evagoras specimens which were collected from the same location (Table 2). These STs are results of point mutations in gene hcpA (base 1004) and fbpA (base 1890). Interestingly, none of these hosts showed multiple Wolbachia infection. This suggests that same butterfly hosts within same locality harbor different Wolbachia strains. Hypolimnas bolina (ST125) and Talicada nyseus (ST147) showed mutations at only two positions (base 1503 of ftsZ and base 1709 of fbpA). Surprisingly, the Hypolimnas bolina host of Wolbachia (ST125) was collected from French Polynesia, while the later one, Talicada nyseus (ST147), was collected from India. Although these two places are geographically completely separated, the two distinct butterfly host species harbor very closely related Wolbachia. ST41 was observed among a wide range of butterfly hosts (42.31% of the infected specimens) from three different families and different localities. This ST showed point mutations in gatB at positions 120 and 359 relative to ST150 from Colotis amata. From the data presented in this study, it can be inferred that point mutations within MLST genes play a crucial role in generating new Wolbachia STs.
Butterfly host-Wolbachia relationships.
Butterfly Wolbachia phylogenies revealed a very distinct pattern of distribution. Wolbachia strains from same or different butterfly hosts from this study were closely related to each other and to representatives of lepidopteran symbionts reported in the MLST database (Table 2; Fig. 1). At the same time, the same host species, Eurema hecabe (ST40, ST41, ST156, and ST157), Colotis amata (ST147 and ST150), Hypolimnas bolina (ST91, ST125, and ST148), Talicada nyseus (ST125 and ST146), and Jalmenus evagoras (ST154 and ST155) carried distinctly different Wolbachia isolates (Table 2 and Fig. 1). A strict geographical congruence between the Wolbachia from butterfly species was not observed (Fig. 1). In terms of geography, Wolbachia strains have been recovered from lepidopteran host species in Ecuador, French Polynesia, Ghana, Japan, Malaysia, Russia, South Africa, Taiwan, Tanzania, and the United States. Country-wise relatedness was not observed for butterfly Wolbachia isolates, since distantly related hosts from different countries shared closely related strains (Fig. 1).
There are different possibilities for scenarios describing the evolution of the distribution and transfer of butterfly Wolbachia isolates. As butterflies share Wolbachia variants with divergent host species, the scenario of long-term cocladogenesis of Wolbachia and butterfly as in the case of clade C and D Wolbachia strains and filarial nematodes looks unfeasible. Alternatively, a scenario of Wolbachia invasion before differentiation of butterfly host species could be possible. In such scenario, the common ancestor of the butterfly host complex could have been originally infected with multiple Wolbachia strains, and loss and or acquisition of Wolbachia might have occurred during species differentiation. Horizontal transfer of divergent Wolbachia from outside the butterfly host genus in already genetically differentiated species might be other possibility. Strict association of one Wolbachia strain with one butterfly species appears to be an impractical explanation, as similar strains are shared by different host species. Point mutation and recombination within Wolbachia strains after the acquisition of Wolbachia in host species could be the other possibility, as observed in this study.
Phylogenetically diverse types of Wolbachia (supergroups B and A) have been reported from butterfly hosts in studies carried out so far (12, 14, 18, 23, 41, 48). Currently, the MLST database has a record for Wolbachia strains from the families Nymphalidae, Lycaenidae, and Pieridae. Wolbachia strains from supergroups A and B have been found in members of the Nymphalidae and Lycaenidae, while supergroup B strains have been found in the Pieridae. Russell et al. (41) reported the presence of supergroup A and B Wolbachia isolates in the Lycaenidae. The survey carried out by Tagami and Miura (48) reported the presence of supergroups A and B in the family Nymphalidae and supergroup B in the rest of the butterfly families except the family Papilionidae, in which they did not find Wolbachia infection. The present study suggests the presence of only supergroup B Wolbachia in the sampled populations of India of five different families with first detection in family Papilionidae. It is worthwhile adding here that different Wolbachia strains infecting the same or closely related butterfly species share close genetic relatedness with strains infecting other lepidopteran or insects. This advocates possibility of horizontal movement of Wolbachia to species of the complex, or to their last common ancestor.
The prevalence of the Wolbachia was high in some of the butterfly populations in this study. When the butterfly populations with a sample size of more than three are considered, Wolbachia prevalences are 100% in Talicada nyseus, 45% in Eurema hecabe, and 43% in Hypolimnas bolina. The prevalence and distribution of the symbionts in these species give an indication of the impact of Wolbachia on butterfly populations and merit further study. Although the Wolbachia phenotype in some of the butterflies is currently known, our study lays the groundwork for further biological investigations of the effects of Wolbachia on Indian butterfly populations and the relevance of Wolbachia in the evolutionary process of their butterfly hosts.
ACKNOWLEDGMENTS
Financial assistance in the form of project grant from the Department of Biotechnology, Government of India, is gratefully acknowledged.
We are thankful to the Director, NCCS, for providing the necessary infrastructure. Support for J.H.W. came from U.S. NSF grants EF-0328363 and DEB-0821936 and an ASM Indo-US Professorship travel award.
Footnotes
Published ahead of print 13 April 2012
REFERENCES
- 1. Baldo L, et al. 2008. Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol. Ecol. 17:557–569 [DOI] [PubMed] [Google Scholar]
- 2. Baldo L, et al. 2006. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72:7098–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baldo L, Prendini L, Corthals A, Werren JH. 2007. Wolbachia are present in Southern African scorpions and cluster with supergroup F. Curr. Microbiol. 55:367–373 [DOI] [PubMed] [Google Scholar]
- 4. Baldo L, Lo N, Werren JH. 2005. The mosaic nature of the Wolbachia surface protein (wsp). J. Bacteriol. 187:5406–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. 2006. Widespread recombination throughout Wolbachia genomes. Mol. Biol. Evol. 23:437–449 [DOI] [PubMed] [Google Scholar]
- 6. Bandi C, Anderson TJC, Genchi C, Blaxter ML. 1998. Phylogeny of Wolbachia in filarial nematodes. Proc. Biol. Sci. 265:2407–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bordenstein SR, Rosengaus RB. 2005. Discovery of a novel Wolbachia supergroup in Isoptera. Curr. Microbiol. 51:393–398 [DOI] [PubMed] [Google Scholar]
- 8. Borges RM, Gowda V, Zacharias M. 2003. Butterfly pollination and high contrast visual signals in a low-density distylous plant. Oecologia 136:571–573 [DOI] [PubMed] [Google Scholar]
- 9. Braig HR, Zhou W, Dobson SL, O'Neill SL. 1998. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J. Bacteriol. 180:2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cano JM, Gurrea P, Montalban B, Urena L, Iglesias J. 2009. Is the butterfly Tomares ballus (Lepidoptera: Lycaenidae) a potential pest of Lens culinaris (Leguminosae)? Rev. Biol. Trop. 57:623–634 [DOI] [PubMed] [Google Scholar]
- 11. Casiraghi M, et al. 2005. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology 151:4015–4022 [DOI] [PubMed] [Google Scholar]
- 12. Charlat S, et al. 2005. Prevalence and penetrance variation of male killing Wolbachia across Indo-Pacific populations of the butterfly Hypolimnas bolina. Mol. Ecol. 14:3525–3530 [DOI] [PubMed] [Google Scholar]
- 13. Chen D, et al. 2012. Phylogenetic characterization of a microsporidium (Nosema sp. MPr) isolated from the Pieris rapae. Parasitol. Res. [Epub ahead of print.] doi:10.1007/s00436-012-2829-6 [DOI] [PubMed] [Google Scholar]
- 14. Dyson EA, Kamath MK, Hurst GD. 2002. Wolbachia infection associated with all-female broods in Hypolimnas bolina (Lepidoptera: Nymphalidae): evidence for horizontal transmission of a butterfly male-killer. Heredity 88:166–171 [DOI] [PubMed] [Google Scholar]
- 15. Fiedler K. 1991. Systematic, evolutionary, and ecological implications of myrmecophily within the Lycaenidae (Insecta: Lepidoptera: Papilionoidea). Bonn Zool. Monogr. 31:1–210 [Google Scholar]
- 16. Gaonkar H. 1996. Butterflies of the Western Ghats, India (including Sri Lanka): biodiversity assessment of a threatened mountain system. Centre for Ecological Sciences, IISc, Bangalore, India, and the Natural History Museum, London, United Kingdom [Google Scholar]
- 17. Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. 2008. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol. Lett. 281:215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiroki M, Tagami Y, Miura K, Kato Y. 2004. Multiple infection with Wolbachia inducing different reproductive manipulations in the butterfly Eurema hecabe. Proc. Biol. Sci. 271:1751–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hornett EA, Charlat S, Wedell N, Jiggins CD, Hurst GD. 2009. Rapidly shifting sex ratio across a species range. Curr. Biol. 19:1628–1631 [DOI] [PubMed] [Google Scholar]
- 20. Huelsenbeck JP, Ronquist F. 2001. MRBAYES 3: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 21. Huigens ME, de Almeida RP, Boons PA, Luck RF, Stouthamer R. 2004. Natural interspecific and intraspecific horizontal transfer of parthenogenesis-inducing Wolbachia in Trichogramma wasps. Proc. Biol. Sci. 271:509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiggins FM, von Der Schulenburg JH, Hurst GD, Majerus ME. 2001. Recombination confounds interpretations of Wolbachia evolution. Proc. Biol. Sci. 268:1423–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiggins FM, Hurst GD, Dolman CE, Majerus ME. 2000. High prevalence male-killing Wolbachia in the butterfly Acraea encedana. J. Evol. Biol. 13:495–501 [Google Scholar]
- 24. Kageyama D, Narita S, Noda H. 2008. Transfection of feminizing Wolbachia endosymbionts of the butterfly, Eurema hecabe, into the cell culture and various immature stages of the silk moth, Bombyx mori. Microb. Ecol. 56:733–741 [DOI] [PubMed] [Google Scholar]
- 25. Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 26. Kunte KJ. 2000. Butterflies of peninsular India. Indian Academy of Sciences, Bangalore, and Universities Press, Hyderabad, India [Google Scholar]
- 27. Kunte K, Joglekar A, Ghate U, Pramod P. 1999. Patterns of butterfly, bird and tree diversity in the Western Ghats. Curr. Sci. 77:577–586 [Google Scholar]
- 28. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 29. Linares MC, Soto-Calderón ID, Lees DC, Anthony NM. 2009. High mitochondrial diversity in geographically widespread butterflies of Madagascar: a test of the DNA barcoding approach. Mol. Phylogenet. Evol. 50:485–495 [DOI] [PubMed] [Google Scholar]
- 30. Lo N, Casiraghi M, Salati E, Bazzocchi C, Bandi C. 2002. How many Wolbachia supergroups exist? Mol. Biol. Evol. 19:341–346 [DOI] [PubMed] [Google Scholar]
- 31. Lohman DJ, Peggie D, Pierce NE, Meier R. 2008. Phylogeography and genetic diversity of a widespread Old World butterfly, Lampides boeticus (Lepidoptera: Lycaenidae). BMC Evol. Biol. 8:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin DP, et al. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Narita S, Kageyama D, Nomura M, Fukatsu T. 2007. Unexpected mechanism of symbiont-induced reversal of insect sex: feminizing Wolbachia continuously acts on the butterfly Eurema hecabe during larval development. Appl. Environ. Microbiol. 73:4332–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nylander JAA. 2002. MrModeltest 2.2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden [Google Scholar]
- 35. Padidam M, Sawyer S, Fauquet CM. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225 [DOI] [PubMed] [Google Scholar]
- 36. Paraskevopoulos C, Bordenstein SR, Wernegreen JJ, Werren JH, Bourtzis K. 2006. Toward a Wolbachia multilocus sequence typing system: discrimination of Wolbachia strains present in Drosophila species. Curr. Microbiol. 53:388–395 [DOI] [PubMed] [Google Scholar]
- 37. Pierce NE, et al. 2002. The ecology and evolution of ant association in the Lycaenidae (Lepidoptera). Annu. Rev. Entomol. 47:733–771 [DOI] [PubMed] [Google Scholar]
- 38. Ros VID, Fleming VM, Feil EJ, Breeuwer JAJ. 2009. How diverse is Wolbachia? Multiple gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae). Appl. Environ. Microbiol. 75:1036–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rowley SM, Raven RJ, McGraw EA. 2004. Wolbachia pipientis in Australian spiders. Curr. Microbiol. 49:208–214 [DOI] [PubMed] [Google Scholar]
- 40. Rozas J, Rozas R. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 [DOI] [PubMed] [Google Scholar]
- 41. Russell JA, et al. 2009. Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution 63:624–640 [DOI] [PubMed] [Google Scholar]
- 42. Sakamoto Y, Hirai N, Tanikawa T, Yago M, Ishii M. 2011. Infection by two strains of Wolbachia and sex ratio distortion in a population of the endangered butterfly Zizina emelina (Lepidoptera: Lycaenidae) in northern Osaka Prefecture, central Japan. Ann. Entomol. Soc. Am. 104:483–487 [Google Scholar]
- 43. Salunke BK, et al. 2010. Diversity of Wolbachia in Odontotermes spp. (Termitidae) and Coptotermes heimi (Rhinotermitidae) using the multigene approach. FEMS Microbiol. Lett. 307:55–64 [DOI] [PubMed] [Google Scholar]
- 44. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Smith JM. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126–129 [DOI] [PubMed] [Google Scholar]
- 46. Stouthamer R, Breeuwer JA, Luck RF, Werren JH. 1993. Molecular identification of microorganisms associated with parthenogenesis. Nature 361:66–68 [DOI] [PubMed] [Google Scholar]
- 47. Sugimoto TN, Fujii T, Kayukawa T, Sakamoto H, Ishikawa Y. 2010. Expression of a doublesex homologue is altered in sexual mosaics of Ostrinia scapulalis moths infected with Wolbachia. Insect Biochem. Mol. Biol. 40:847–854 [DOI] [PubMed] [Google Scholar]
- 48. Tagami Y, Miura K. 2004. Distribution and prevalence of Wolbachia in Japanese populations of Lepidoptera. Insect Mol. Biol. 13:359–364 [DOI] [PubMed] [Google Scholar]
- 49. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 50. Werren JH. 1997. Biology of Wolbachia. Annu. Rev. Entomol. 42:587–609 [DOI] [PubMed] [Google Scholar]
- 51. Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6:741–751 [DOI] [PubMed] [Google Scholar]
- 52. Werren JH, Bartos JD. 2001. Recombination in Wolbachia. Curr. Biol. 11:431–435 [DOI] [PubMed] [Google Scholar]
- 53. Werren JH, Windsor D, Guo L. 1995. Distribution of Wolbachia among neotropical arthropods. Proc. R. Soc. Lond. B 262:197–204 [Google Scholar]
- 54. Yun Y, et al. 2011. Wolbachia strains typing in different geographic population spider, Hylyphantes graminicola (Linyphiidae). Curr. Microbiol. 62:139–145 [DOI] [PubMed] [Google Scholar]
- 55. Zhang X, Norris DE, Rasgon JL. 2011. Distribution and molecular characterization of Wolbachia endosymbionts and filarial nematodes in Maryland populations of the lone star tick (Amblyomma americanum). FEMS Microbiol. Ecol. 77:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]