Abstract
Pseudomonas spp. adapt rapidly to environmental fluctuations. Loss or overproduction of polyphosphate reduces the fitness of Pseudomonas fluorescens Pf0-1, indicating the importance of the fine-tuning of polyphosphate production. An antisense RNA was investigated and shown to regulate the polyphosphate kinase gene (ppk) by a posttranscriptional mechanism reducing ppk transcript abundance.
TEXT
Natural environments present a multitude of challenges to which bacteria have robust and efficient solutions. In soils, bacteria must respond to changing environmental conditions, including temperature, moisture, pH, nutrient levels, and populations of competing species. Studies of growth in soil and plant environments have shown that numerous genes are upregulated relative to their levels in in vitro culture (8, 24, 28, 32, 35), indicating that a complex and mostly unexplored suite of genetic mechanisms are utilized for optimal growth in nonlaboratory settings. We have been exploring the genetic basis underlying the fitness of Pseudomonas fluorescens Pf0-1 in a loam soil (for examples, see references 3, 13, 15, and 31). Such studies give insight into ecologic success and are of importance for the optimization of applications such as the biologic control of plant pathogens and the bioremediation of contaminated sites. These applications require that useful bacteria be able to compete with indigenous microbes and persist for a sufficiently long time to be successful.
Several different systems have been implicated in the maintenance of the optimal fitness of P. fluorescens Pf0-1 in soil. For example, the transcriptional regulator AdnA is required for growth and spread under field conditions (15) and the gene cosA is required for efficient colonization of sterile soil (31). We recently showed that inorganic polyphosphate (PolyP) is important for the competitive fitness of P. fluorescens Pf0-1 (33). PolyP is a chain of inorganic phosphate (Pi) molecules joined by phosphoanhydride bonds, the formation of which in bacteria is catalyzed by the enzyme PolyP kinase. Molecules of PolyP, which can be greater than 100 monomers, are found in all branches of life on earth (14). In a range of bacteria, PolyP has been associated with numerous phenotypes, including virulence (12, 27), quorum sensing (27), motility (20, 26, 30, 37), and survival (12, 25, 33). Furthermore, absence of PolyP has often been associated with pleiotropic effects but these phenotypes vary greatly among different species (2).
The gene ppk, specifying PolyP kinase, is part of the Pho regulon in P. fluorescens and is therefore upregulated in response to low levels of available Pi (33). The Pho regulon is controlled by the two-component regulatory pair PhoB and PhoR in response to low extracellular levels of Pi. When the environmental Pi level is low, the histidine kinase PhoR phosphorylates PhoB, which in turn binds promoters of Pho regulon genes, thus activating the response to phosphate starvation. Critical to this system is the high-affinity transport complex PstSCAB-PhoU, which imports Pi at times of Pi starvation and also negatively regulates PhoR phosphorylation of PhoB (36), thus repressing the entire Pho regulon. In our experiments with a ppk mutant of P. fluorescens Pf0-1, we observed that loss of PolyP production led to a reduction in competitive fitness (33). However, our prediction that overproduction of PolyP in a pst mutant would relieve the fitness defect proved to be incorrect. In fact, the pst mutant had a greater competitive fitness defect than the ppk mutant. A pst ppk double mutant had the same competitive fitness phenotype as a ppk single mutant; the ppk deletion rescued the more severe pst-related phenotype, indicating that both the absence and the overproduction of PolyP are deleterious, suggesting that precise control of PolyP production is important for survival in soil.
In a previous study which aimed to identify soil-induced genes in P. fluorescens Pf0-1, a transcriptionally active sequence antisense to ppk (termed iiv8) was discovered (32). Given the apparent need for the fine-tuning of ppk expression, we examined whether this sequence codes for a protein or antisense RNA (asRNA) that is capable of modulating PolyP production and determined the mode of asRNA action.
Construction of an iiv8 expression vector.
The gene iiv8 is upregulated during growth in soil (32), but specific factors influencing the expression of iiv8 are currently unknown. Therefore, to allow induced expression above basal levels we constructed a plasmid carrying iiv8 under the control of the PBAD promoter. The 658-bp iiv8 transcribed region previously determined by 5′ and 3′ rapid amplification of cDNA ends (33) was amplified by PCR using primers iiv8BAD-F (5′-GAGGAATTCGCTGTTTTGCAGCAGTTTCG-3′ [EcoRI restriction site is in italics]) and iiv8BAD-R (5′-GAGAAGCTTGCGCTTCATCCGTCGTCG-3′ [HindIII restriction site is in italics]), resulting in a 658-bp amplicon spanning positions 6131885 to 6132542 of the Pf0-1 genome. Using the EcoRI and HindIII restriction sites included in the primers, the iiv8 amplicon was cloned with pHERD26T (Tetr) (23), resulting in the plasmid pHERDiiv8, in which iiv8 is controlled by PBAD. Induction of iiv8 expression by arabinose was confirmed by reverse transcription (RT)-PCR (not shown).
Construction of P. fluorescens Pf0-1ΔrecA.
To test the impact of iiv8 on ppk, our experiments depend on the expression of iiv8 in trans. Interpretation of the data could be complicated if recombination between pHERDiiv8 and chromosomal ppk occurred, as a ppk null phenotype would result. If some cells in the population carried pHERDiiv8 as an integrated element, PolyP produced by the population would be reduced because of the subset of cells carrying a plasmid integrated in the middle of ppk. To avoid recombination-mediated effects, we constructed a recA deletion mutant of P. fluorescens Pf0-1 which was used in all experiments. Sequences flanking recA were amplified by PCR using primer pairs recD5f and recD5r (5′-CAGCAGCTGAATGTACCGAC and 5′-GGTTGTCATCGGTGCAATCAGTCCTCACGTAATCAATAAGG, respectively) and recD3f and recD3r (5′-CCTTATTGATTACGTGAGGACTGATTGCACCGATGACAACC and 5′-GAACCGTCGAAGGTAACGTG, respectively). The products were subsequently joined by splicing by overlap extension PCR (10). The amplicon in which recA was absent was cloned into pGEM-T Easy (Promega) and then cloned into pSR47s (16) using the NotI restriction sites of pGEM-T Easy. The resulting clone was used to produce the recA deletion in P. fluorescens Pf0-1 by allele exchange as described previously (32). Deletion of recA from the P. fluorescens Pf0-1ΔrecA strain (bases 1346268 to 1347327) was confirmed by PCR using a primer which anneals outside the recA flanking region used to construct the deletion mutant.
PolyP accumulation is influenced by iiv8.
We reasoned that the gene iiv8, found antisense to ppk on the chromosome (33), may influence the production of PolyP. The iiv8 transcript might interact with ppk mRNA such that translation is reduced or enhanced. To examine this possibility, we transferred plasmids pHERD26 and pHERDiiv8 into P. fluorescens Pf0-1ΔrecA by conjugation from Escherichia coli S17-1 and tested the effect of induced iiv8 transcription on PolyP accumulation. Because they are transcribed from the same stretch of DNA, it is not possible to separate iiv8 and ppk. Therefore, these experiments tested the effect of plasmid-based iiv8 in the background of Pf0-1, which has a functional single copy of iiv8.
Extraction and quantification of PolyP were carried out as described previously (33), with modifications to allow induction of expression from PBAD. Test strain Pf0-1ΔrecA(pHERDiiv8) and vector control strain Pf0-1ΔrecA(pHERD26) were grown for 16 h in high-phosphate morpholinepropanesulfonic acid (MOPS) medium (MOPS-H), which contains 2 mM K2HPO4 (33), at 30°C, after which the bacteria from 1 ml of culture were collected by centrifugation, washed twice in MOPS-H, and finally suspended in 1 ml of MOPS-H. Two flasks of MOPS-H were inoculated with each strain (1:50 dilution) and incubated at 30°C. After 6 h of growth, arabinose was added (final concentration of 0.25%) to one culture of each strain to induce expression from PBAD. After a further 2 h of growth, PolyP was extracted from induced and noninduced cultures of each strain.
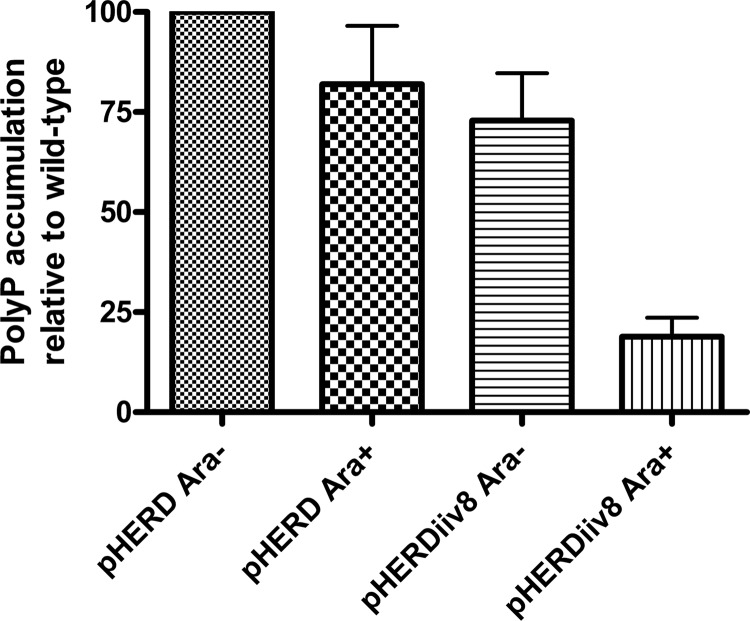
Induction of transcription from PBAD in Pf0-1ΔrecA (pHERDiiv8) resulted in a significant reduction in PolyP accumulation relative to that in the noninduced culture and the vector controls (P < 0.01; t test). When iiv8 was induced, the amount of accumulated PolyP was approximately 25% of that in the Pf0-1ΔrecA vector control strain (Fig. 1). The probable major regulator of ppk is PhoR, which, when derepressed in a pstSCAB mutant of Pf0-1, caused a 6-fold increase in PolyP (33). The 4-fold reduction in PolyP accumulation observed here supports the hypothesis that iiv8 is a minor regulator of ppk and plays a role in modulating the fine-tuning of PolyP production or accumulation. There was a small (but not significant) decrease in PolyP accumulation in two controls. Uninduced pHERDiiv8 appears to be leaky, which could explain the reduced PolyP accumulation by uninduced Pf0-1(pHERDiiv8). The reason for a slight reduction in PolyP accumulation by induced Pf0-1 carrying the pHERD vectors is unclear.
Fig 1.
Accumulation of PolyP in P. fluorescens Pf0-1ΔrecA carrying pHERDiiv8 or the vector (pHERD). Induction of iiv8 transcription from the PBAD promoter in pHERDiiv8 resulted in PolyP accumulation that was approximately 25% of that obtained with the uninduced vector control (P < 0.01). The y axis shows PolyP accumulation as a percentage of that in uninduced Pf0-1 carrying the pHERD26 vector after normalization for cell number. Data are the average of at least three independent experiments. Error bars show the standard deviations.
iiv8 does not affect the transcriptional activity of the ppk promoter.
PhoR regulation of ppk is at the level of transcription, as is the case for PhoR regulation of other Pho regulon genes. PhoR recognizes and binds “Pho boxes” associated with phosphate-regulated promoters, causing increased promoter activity. While iiv8 is antisense to ppk, a mechanism to control ppk other than antisense interference with ppk mRNA was also possible. One such possibility was negative regulation of transcription from Pppk.
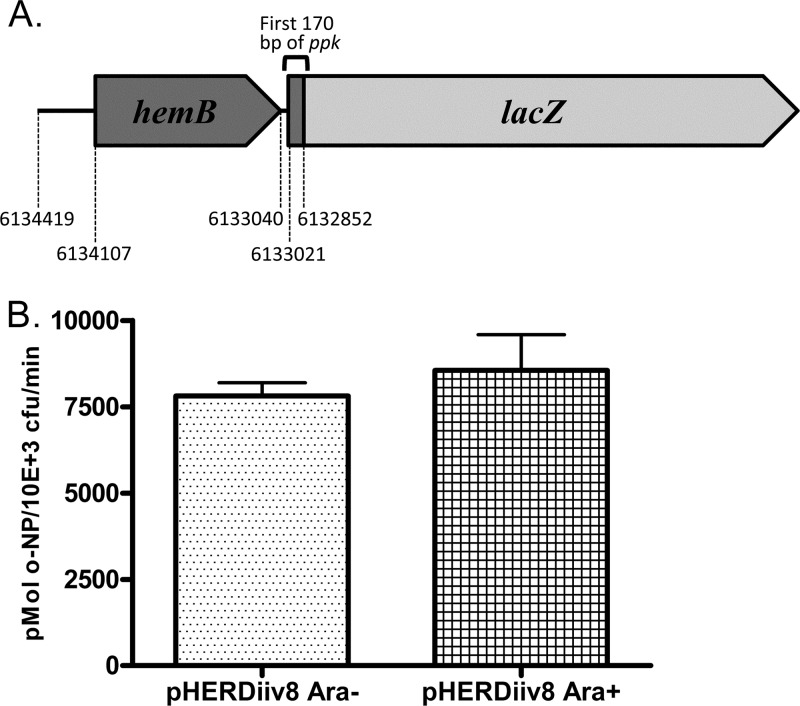
We constructed a hemB-ppk-lacZ transcriptional fusion and tested whether the induction of iiv8 expression would alter transcription. First, promoterless lacZ was cloned from pHRP309 (22) into the SmaI/HindIII sites in mini-Tn7 carried by pHRB2 (18). The Pf0-1 sequence spanning bases 6132852 to 6134419 was amplified and cloned into the XbaI restriction site upstream of lacZ′. The correct orientation of the hemB-ppk insert relative to lacZ was verified by PCR. It is not known whether ppk is transcribed independently or together with hemB, which is immediately upstream and possesses a probable Pho box (18). Thus, our fusion clone includes hemB to account for the possibility that Pppk is upstream of the hemB open reading frame (Fig. 2A). The fusion clone was mobilized into P. fluorescens Pf0-1ΔrecA by conjugation along with transposase-carrying helper plasmid pUX-BF13 (1), resulting in the insertion of Tn7 carrying hemB-ppk-lacZ into the Tn7 site downstream of glmS in P. fluorescens. For each experimental replicate, Pf0-1ΔrecA carrying pHERDiiv8 was grown in MOPS-H for 16 h. The cells were recovered by centrifugation and washed twice in low-phosphate (0.14 mM) MOPS-L (33). Washed cells were used to inoculate MOPS-L (1:50 of the original culture). These cultures were grown for 6 h, and then one was induced by the addition of arabinose. β-Galactosidase activity was measured (17) after a further 2 h of growth. Induction of iiv8 expression had no significant effect on the transcriptional activity of the fusion construct (P = 0.537; unpaired t test) (Fig. 2B). These data demonstrate that the mechanism by which iiv8 reduces the accumulation of PolyP is unrelated to promoter activity, supporting the suggestion of a direct posttranscriptional mechanism of action. The iiv8 transcript does not act in a manner antagonistic to the Pho regulon upregulation of ppk transcription. Thus, ppk expression can increase during growth in Pi-limited environments, and rather than expression being shut down by iiv8 when PolyP is too high, the amount of Ppk is likely fine-tuned to the appropriate level.
Fig 2.
(A) Diagrammatic representation of the transcriptional fusion construct used to assess ppk transcription in the presence or absence of induced iiv8 expression. Numbers indicate the coordinates in the P. fluorescens Pf0-1 genome sequence (GenBank accession number CP000094). This construct, carried by a mini-Tn7 element, was used to create a single-copy transcriptional fusion in Pf0-1. (B) Expression of iiv8 does not reduce hemB/ppk promoter activity. The activity of the hemB/ppk promoter fused to lacZ was measured using the method of Miller (17). The promoter activity in Pf0-1 carrying pHERDiiv8 did not differ significantly between arabinose-induced and noninduced cultures (P = 0.537). The data shown are averages from three independent experiments. Error bars show the standard deviations.
iiv8 affects the abundance of ppk mRNA.
The alternative hypothesis to iiv8 reduction of PolyP by reduced ppk transcription is that iiv8 RNA acts in a posttranscriptional manner to control PolyP accumulation. Because iiv8 is a cis gene with respect to ppk, an antisense mechanism of action in which iiv8 and ppk transcripts interact is likely. The iiv8 transcript could act by base pairing with ppk transcript and blocking translation or by base pairing causing targeted degradation of ppk mRNA. The latter idea leads to the prediction that induction of iiv8 expression would be associated with a reduced abundance of ppk mRNA. To test this prediction, after 20 h of growth, we induced the expression of iiv8 from pHERDiiv8 in P. fluorescens Pf0-1ΔrecA under low-Pi conditions previously shown to result in high Pho regulon activity (19) and ppk transcription (33). Two hours after induction began, RNA from experimental and control cultures was stabilized using RNAprotect Bacteria Reagent (Qiagen) and extracted using an RNeasy minikit, including the on-column DNase treatment (Qiagen), followed by an additional DNase treatment with RQ1 DNase (Promega). RNA concentration and purity were assessed using a NanoDrop spectrophotometer. The abundance of ppk mRNA was quantified by quantitative RT-PCR (qRT-PCR). Primers for ppk (ppk-qRT-F, 5′-CGAAACACCTGTCGGACTAC; ppk-qRT-R, 5′-GCTGATCACGCTGAAAATGT) were designed using recommended parameters (4), and rplU (gene for 50S ribosomal protein L51; Pf0-1 locus tag Pfl01_4860) was chosen as a reference based on observations that its expression was unchanged under high- and low-Pi conditions (18).
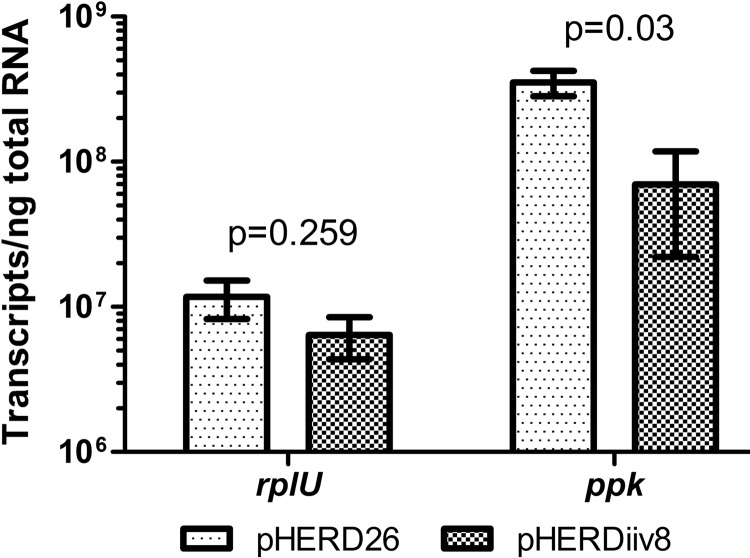
RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System from Invitrogen. The cDNAs obtained were quantified by qPCR using QuantiTect SYBR green PCR for qPCR mix (Qiagen) and a Stratagene Mx3000P detection system. The 25-μl reaction mixtures included 12.5 μl of SYBR reaction mix, 4 μl of 1:10-diluted cDNA, and primers at a final concentration of 0.3 μM, as used previously in our laboratory (29). To create standard curves, genomic DNA was PCR amplified using rplU and ppk primers. The resulting amplicons were gel purified, quantified (NanoDrop), and diluted in 10-fold steps from 100 ng to 0.0001 ng prior to qPCR performed as described above. All samples and standards were processed with three technical replicates each, as well as three biological replicates. Template-free controls were included in the plate to verify that the reaction mixtures were free of contaminating DNA, and +RNA/−reverse transcriptase reaction mixtures were included to illustrate the higher cycle threshold values expected with an enzyme-free reaction mixture. The amplification cycle was 95°C for 15 min; 45 rounds of 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min; and finally a dissociation curve of 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s to make sure that no primer dimers were interfering with the fluorescence in the reaction mixture. Raw cycle threshold values were obtained with MxPro-Mx3000P software and used to calculate absolute transcript numbers using the corresponding standard curves. The analysis of ppk transcript abundance revealed that when iiv8 expression was increased, the ppk transcript levels were significantly (P = 0.03; unpaired t test) reduced to 1/5 of the level found in cells in which iiv8 expression was not elevated, whereas rplU transcript levels were not significantly affected (Fig. 3). These data are not greatly different from the change in PolyP accumulation observed with the induction of iiv8 (Fig. 1).
Fig 3.
Absolute numbers of transcripts per nanogram of total RNA from rplU (control) and ppk in Pf0-1 carrying either the vector pHERD26 or iiv8 asRNA-expressing pHERDiiv8. Induction of iiv8 significantly reduced transcript ppk abundance (t test) relative to that in the control sample. These data are the average and standard error of the mean from three biological replicate experiments, each of which consisted of three technical replicates.
Taken together, these data support the conclusion that a cis-acting asRNA is involved in the control of PolyP production and acts by reducing the amount of ppk mRNA. Whether iiv8 directly senses the PolyP concentration or is regulated by another system which responds to the PolyP level is unknown. The existence of asRNAs in phage and plasmids is well established, but until recently, their importance as regulators of bacterial chromosomal genes had been underappreciated. New details of asRNA have emerged from experimental studies and from transcriptome profiling using methods that permit the discrimination of coding strands of DNA. For example, a gene specifying a component of the photosynthesis machinery in Synechocystis is regulated by an asRNA (6). High-throughput methods have identified widespread asRNA molecules in E. coli (5) and Pseudomonas syringae (7). Despite these and other studies which reveal the existence of asRNA (reviewed in reference 9), its functional characterization is somewhat limited. The role of iiv8 in the modulation of ppk expression adds to the growing body of knowledge on asRNA.
Alongside elucidation of the importance of asRNA in bacteria, the question of mechanistic action requires further research. While the mechanism of many trans-acting sRNAs is clear, cis-encoded asRNAs have received less attention. One of the earliest and best-described asRNA regulation mechanisms encoded in a bacterial genome (as opposed to a phage or plasmid) is the isiA-IsrR system in Synechocystis strain PCC6803 (6). The IsrR asRNA binds isiA mRNA, resulting in the degradation of both RNA species. An alternative mechanism, which is used in the posttranscriptional control of gadX by GadY in E. coli, is stabilization of the mRNA (34). GadY base pairs with the intergenic sequence between gadX and gadW in the bicistronic mRNA, and RNase-dependent processing at this site of interaction increases the stability of the gadX transcript (21). asRNAs which overlap the 5′ untranslated region of a protein-coding gene may act to inhibit translation by blocking the access of the ribosome to the Shine-Dalgarno sequence and/or the initiation codon. For example, in E. coli, the small asRNA SymR represses the translation of the symE transcript, most likely by blocking ribosomal access to the Shine-Dalgarno sequence (11).
Based on our qRT-PCR data which show a large decrease in ppk transcript abundance associated with iiv8 overexpression, we suggest that a mechanism involving destabilization of the mRNA is at work in the ppk-iiv8 system. The fact that iiv8 is upregulated during growth in soil (32) indicates that fine-tuning of ppk expression is necessary for survival under natural conditions and suggests that responsiveness to fluctuating Pi levels is important. The discovery of a new mechanism for the regulation of ppk increases our understanding of PolyP and its production but does not give further insight into the function of PolyP. However, the existence of both global (via the Pho regulon) and specific regulatory mechanisms highlights the importance of the control of PolyP production, which may be crucial in the coordination of the multitude of roles that PolyP appears to play in bacteria.
ACKNOWLEDGMENTS
We thank Cristian Ruiz for advice on the analysis and interpretation of qRT-PCR data.
This work was supported by Agriculture and Food Research Initiative competitive grant 2010-65110-20392 from the USDA National Institute of Food and Agriculture Microbial Functional Genomics Program.
Footnotes
Published ahead of print 6 April 2012
REFERENCES
- 1. Bao Y, Lies DP, Fu H, Roberts GP. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167–168 [DOI] [PubMed] [Google Scholar]
- 2. Brown MRW, Kornberg A. 2008. The long and short of it—polyphosphate, PPK and bacterial survival. Trends Biochem. Sci. 33:284–290 [DOI] [PubMed] [Google Scholar]
- 3. Deflaun MF, Marshall BM, Kulle EP, Levy SB. 1994. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl. Environ. Microbiol. 60:2637–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dorak MT. 2006. Real-time PCR (BIOS advanced methods). Taylor & Francis Group, New York, NY [Google Scholar]
- 5. Dornenburg JE, DeVita AM, Palumbo MJ, Wade JT. 2010. Widespread antisense transcription in Escherichia coli. mBio 1(1): e00024–10 doi:10.1128/mBio.00024-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dühring U, Axmann IM, Hess WR, Wilde A. 2006. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. U. S. A. 103:7054–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Filiatrault MJ, et al. 2010. Transcriptome analysis of Pseudomonas syringae identifies new genes, ncRNAs, and antisense activity. J. Bacteriol. 192:2359–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gal M, Preston GM, Massey RC, Spiers AJ, Rainey PB. 2003. Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol. Ecol. 12:3109–3121 [DOI] [PubMed] [Google Scholar]
- 9. Georg J, Hess WR. 2011. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 75:286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68 [DOI] [PubMed] [Google Scholar]
- 11. Kawano M, Aravind L, Storz G. 2007. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 64:738–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim K-S, Rao NN, Fraley CD, Kornberg A. 2002. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. U. S. A. 99:7675–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim W, Levy SB. 2008. Increased fitness of Pseudomonas fluorescens Pf0-1 leucine auxotrophs in soil. Appl. Environ. Microbiol. 74:3644–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kornberg A, Rao NN, Ault-Riche D. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89–125 [DOI] [PubMed] [Google Scholar]
- 15. Marshall B, et al. 2001. The adnA transcriptional factor affects persistence and spread of Pseudomonas fluorescens under natural field conditions. Appl. Environ. Microbiol. 67:852–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matthews M, Roy CR. 2000. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect. Immun. 68:3971–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 18. Monds RD, Newell PD, Gross RH, O'Toole GA. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63:656–679 [DOI] [PubMed] [Google Scholar]
- 19. Monds RD, Newell PD, Schwartzman JA, O'Toole GA. 2006. Conservation of the Pho regulon in Pseudomonas fluorescens Pf0-1. Appl. Environ. Microbiol. 72:1910–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogawa N, Tzeng C-M, Fraley CD, Kornberg A. 2000. Inorganic polyphosphate in Vibrio cholerae: genetic, biochemical, and physiologic features. J. Bacteriol. 182:6687–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Opdyke JA, Fozo EM, Hemm MR, Storz G. 2011. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J. Mol. Biol. 406:29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parales RE, Harwood CS. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram− bacteria. Gene 133:23–30 [DOI] [PubMed] [Google Scholar]
- 23. Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. pBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74:7422–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rainey PB. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243–257 [DOI] [PubMed] [Google Scholar]
- 25. Rao NN, Kornberg A. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:4885–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rashid MH, et al. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rediers H, Rainey PB, Vanderleyden J, De Mot R. 2005. Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 69:217–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruiz C, Levy SB. 2010. Many chromosomal genes modulate MarA-mediated multidrug resistance in Escherichia coli. Antimicrob. Agents Chemother. 54:2125–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi X, Rao NN, Kornberg A. 2004. Inorganic polyphosphate in Bacillus cereus: motility, biofilm formation, and sporulation. Proc. Natl. Acad. Sci. U. S. A. 101:17061–17065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Silby MW, Levy SB. 2008. Overlapping protein-encoding genes in Pseudomonas fluorescens Pf0-1. PLoS Genet. 4:e1000094 doi:10.1371/journal.pgen.1000094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silby MW, Levy SB. 2004. Use of in vivo expression technology to identify genes important in growth and survival of Pseudomonas fluorescens Pf0-1 in soil: discovery of expressed sequences with novel genetic organization. J. Bacteriol. 186:7411–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Silby MW, Nicoll JS, Levy SB. 2009. Requirement of polyphosphate by Pseudomonas fluorescens Pf0-1 for competitive fitness and heat tolerance in laboratory media and sterile soil. Appl. Environ. Microbiol. 75:3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tramonti A, De Canio M, De Biase D. 2008. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 70:965–982 [DOI] [PubMed] [Google Scholar]
- 35. Wang Y, Morimoto S, Ogawa N, Fujii T. 2011. A survey of the cellular responses in Pseudomonas putida KT2440 growing in sterilized soil by microarray analysis. FEMS Microbiol. Ecol. 78:220–232 [DOI] [PubMed] [Google Scholar]
- 36. Wanner BL. 1996. Phosphorus assimilation and control of the phosphate regulon, p 1357–1381 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 ASM Press, Washington DC [Google Scholar]
- 37. Zhang H, Rao NN, Shiba T, Kornberg A. 2005. Inorganic polyphosphate in the social life of Myxococcus xanthus: motility, development, and predation. Proc. Natl. Acad. Sci. U. S. A. 102:13416–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]