Abstract
In our previous study, Bacillus subtilis strain BSK3S, containing a polymyxin biosynthetic gene cluster from Paenibacillus polymyxa, could produce polymyxin only in the presence of exogenously added l-2,4-diaminobutyric acid (Dab). The dependence of polymyxin production on exogenous Dab was removed by introducing an ectB gene encoding the diaminobutyrate synthase of P. polymyxa into BSK3S (resulting in strain BSK4). We found, by observing the complete inhibition of polymyxin synthesis when the spo0A gene was knocked out (strain BSK4-0A), that Spo0A is indispensable for the production of polymyxin. Interestingly, the abrB-spo0A double-knockout mutant, BSK4-0A-rB, and the single abrB mutant, BSK4-rB, showed 1.7- and 2.3-fold increases, respectively, in polymyxin production over that of BSK4. These results coincided with the transcription levels of pmxA in the strains observed by quantitative real-time PCR (qRT-PCR). The AbrB protein was shown to bind directly to the upstream region of pmxA, indicating that AbrB directly inhibits the transcription of polymyxin biosynthetic genes. The BSK4-rB strain, producing high levels of polymyxin, will be useful for the development and production of novel polymyxin derivatives.
INTRODUCTION
Humans currently face a very serious threat from multiple antibiotic-resistant bacteria, commonly called superbugs (20). Multidrug resistance is an important issue for public health and is frequently found in pathogenic Gram-negative bacteria, such as Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae (20, 21).
Polymyxin, a long-known lipopeptide antibiotic having bactericidal activity against Gram-negative bacteria (1), has recently been reintroduced in clinical practice because it is sometimes the only available antibiotic for the treatment of multidrug-resistant Gram-negative pathogenic bacteria (15). However, adverse effects, such as nephrotoxicity and neurotoxicity, have been reported during the early clinical administration of polymyxin, and the clinical use of polymyxin has been limited, although it has excellent antisuperbug activity (9, 33). Therefore, it will be another challenge to develop new polymyxin derivatives having improved pharmacological and toxicological characteristics. During the last few decades, many studies have been conducted to generate polymyxin derivatives by chemical synthesis or enzymatic modifications and to analyze structure-function relationships to develop improved polymyxins having higher activity and lower toxicity (32). Although some interesting results have been obtained from these trials, the efficiency of generating derivatives using these approaches has been low (30).
We recently completed whole-genome sequencing of Paenibacillus polymyxa E681 (13), located nonribosomal peptide synthetase genes responsible for the synthesis of polymyxin and fusaricidin, and analyzed their functions (2, 3). We then succeeded in transferring the entire polymyxin synthetase gene cluster (pmxABCDE) into the chromosome of the surrogate host (Bacillus subtilis 168) and producing polymyxin in the recombinant strain. Molecular tools and technologies for genetic manipulation and strain improvement have been highly developed for use in B. subtilis, but those tools are very poor for use in P. polymyxa. Therefore, the Bacillus expression system will be very useful for conducting studies to analyze structure-function relationships, to efficiently generate polymyxin and polymyxin derivatives, and to improve polymyxin purity. Polymyxin synthesis by the recombinant B. subtilis strain was possible, however, only with the exogenous addition of l-2,4-diaminobutyric acid (Dab), a main substrate for polymyxin, and the production level was low (2). Therefore, it was necessary to solve these problems to increase the usefulness of the system.
In this study, we attempted to enable the recombinant B. subtilis strain to efficiently produce polymyxin without the addition of Dab by insertion of a gene necessary for the synthesis of Dab and to increase the production level of polymyxin by control of genes affecting the expression of the polymyxin biosynthetic gene cluster. We also attempted to reveal the mechanism for regulation of polymyxin production by analyzing the effect of null mutations of spo0A or abrB on the expression of pmxA and by observing the mode of interaction between AbrB and the promoter region of pmxA.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and genetic material.
The bacterial strains, plasmids, and primers used in this study are described in Table 1. Escherichia coli DH5α was used for construction of recombinant plasmids and for monitoring of polymyxin activity. B. subtilis strains were grown in LB broth or LB agar medium at 37°C for general purposes and in Cal18 medium (10) with shaking at 37°C for studies of polymyxin production. A supplemental solution of antibiotics contained 5 μg/ml chloramphenicol, 1 μg/ml erythromycin, 100 μg/ml spectinomycin, 10 μg/ml tetracycline, 8 μg/ml neomycin, and 100 μg/ml ampicillin. l-2,4-diaminobutyric acid was purchased from Sigma-Aldrich (St. Louis, MO).
Table 1.
Bacterial strains, plasmids, and primers used in the study
| Strain, plasmid, or primer | Relevant genotype/characteristics/sequence (5′ to 3′)a | Reference/source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Strain for general DNA manipulation and monitoring of polymyxin activity | 8 |
| BL21(DE3) | Host strain for overexpression under controlled T7 promoter | 29 |
| B. subtilis | ||
| BSK3S | Strain containing the entire polymyxin synthetase gene cluster on amyE locus, functional sfp gene | 2 |
| BSK4 | srfC::etcB-tet of BSK3S | This study |
| BSK4-0A | spo0A::erm of BSK4 | This study |
| BSK4-rB | abrB::neo of BSK4 | This study |
| BSK4-0A-rB | spo0A::erm of BSK4-rB | This study |
| P. polymyxa E681 | Wild type; produces polymyxin A | 23 |
| Plasmids | ||
| pHPS9 | E. coli-B. subtilis shuttle vector, cm, erm | 6 |
| pGEM-T Easy | Plasmid for cloning of PCR products, Ap | Promega |
| pET22b | Plasmid for expression of His-tagged protein, Ap | Novagen |
| pET22-bsabrB | Plasmid containing abrB gene of B. subtilis 168 | This study |
| Primers | ||
| ectF | CCTGTTGATAGAGTAAACTCC | |
| ectR | ATGGCCCGTTTGTTGATGAGCAACTAGAGCAATCC | |
| tetF | AACAAACGGGCCATATTGTTG | |
| tetR | GATACAAGAGAGGTCTCTCG | |
| srfC1 | AGATATGTATTACCTATCGCC | |
| srfC2 | TTACTCTATCAACAGGGATTGGCGTTCACTGCTTCCTTG | |
| Srf3 | ACGAGAGACCTCTCTTGTAGGATTGGATGAAGGGGCTTCGCT | |
| Srf4 | TCAACGCTTCGACATCACTTTC | |
| mlkneoF | TCGAGATCAGGGAATGAGTTT | |
| mlkneoR | AATAAATACGTAACCAACATG | |
| abrff | CTGTACCAGCCTTCATACTC | |
| abrfr-neo | AAACTCATTCCCTGATCTCGAACACGTCCTAATTCATCAAC | |
| abrrf-neo | CATGTTGGTTACGTATTTATTCATCAGCGAAATCCAAAACC | |
| abrrr | CAAATCGAGCCGAAACGTGTAC | |
| Bsabr-nde | TACATATGTTTATGAAATCTACTGGTATTG | |
| Bsabr-xho | TGCTCGAGTTTAAGGTTTTGAAGCTGG | |
| pmxAF/pmxAF-biotin | AGCATATTGAAGCAGAGGAG | |
| pmxAR/pmxAR-biotin | TCATCGGTAAAAGCAATCCGG | |
| BSF349 | AGGCAGCAGTGGGGAAT | |
| 16S-786R | CTACCAGGGTATCTAATC |
Resistance gene or phenotype against antibiotics: tet (tetracycline), erm (erythromycin), neo (neomycin), cm (chloramphenicol), and Ap (ampicillin). The restriction sites of endonucleases in nucleotide sequences are underlined.
Construction of recombinant strains.
B. subtilis BSK4 was constructed by the following procedures. The putative ectB gene (PPE_02294 in GenBank accession no. CP000154) was obtained by PCR with the primer set ectF and ectR from chromosomal DNA of P. polymyxa E681. The PCR product was fused to a tetracycline resistance gene amplified from plasmid pBC16 (19) with primers tetF and tetR, using a fusion PCR method to construct an ectB-tet cassette. The N-terminal region and the C-terminal region of srfC were amplified from the chromosomal DNA of B. subtilis with primer sets consisting of srfC1 and srfC2, and srf3 and srf4, respectively. The two PCR fragments were fused with the ectB-tet cassette by a second fusion PCR and then introduced into the srfC locus of strain BSK3S by homologous recombination to construct strain BSK4 (Fig. 1A).
Fig 1.
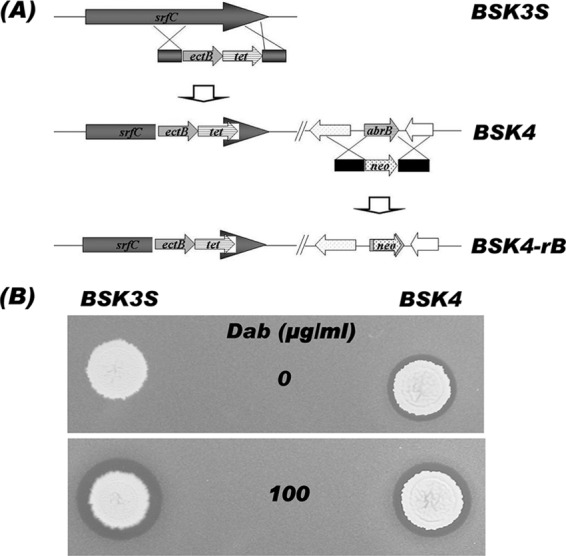
Schematic diagram showing the strategy for construction of BSK4 and BSK4-rB from BSK3S (A) and antibacterial activities of BSK3S and BSK4 with and without addition of exogenous Dab (B). The BSK4 strain was constructed by introducing the ectB gene of P. polymyxa E681 into the srfC locus of strain BSK3S. Then, the abrB gene was disrupted to produce the BSK4-rB strain. To analyze the antimicrobial activities of the recombinant strains, E. coli plates were prepared with and without Dab, as described by Choi et al. (2), and the cells of the strains were inoculated and incubated at 37°C for 24 h to observe the growth inhibition effect.
The strain BSK4-0A was constructed by homologous recombination using BS9903 chromosomal DNA (24) and then selecting erythromycin-resistant transformants. The abrB mutant strain, BSK4-rb, was constructed by a procedure shown in Fig. 1A. Two DNA fragments containing an N-terminal and upstream region and a C-terminal and downstream region of abrB, respectively, were amplified by PCR with primer sets consisting of abrff and abrfr-neo, and abrrf-neo and abrrr, respectively. The neomycin resistance gene was obtained from the pMLK83 plasmid (11) by PCR with primers mlkneoF and mlkneoR. The three PCR products were joined by a second PCR, and the resulting product was introduced into the strains BSK4 and BSK4-0A to construct BSK4-rB and BSK4-0A-rB, respectively, by homologous recombination.
Antibacterial activity assay and ESI-LC/MS analysis.
Recombinant B. subtilis strains were grown in Cal18 medium at 37°C with vigorous shaking (220 rpm), and the cell-free supernatants were harvested at different times (18, 24, 30, and 36 h). A 5-μl aliquot of each culture was dropped onto E. coli plates for antibacterial assay using a previously described method (2). Electrospray ionization-liquid chromatography/mass spectrometry (ESI-LC/MS) analysis was performed using a method previously described by Choi et al. (2). For quantitative analysis of polymyxin produced by B. subtilis strains, high-performance liquid chromatography (HPLC) separation was performed on a Finnegan Surveyor Modular HPLC System (Thermo Fisher Scientific, Inc., Waltham, MA) using an XTerra MS C18 column (5 μm; 2.1 by 150 mm; Waters, Milford, MA) with a BetaBasic-18 guard column (2.1 by 10 mm; Finnigan, Thermo Scientific). Mobile phase A was water, and mobile phase B was acetonitrile, both containing 0.1% formic acid. The gradient elution at a flow rate of 0.3 ml/min was performed as follows: 0 to 15 min, 15 to 80% B (linear gradient), and 15 to 20 min, 100% B (isocratic). Selected ion-monitoring (SIM) mass spectra were obtained at m/z 579.5 [M + 2H]2+, with 3 microscans and a maximum ion injection time of 200 ms.
Quantitative real-time PCR (qRT-PCR) analysis.
Total RNA was extracted from bacterial cells using an RNeasy minikit (Qiagen, Hilden, Germany), and 500 ng of DNA-free RNA was used for the first-strand synthesis with primers pmxAR and 16S-786R. One hundred-fold-diluted cDNA was used for amplification of the pmxA promoter region and the 16S rRNA gene, with the primer sets pmxAF and pmxAR, and BSF349 and 16S-786R, respectively. Real-time monitoring of amplification was performed using the iQ SYBR green Supermix kit and the CFX96 Real-Time PCR detection system (Bio-Rad, Hercules, CA).
Production and purification of AbrB.
The abrB gene of B. subtilis 168 was amplified with the primer set Bsabr-nde and Bsabr-xho. The amplified product was cloned into plasmid pET22b using NdeI and XhoI sites and introduced into E. coli BL21(DE3). The His-tagged AbrB protein was purified from the E. coli cells using an Ni-nitrilotriacetic acid (NTA) purification system (Qiagen, Hilden, Germany).
EMSA.
An electrophoretic mobility shift assay (EMSA) was performed using biotin-labeled DNA as a probe. The promoter region of pmxA (PpmxA; 250 bp) was amplified with a 5′-biotinylated oligomer set, pmxAF-biotin and pmxAR-biotin. The PCR product was purified by agarose gel extraction. The non-biotin-labeled PpmxA was obtained by PCR with primers pmxAF and pmxAR and used as a specific competitor to perform EMSA. The binding reaction was performed similarly to the one described by Strauch (27), except that 50 μg of poly(dI-dC) per ml was used as the nonspecific competitor. Biotinylated PpmxA (10 ng) was mixed with 500 nM AbrB protein or with 5- or 10-fold amounts of unlabeled PpmxA (specific competitor) in a final volume of 20 μl. The mixture was incubated for 40 min at room temperature, and then, 10-μl aliquots were electrophoresed in a 5% native polyacrylamide gel. The image of electrophoretic mobility shift was detected by using a Light Shift Chemiluminescent EMSA Kit (Thermo Scientific) under a LAS 3000 Chemiluminescent Imaging Analyzer (Fujifilm, Tokyo, Japan).
RESULTS AND DISCUSSION
Production of polymyxin without external addition of Dab by introducing the ectB gene into B. subtilis.
Heterologous production of polymyxin facilitated by transferring the pmx genes (pmxABCDE) responsible for the biosynthesis of polymyxin in P. polymyxa E681 into B. subtilis was dependent on the exogenous addition of Dab, because B. subtilis 168 was not able to produce Dab due to the lack of a gene coding for an enzyme responsible for Dab synthesis (2). The biosynthesis of Dab is mediated by 2,4-diaminobutyrate aminotransferase, encoded by ectB. It is known that ectB is composed of an operon structure with ectA and ectC and is involved in ectoine synthesis in halophilic bacteria, such as Halobacillus halophilus (14, 34). A gene encoding a homolog of EctB found in the genome sequence of P. polymyxa E681 (PPE_02294 in GenBank accession no. CP000154) showed 51% amino acid identity with that of H. halophilus, and interestingly, it was monocistronic.
To produce polymyxin in B. subtilis without an exogenous supply of Dab, we introduced the putative ectB gene of P. polymyxa into the srfC region of B. subtilis BSK3S (2) harboring entire pmx genes. As shown in Fig. 1B, the strain BSK4 exhibited clear antibacterial activity against E. coli DH5α on LB agar plates without an exogenous supply of Dab. This was in contrast with BSK3S, which could inhibit E. coli growth only in the presence of Dab. This result indicated that a protein encoded by the putative ectB gene of P. polymyxa was produced and successfully played the role of 2,4-diaminobutyrate aminotransferase to synthesize Dab in B. subtilis, which then produced polymyxin in the absence of exogenous Dab (Fig. 1B). The biosynthesis of surfactin in the strain BSK4 was completely blocked by knockout of the srfC gene, part of the surfactin biosynthetic gene cluster, during the construction of this strain. The deficiency of surfactin production by B. subtilis BSK4 in DSM-GGTris medium (4) was confirmed by measuring surface tension and the ESI-LC/MS configuration (data not shown). The elimination of surfactin production, a cyclic lipopeptide having antimicrobial activity, will be an advantage for producing polymyxin or its derivatives in pure form.
The dependence of polymyxin production on functional Spo0A.
In our preliminary study, we found that a sporulation mutant of P. polymyxa E681 that was obtained by knocking out spo0A could not produce polymyxin (data not shown). There are reports showing that spontaneous asporogenic mutants of P. polymyxa do not produce polymyxin, and these reports also support the dependence of polymyxin production on sporulation (12, 18, 31).
To investigate whether polymyxin production is also dependent on sporulation in B. subtilis, the spo0A mutant of BSK4 was constructed as described in Materials and Methods. The mutant strain, BSK4-0A, showed no antibacterial activity against E. coli (Fig. 2A) and also showed no polymyxin peak in LC analysis of the culture supernatant (data not shown). These results indicate that functional Spo0A, a master transcriptional regulator for entrance into sporulation, is also required to produce polymyxin in B. subtilis.
Fig 2.
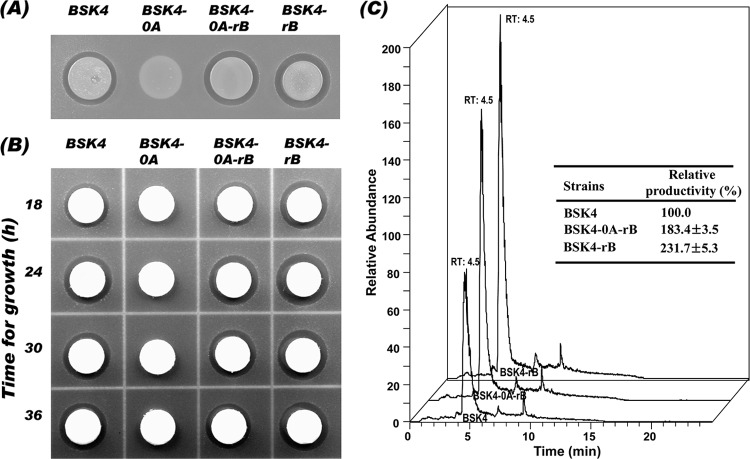
Antibacterial activities of spo0A or abrB mutant strains derived from BSK4 against E. coli (A and B) and quantitative HPLC analysis of polymyxin produced by the strains after growing for 24 h (C). The B. subtilis strains BSK4, BSK4-0A, BSK4-0A-rB, and BSK4-rB were grown in Cal18 medium for 18, 24, 30, and 36 h. A culture aliquot (5 μl) of each strain grown for 24 h was dropped directly onto E. coli plates containing E. coli cells (∼106 CFU/ml) for antibacterial assay (A), and 50 μl of cell-free supernatant of each strain harvested at different growth times was loaded onto a paper disk and transferred to the E. coli plates (B). The growth inhibition was observed after 24 h of incubation at 37°C. (C) The 24-h culture samples were used for quantitative analysis of polymyxin by ESI-LC/MS, and the [M + 2H]2+ ion peaks of 579.5 m/z were obtained. The areas of the ion peaks were determined for comparison with each other.
The regulation of polymyxin production by AbrB.
The DNA-binding protein Spo0A regulates, directly or indirectly, the expression of >500 genes in B. subtilis. Among them, 40 genes are directly activated and 81 are directly repressed by phosphorylated Spo0A (17). Spo0A also regulates many genes indirectly through repression of abrB (5, 7, 26), and when Spo0A is nonfunctional in a cell, it leads to an increase in abrB expression. It was reported that AbrB repressed the expression of tycA, a Bacillus brevis gene that encodes tyrocidine synthetase I, which is an enzyme involved in the biosynthesis of the cyclic decapeptide tyrocidine (22). In the case of Bacillus cereus, production of cereulide, which is a cyclic dodecadepsipeptide synthesized by a nonribosomal peptide synthetase encoded by the ces genes, was regulated by Spo0A and AbrB (16). It was also reported that deletion of the abrB gene strongly increased the production of the lantibiotic subtilin in B. subtilis (25). In this context, it was also expected that the necessity for functional Spo0A in polymyxin production might be related to repression of abrB. To test this hypothesis, we constructed abrB single and spo0A abrB double mutants of BSK4. As shown in Fig. 2A, BSK4-0A-rB with spo0A abrB double mutations recovered the antibacterial activity against E. coli in an agar plate assay. The antibacterial activity seemed to be even higher than that of BSK4. Another mutant strain, BSK4-rB, with the abrB single mutation, also showed higher antibacterial activity than BSK4 (Fig. 2A). For the quantitative analysis of polymyxin produced by the recombinant B. subtilis strains, Cal18 medium was selected for growing bacterial cells in a preliminary test for comparing 20 kinds of media (data not shown), and the cells grown in Cal18 medium for 24 h, which showed the highest antibacterial activity among samples harvested at different times, was used for the quantitative analysis of polymyxin (Fig. 2B). LC/MS results revealed that strains BSK4-rB and BSK4-0A-rB produced 2.3- and 1.8-fold more polymyxin, respectively, than BSK4 (Fig. 2C). This suggests that polymyxin production is repressed by AbrB and that Spo0A regulates polymyxin production in a positive manner by repression of abrB.
Effect of spo0A and/or abrB mutation on expression of pmxA.
As in the case of Spo0A, AbrB, a transition state regulator, binds to DNA and regulates the expression of downstream genes (28). In this study, we investigated the effects of null mutations of spo0A or abrB on the expression of pmxA, and the transcription level of pmxA was analyzed by qRT-PCR. As shown in Fig. 3, the expression of pmxA was completely repressed in BSK4-0A cells; however, it was recovered in BSK-0A-rB, and the expression level reached 1.7 times that of BSK4. This result suggests that AbrB is a negative regulator of pmxA expression, and null mutation of spo0A, whose product is a repressor of the transcription of abrB, caused overexpression of abrB and then repression of pmxA. In BSK4-0A-rB with double mutations in spo0A and abrB, the repression of pmxA by AbrB could be relieved. In BSK4-rB with an abrB single mutation, the expression of pmxA was increased and reached a 2.3-times-higher level than in BSK4 (Fig. 3). These qRT-PCR results coincided well with the results of quantitative analysis of polymyxin production in the strains (Fig. 2C).
Fig 3.
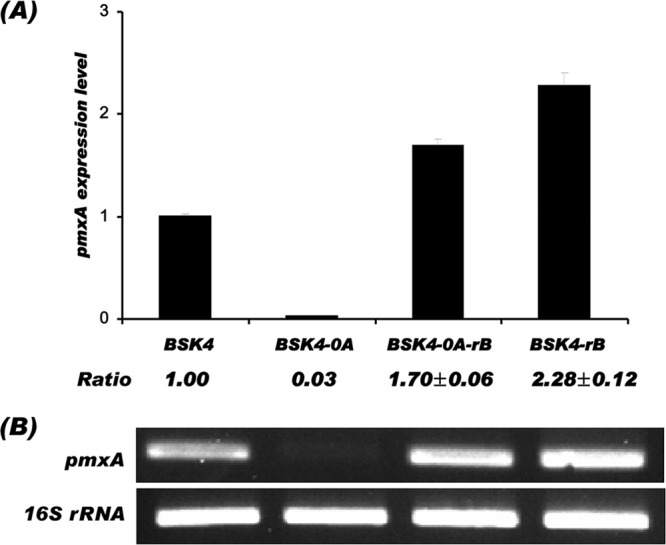
Effects of spo0A or abrB mutations on expression of pmxA in B. subtilis analyzed by qRT-PCR (A) and the gel electrophoresis image (B). The relative expression ratios of pmxA for the three mutant strains, BSK4-0A, BSK4-0A-rB, and BSK4-rB, to their parent strain, BSK4, were calculated from the real-time PCR efficiencies. The error bars indicate standard errors of the mean (SEM); n = 3 in all RT-PCR analyses; P < 0.05.
Direct binding of AbrB to the promoter region of pmxA.
To find out whether AbrB represses the expression of polymyxin biosynthetic genes directly or indirectly, we investigated the binding of AbrB to the upstream region of the pmxA gene. As shown in Fig. 4, the movement of biotin-labeled PpmxA was retarded in the presence of B. subtilis AbrB. However, the addition of 5- or 10-fold excess of unlabeled PpmxA, the specific competitor, restored the movement of biotin-labeled PpmxA, possibly by hindering the interaction between the biotin-labeled DNA and AbrB. From these results, it was found that pmxA gene expression is controlled by direct binding of AbrB to the pmxA promoter in B. subtilis.
Fig 4.
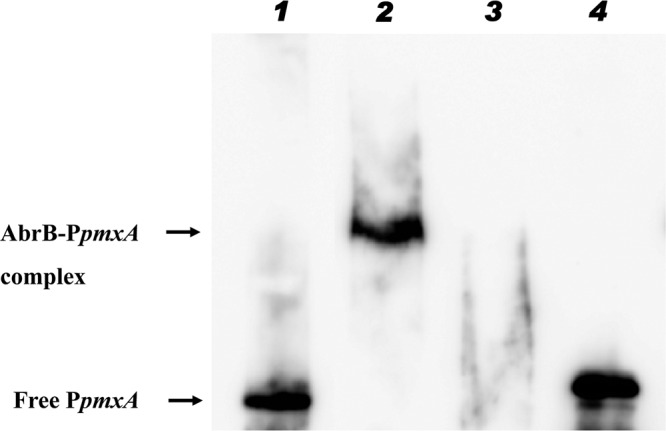
Binding of AbrB to the promoter region of pmxA. The promoter region of pmxA (PpmxA; 250 bp) was labeled with biotin. The biotin-labeled PpmxA (10 ng) interacted with 500 nM AbrB in a binding buffer containing 50 μg of poly(dI-dC) per ml as the nonspecific competitor. Lanes: 1, biotin-labeled PpmxA only; 2, mixture of biotin-labeled PpmxA and AbrB; 3 and 4, mixture of biotin-labeled PmxA, AbrB, and non-biotin-labeled PpmxA (5- and 10-fold for each lane).
Conclusions.
A B. subtilis strain, BSK4, that can produce polymyxin without addition of exogenous Dab was constructed in this study by introducing the ectB gene of P. polymyxa. We found that two important transcription factors, Spo0A and AbrB, are involved in the regulation of biosynthesis of polymyxin in B. subtilis. AbrB was shown to bind directly to the upstream region of pmxA, and it functions as a negative transcriptional regulator. A functional Spo0A was found to be indispensable for polymyxin production. The polymyxin productivity of BSK4 was improved 2.3-fold by null mutation in abrB. This improved system for the production of polymyxin, using the surrogate host B. subtilis, may accelerate structure-function studies and engineering of pmx genes for the generation of novel polymyxin derivatives.
ACKNOWLEDGMENTS
This work was supported by the KRIBB Research Initiative Program, the 21C Frontier Microbial Genomics and Applications Center Program, and the Intelligent Synthetic Biology Center of Global Frontier Project funded by the Ministry of Education, Science and Technology (2011-0031947).
Footnotes
Published ahead of print 30 March 2012
REFERENCES
- 1. Ainsworth GC, Brown AM, Brownlee G. 1947. Aerosporin, an antibiotic produced by Bacillus aerosporus Greer. Nature 160:263. [DOI] [PubMed] [Google Scholar]
- 2. Choi S-K, et al. 2009. Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis. J. Bacteriol. 191:3350–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choi S-K, et al. 2008. Identification and functional analysis of the fusaricidin biosynthetic gene of Paenibacillus polymyxa E681. Biochem. Biophys. Res. Commun. 365:89–95 [DOI] [PubMed] [Google Scholar]
- 4. Cosby WM, Vollenbroich D, Lee OH, Zuber P. 1998. Altered srf expression in Bacillus subtilis resulting from changes in culture pH is dependent on the Spo0K oligopeptide permease and the ComQX system of extracellular control. J. Bacteriol. 180:1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greene EA, Spiegelman GB. 1996. The Spo0A protein of Bacillus subtilis inhibits transcription of the abrB gene without preventing binding of the polymerase to the promoter. J. Biol. Chem. 271:11455–11461 [DOI] [PubMed] [Google Scholar]
- 6. Haima P, van Sinderen D, Schotting H, Bron S, Venema G. 1990. Development of a beta-galactosidase alpha-complementation system for molecular cloning in Bacillus subtilis. Gene 86:63–69 [DOI] [PubMed] [Google Scholar]
- 7. Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52:847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 9. Holmes KK. 1964. Toxicity of colistin and polymixin B. N. Engl. J. Med. 271:633–634 [DOI] [PubMed] [Google Scholar]
- 10. Jensen K, Ostergaard PR, Wilting R, Lassen SF. 2010. Identification and characterization of a bacterial glutamic peptidase. BMC Biochem. 11:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karow ML, Piggot PJ. 1995. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene 163:69–74 [DOI] [PubMed] [Google Scholar]
- 12. Kaur S, Balakrishnan R, Jayaraman K. 1978. The correlation between antibiotic synthesis, transcription and sporulation in Bacillus polymyxa. Biochem. Biophys. Res. Commun. 81:50–57 [DOI] [PubMed] [Google Scholar]
- 13. Kim JF, et al. 2010. Genome sequence of the polymyxin-producing plant-probiotic rhizobacterium Paenibacillus polymyxa E681. J. Bacteriol. 192:6103–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuhlmann AU, Bremer E. 2002. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 68:772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J, et al. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6:589–601 [DOI] [PubMed] [Google Scholar]
- 16. Lucking G, Dommel MK, Scherer S, Fouet A, Ehling-Schulz M. 2009. Cereulide synthesis in emetic Bacillus cereus is controlled by the transition state regulator AbrB, but not by the virulence regulator PlcR. Microbiology 155:922–931 [DOI] [PubMed] [Google Scholar]
- 17. Molle V, et al. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683–1701 [DOI] [PubMed] [Google Scholar]
- 18. Nefelova NV, Filippova MS, Egorov NS. 1980. Effect of polymyxins on Bacillus polymyxa sporogenesis. Mikrobiologiia 49:294–297 [PubMed] [Google Scholar]
- 19. Palva A, Vigren G, Simonen M, Rintala H, Laamanen P. 1990. Nucleotide sequence of the tetracycline resistance gene of pBC16 from Bacillus cereus. Nucleic Acids Res. 18:1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29–40 [DOI] [PubMed] [Google Scholar]
- 21. Peterson LR. 2009. Bad bugs, no drugs: no ESCAPE revisited. Clin. Infect. Dis. 49:992–993 [DOI] [PubMed] [Google Scholar]
- 22. Robertson JB, Gocht M, Marahiel MA, Zuber P. 1989. AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation gene and an antibiotic biosynthesis gene. Proc. Natl. Acad. Sci. U. S. A. 86:8457–8461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ryu C-M, et al. 2005. Nature of a root-associated Paenibacillus polymyxa from field-grown winter barley in Korea. J. Microbiol. Biotechnol. 15:984–991 [Google Scholar]
- 24. Shin B-S, Choi S-K, Park S-H. 1999. Regulation of the Bacillus subtilis phosphotransacetylase gene. J. Biochem. 126:333–339 [DOI] [PubMed] [Google Scholar]
- 25. Stein T, et al. 2002. Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol. Microbiol. 44:403–416 [DOI] [PubMed] [Google Scholar]
- 26. Strauch M, Webb V, Spiegelman G, Hoch JA. 1990. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. U. S. A. 87:1801–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Strauch MA. 1995. In vitro binding affinity of the Bacillus subtilis AbrB protein to six different DNA target regions. J. Bacteriol. 177:4532–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strauch MA, et al. 1989. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 8:1615–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 30. Vaara M, Vaara T. 2010. Structure-activity studies on novel polymyxin derivatives that carry only three positive charges. Peptides 31:2318–2321 [DOI] [PubMed] [Google Scholar]
- 31. Vasantha N, Balakrishnan R, Kaur S, Jayaraman K. 1980. Biosynthesis of polymyxin by Bacillus polymyxa. I. The status of the biosynthetic multienzyme complex during active antibiotic synthesis and sporulation. Arch. Biochem. Biophys. 200:40–44 [DOI] [PubMed] [Google Scholar]
- 32. Velkov T, Thompson PE, Nation RL, Li J. 2010. Structure-activity relationships of polymyxin antibiotics. J. Med. Chem. 53:1898–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60:1206–1215 [DOI] [PubMed] [Google Scholar]
- 34. Zhao B, et al. 2006. Cloning and characterization of the genes for biosynthesis of the compatible solute ectoine in the moderately halophilic bacterium Halobacillus dabanensis D-8(T). Curr. Microbiol. 53:183–188 [DOI] [PubMed] [Google Scholar]