Abstract
Endocrocin is a simple anthraquinone frequently identified in extracts of numerous fungi. Several biosynthetic schemes for endocrocin synthesis have been hypothesized, but to date, no dedicated secondary metabolite gene cluster that produces this polyketide as its major metabolite has been identified. Here we describe our biosynthetic and regulatory characterization of the endocrocin gene cluster in Aspergillus fumigatus. This is the first report of this anthraquinone in this species. The biosynthetic genes required for endocrocin production are regulated by the global regulator of secondary metabolism, LaeA, and encode an iterative nonreducing polyketide synthase (encA), a physically discrete metallo-β-lactamase type thioesterase (encB), and a monooxygenase (encC). Interestingly, the deletion of a gene immediately adjacent to encC, termed encD and encoding a putative 2-oxoglutarate-Fe(II) type oxidoreductase, resulted in higher levels of endocrocin production than in the wild-type strain, whereas overexpression of encD eliminated endocrocin accumulation. We found that overexpression of the encA transcript resulted in higher transcript levels of encA-D and higher production of endocrocin. We discuss a model of the enc cluster as one evolutionary origin of fungal anthraquinones derived from a nonreducing polyketide synthase and a discrete metallo-β-lactamase-type thioesterase.
INTRODUCTION
The anthraquinone endocrocin was first described in 1935 from the lichen Nephromopsis endocrocea and since has been isolated from various fungi (16, 26, 30, 37), insects (22), and plants (17). Endocrocin and related anthraquinones are noted for their industrial uses as dyes, food additives, components of paper making, and cosmetics (18) as well as for their medicinal uses, including as laxative, anti-inflammatory, and antitumor agents (17, 19). Some anthraquinones are also known to be precursors to mycotoxins of global economic and medical importance. The most notorious anthraquinones are the aflatoxin intermediates produced by the agricultural fungal pathogens Aspergillus flavus and A. parasiticus.
Despite its long history of prevalence across kingdoms, the first genetic understanding of endocrocin biosynthesis arose only recently with the discovery of a special class of type I nonreducing polyketide synthases (NR-PKS) that consist of canonical ketosynthase (KS), acyltransferase (AT), and acyl carrier protein (ACP) domains but lack a thioesterase (TE) or Claisen cyclase (CLC) domain (11). The TE/CLC domain is necessary for releasing the nascent polyketide product from the enzyme by catalyzing the Claisen cyclization followed by C-C bond formation and cleavage of the thioester bond between the ACP and the polyketide backbone (15, 23). In NR-PKS lacking TE/CLC domains, the polyketide backbone is released from the PKS by a group of bifunctional enzymes belonging to the metallo-β-lactamase (MβL) superfamily that are now termed metallo-β-lactamase-type thioesterases (MβL-TE) (4). These MβL-TEs were shown to catalyze both hydrolysis of the nascent PKS and Claisen cyclization (27). In a recent biochemical study, the MβL-TE (AptB) was shown to require two Mn2+ cations for activity, which distinguishes it from other MβLs, which utilizes Zn2+ (27). To date, four MβL-releasing NR-PKS in the aspergilli have been characterized, namely, the asperthecin and monodictyphenone NR-PKS (AptA and MdpA) in A. nidulans, the atrochrysone carboxylic acid (ACA) NR-PKS (ACAS) in A. terreus, and very recently the naphthacenedione (TAN-1612) NR-PKS (AdaA) in A. niger (4, 9, 27, 40). All four NR-PKS require the activity of adjacent MβL-TEs, i.e., AptB, MdpF, atrochrysone carboxyl ACP thioesterase (ACTE), and AdaB, respectively. Interestingly, both the asperthecin and atrochrysone carboxylic acid clusters produce or are predicted to produce endocrocin and emodin as pathway intermediates (4, 9, 40). The monodictyphenone cluster, on the other hand, produces emodin as a pathway intermediate, but endocrocin was isolated only from an mdp cluster mutant lacking a putative decarboxylase activity. This enzyme, MdpH, was hypothesized to inhibit endocrocin production and, via this action, to facilitate the emodin-to-monodictyphenone biosynthetic route (9). Thus, endocrocin was considered a branch product of the monodictyphenone pathway.
Here we present our finding of a new TE-less NR-PKS located within an uncharacterized secondary metabolite (SM) gene cluster in Aspergillus fumigatus and show that this TE-less NR-PKS is responsible for the production of endocrocin; this is the first report of this anthraquinone in this species. We propose a biosynthetic scheme for the endocrocin gene cluster and further discuss a model of endocrocin production as a minimal biosynthetic unit of fungal anthraquinone gene clusters derived from the coupling of an NR-PKS with an MβL-TE and an anthrone oxidase, which distinguishes this cluster from the asperthecin cluster of A. nidulans.
MATERIALS AND METHODS
Bioinformatic analysis.
Genome-wide in silico prediction of SM clusters in A. fumigatus was obtained from ftp://ftp.jcvi.org/pub/software/smurf/ based on the software Secondary Metabolite Unique Region Finder (SMURF) (21). LaeA-regulated SM clusters have been previously identified in A. fumigatus (29). A position-specific iterated BLAST (PSI-BLAST) was used to identify closely related proteins for the endocrocin cluster genes (2). Protein sequence alignment of AFUA_4G00225 (EncC) to HypC and other related proteins was done using the Clustal W method (42) in the MegAlign module (v. 8.0.2) of the Lasergene sequence analysis package (DNAStar Inc.).
Fungal strains and growth conditions.
Aspergillus fumigatus strains used in this study are listed in Table 1. Strains were maintained as glycerol stocks and activated on solid glucose minimal medium (GMM) at 37°C with appropriate supplements (34). For pyrG auxotrophs, the growth medium was supplemented with 5 mM uridine and uracil. Conidia were harvested in 0.01% Tween 80 and enumerated using a hemocytometer. For endocrocin analysis on solid medium, A. fumigatus strains were point inoculated on either solid GMM or Czapek yeast autolysate medium (CYA) at 1 × 104 conidia/inoculum and cultured at 29°C or 37°C without light selection (20). For endocrocin mutant analysis under liquid shake conditions, A. fumigatus strains were inoculated into liquid GMM at 1 × 106 conidia/ml and cultured at 25°C and 250 rpm without light selection.
Table 1.
A. fumigatus strains used in this study
| Fungal strain(s) | Genotype | Reference |
|---|---|---|
| CEA17 KU80 pyrG− | ΔnkuB::A. fumigatus pyrG, pyrG− | 12 |
| CEA17 KU80 pyrG+ | ΔnkuB::A. fumigatus pyrG, pyrG+ | 12 |
| TJHO3.1 | ΔnkuB::A. fumigatus pyrG; ΔlaeA::A. parasiticus pyrG | This study |
| TFYL6.3 | Δnku::A. fumigatus pyrG; ΔencA::A. parasiticus pyrG | This study |
| TFYL8.1 | ΔnkuB::A. fumigatus pyrG; ΔencB::A. parasiticus pyrG | This study |
| TFYL23.1 | ΔnkuB::A. fumigatus pyrG; ΔencC::A. parasiticus pyrG | This study |
| TFYL16.1, TFYL16.2 | ΔnkuB::A. fumigatus pyrG; ΔencD::A. parasiticus pyrG | This study |
| TFYL9.3 | ΔnkuB::A. fumigatus pyrG; ΔAFUA_4G00200::A. parasiticus pyrG | This study |
| TFYL21.1 | ΔnkuB::A. fumigatus pyrG; ΔAFUA_4G00240::A. parasiticus pyrG | This study |
| TFYL22.1 | ΔnkuB::A. fumigatus pyrG; ΔAFUA_4G00250::A. parasiticus pyrG | This study |
| TFYL1.51 | ΔnkuB::A. fumigatus pyrG; A. parasiticus pyrG::gpdAp::encA | This study |
| TFYL30.1, TFYL30.2 | ΔnkuB::A. fumigatus pyrG; ΔencC::A. parasiticus pyrG encD::hph | This study |
| TFYL31.1, TFYL31.2 | ΔnkuB::A. fumigatus pyrG; A. parasiticus pyrG::gpdAp::encD | This study |
Construction of the endocrocin pathway mutants.
Most of the A. fumigatus mutant strains listed in Table 1 were constructed using a double-joint fusion PCR (DJ-PCR) approach (41). Genomic DNA was isolated from A. fumigatus using standard procedures (32, 34). The construction of DJ-PCR products, protoplast production, and transformation were carried out as previously described (41). For the construction of the encA deletion cassette, two 1,000- to 1,500-bp fragments flanking the targeted 770-bp deletion region internal to the PKS were amplified by PCR from CEA17 KU80 pyrG+ genomic DNA. A. parasiticus pyrG as a selectable marker was amplified from the plasmid pJW24 (8). The three fragments were fused and then amplified using two nested primers (see Table S1 in the supplemental material) to generate the deletion cassette (see Fig. S1A in the supplemental material). encA was deleted from the CEA17 KU80 pyrG- strain (12) to create strain TFYL6.3. Multiplex PCR was used to confirm the absence of the encA open reading frame (ORF) (as assessed by lack of amplification product in the deletion mutant) using internal primers to encA and control internal primers to A. fumigatus gpdA (see Table S1 in the supplemental material). Single integration of the transformation construct was confirmed via Southern analysis using two restriction digest profiles, namely, KpnI and SacI (see Fig. S1B in the supplemental material). Deletion mutants for other putative cluster members were constructed as described above (Table 1). See Fig. S1 and Table S1 in the supplemental material for Southern analysis and primers used in the creation of these mutants, respectively. The encA and encD overexpression strains were created via promoter replacement. The marker gene A. parasiticus pyrG fused to an A. nidulans constitutive glyceraldehyde-3-phosphate dehydrogenase promoter, gpdAp, was amplified from plasmid pJMP9.1 (J. M. Palmer and N. P. Keller, unpublished data). Next, 1,000- to 1,500-bp fragments upstream and downstream from the predicted transcriptional start site were amplified from CEA17 KU80 pyrG+ genomic DNA. The three fragments (pyrG::gpdAp and the up- and downstream flanks) were fused using DJ-PCR to generate the overexpression cassettes (an example of the overexpression cassette construct using encA is depicted in Fig. S1C in the supplemental material) and transformed into CEA17 KU80 pyrG+ as mentioned above. A forward primer homologous to the gpdAp sequence and a reverse primer homologous to sequence outside the transformation cassette were used to screen transformants with integration at the locus of interest, followed by Southern analysis using EcoRI and SacI restriction digests (see Fig. S1D in the supplemental material). The EncD complementation strain was created via ectopic integration of the plasmid pFYL2 (bearing the wild-type [WT] EncD ORF and hygromycin resistance gene) into the EncD deletion strain TFYL16.1. The A. fumigatus EncD complementation cassette (including 1 kb of upstream promoter region and 0.5-kb downstream terminator regions) was amplified from A. fumigatus CEA17 KU80 pyrG+ (Table 1) using primers FY encD comp (NotI) FOR and FY encD comp (XbaI) REV (see Table S1 in the supplemental material), digested with NotI and XbaI, and ligated into pUCH2-8 (1) to generate pFYL2. pUCH2-8 contains the gene encoding hygromycin B phosphotransferase, which was the selectable marker used for transformation of Aspergillus.
Endocrocin extraction and thin-layer chromatography (TLC) analysis. (i) Small-scale whole-culture extraction.
The A. fumigatus wild type and all mutants (Table 1) were cultivated as mentioned above. A 1.2-cm-diameter core was extracted from the middle of the fungal culture and homogenized in 2 ml of 0.01% Tween 80. The homogenized mixture was extracted with an equal volume of ethyl acetate by routine vortexing at room temperature over the course of 30 min. The mixture was then centrifuged for 5 min at 3,500 rpm, and the ethyl acetate layer was removed and allowed to evaporate at room temperature to yield a dried crude extract.
(ii) TLC analysis.
The dried crude extract was reconstituted in 50 to 100 μl of ethyl acetate, and 5 to 10 μl was used for TLC analysis. The crude extracts were spotted onto a 250-μm analytical silica plate (Whatman, catalog no. 4410-222) and resolved in toluene-ethyl acetate-formic acid (5:4:0.8, vol/vol/vol). Plates were visualized at 254 nm and 366 nm using a FOTO/Analyst Investigator gel imaging system (Fotodyne Inc.).
Purification and identification of endocrocin. (i) Large-scale extraction and fractionation.
A. fumigatus wild-type and ΔencA strains were point inoculated on solid GMM at 1 × 104 conidia/inoculum. Approximately 120 plates were cultivated at 29°C for 10 days. The culture was homogenized and then extracted twice with ethyl acetate. The combined ethyl acetate fractions were filtered through a 5- to 10-μm-particle-retention-size filter paper (Fisherbrand, P5 grade) to remove residual agar and hyphal material and later through a 1- to 5-μm-particle-retention-filter paper (Fisherbrand, P2 grade) to remove residual conidia. The filtered extract was then evaporated in vacuo to yield a dried residue of crude extract. The dried extract was reconstituted in approximately 5 ml of a 1:1 (vol/vol) chloroform-methanol mixture and then fractionated via gel filtration chromatography using Sephadex LH-20 (Sigma, 25 to 100 m). A total of 15 fractions (FY01A to FY01O) were obtained, and each fraction was subjected to TLC and visualized as described above (see Fig. S3 in the supplemental material).
(ii) HPLC analysis.
The fractions (FY01J and FY01K) containing the compound of interest as assessed by TLC were combined, reconstituted in methanol (1 mg/ml), and subjected to high-performance liquid chromatography (HPLC) on a Shimadzu Prominence HPLC system equipped with a diode array detector (DAD). A gradient system of methanol and water (containing 0.1% acetic acid) was employed with a flow rate of 3 ml/min on a reverse-phase C18 column (Phenomenex; Onyx monolithic C18, 100 by 4.6 mm). The gradient started from methanol-water (1%:99%), followed by a linear gradient to reach methanol (100%) in 4 min and a hold for 2.5 min. Each peak was collected and subjected to TLC analysis to identify the compound of interest. The purified red metabolite of interest was obtained at retention time (tR) equal to 3.9 min.
(iii) Compound identification.
Nuclear magnetic resonance (NMR) data were recorded on a Varian 500 MHz NMR spectrometer equipped with an HCX 5-mm probe and on a Varian 600-MHz NMR spectrometer with a 5-mm 1H(13C/15N) cryogenic probe. The high-resolution mass spectrometry (HRMS) data were recorded on a Bruker maXis high-resolution electrospray ionization-quadrupole time of flight (HRESI-Q-TOF) mass spectrometer. The structure of endocrocin was determined by NMR (1H NMR and heteronuclear single-quantum coherence [HSQC]), HRMS data, and comparison with data reported in the literature. The compound was identical to an authentic endocrocin sample provided by Clay Wang at the University of Southern California: 1H NMR (600 MHz, CD3OD) δ 7.59 (1H, s, H-4), 7.17 (1H, d, J = 2.4 Hz, H-5), 6.55 (1H, d, J = 2.4 Hz, H-7), 2.46 (3H, s, 3-CH3); HRESI-MS m/z 315.0494 (M + H)+ (calculated for C10H16O7, 315.0499).
RNA extraction and Northern analysis.
Unless otherwise noted, 1 × 106 conidia/ml of A. fumigatus strains was inoculated into 250 ml of liquid GMM and grown for 24 h at 30°C and 250 rpm shaking. Mycelia were harvested by filtration through Miracloth (Calbiochem) and transferred onto solid GMM. Total RNA was extracted with TRIzol reagent (Invitrogen) from freeze-dried mycelia harvested at various time points after transfer, following the manufacturer's protocol. Probes for Northern analysis were constructed at regions internal to the gene of interest using primers listed in Table S1 in the supplemental material. For Northern analysis of the encA overexpression strain, 1 × 106 conidia/ml from both wild-type and mutant strains were inoculated into 50 ml of liquid GMM and grown for 54 h at 25°C and 250 rpm shaking. Mycelia were harvested by filtration through Miracloth (Calbiochem), freeze-dried, and extracted as mentioned above.
Endocrocin stability analysis.
An endocrocin solution in methanol was incubated at 37°C for a total of 15 days and analyzed on a Shimadzu Prominence HPLC system equipped with a diode array detector (DAD) at days 0, 3, and 15th. A gradient system of acetonitrile and water (containing 0.1% acetic acid) was employed with a flow rate of 1 ml/min on a reverse-phase phenyl-hexyl column (Phenomenex; Luna, 5 μm, 250 by 4.6 mm). The gradient started from acetonitrile-water (20%:80%), followed by a linear gradient to reach acetonitrile (100%) in 20 min and a hold for 5 min. Emodin and asperthecin solutions in methanol were analyzed under the same HPLC conditions.
RESULTS
Identification of A. fumigatus nonreducing polyketide synthases lacking C-terminal thioesterase domains.
A position-specific iterated BLAST (PSI-BLAST) search of the A. fumigatus genome using two iterations and a query protein sequence of MdpG, a nonreducing polyketide synthase (NR-PKS) from A. nidulans that is responsible for monodictyphenone biosynthesis, revealed three NR-PKS that lack a thioesterase/Claisen cyclase (TE/CLC) domain. These were AFUA_4G14560 (66% identity), AFUA_4G00210 (51% identity), and AFUA_7G00160 (49% identity). All three PKS are found in putative SM clusters, with each cluster containing a putative metallo-β-lactamase domain-encoding gene.
Further examination of the SMURF-predicted secondary metabolite (SM) clusters of each NR-PKS suggested that AFUA_4G00210 is located in a canonical anthraquinone cluster (Fig. 1) whereas both AFUA_4G14560 and AFUA_7G00160 are located in clusters suggestive of hybrid metabolites of higher chemical complexity. AFUA_4G14560 is located in a cluster that putatively consists of 32 ORFs, one of which encodes a putative nonribosomal peptide synthetase (NRPS)-like enzyme possibly indicative of a hybrid PKS-NRPS product. The AFUA_7G00160 cluster contains a putative dimethylallyl tryptophan synthase (DMAT) gene, suggesting a hybrid PKS-DMAT-derived product. Here we focused on AFUA_4G00210 (here called encA) and found that deletion of this gene resulted in the loss of one major and two minor visible UV-active metabolites (Fig. 2A).
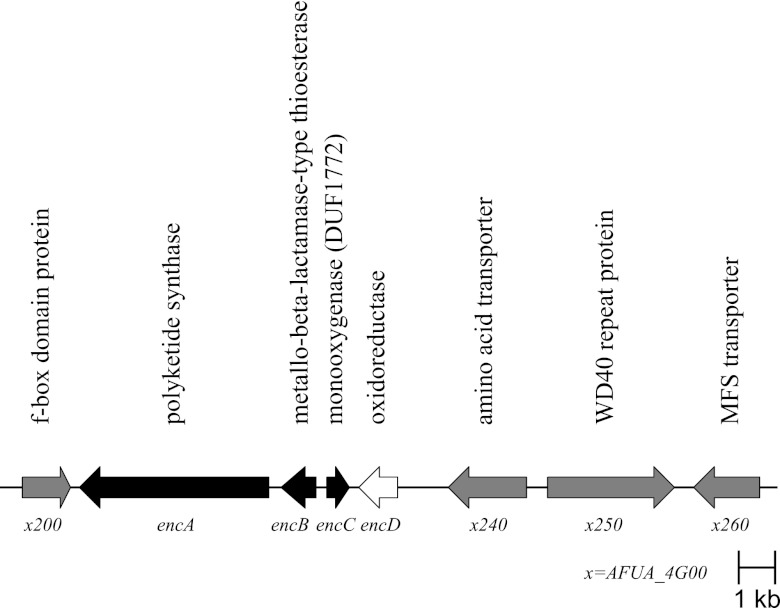
Fig 1.
Endocrocin gene cluster organization. Black arrows denote genes that are required for endocrocin production. The white arrow denotes a gene that increases the level of endocrocin production. Gray arrows denote genes that are not part of the enc cluster as assessed by lack of coregulation and effect on endocrocin production. MFS, major facilitator superfamily.
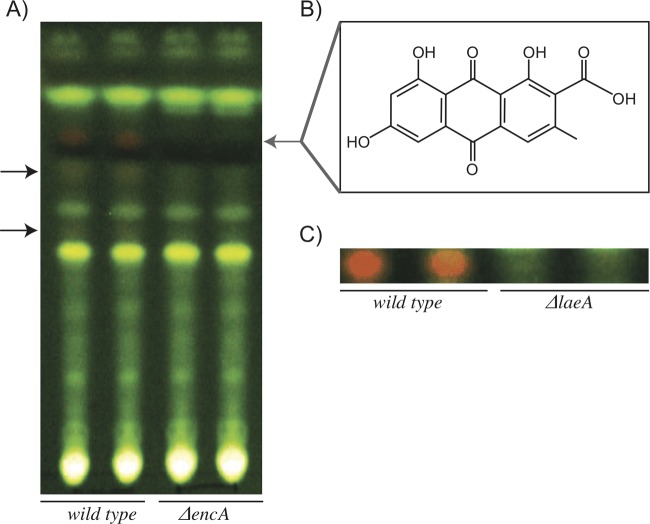
Fig 2.
(A) TLC analysis of crude extracts, comparing the encA deletion mutant to the CEA17 ΔnkuB wild-type strain of A. fumigatus. The gray arrow marks endocrocin, while black arrows mark two minor UV-active metabolites that are absent in the deletion strain. (B) Chemical structure of endocrocin. (C) TLC analysis of crude extracts, comparing the laeA deletion mutant to the CEA17 ΔnkuB wild-type strain of A. fumigatus.
Purification and identification of endocrocin in A. fumigatus.
Despite the visible difference in metabolite production in the ΔencA mutant (Fig. 2A), the total HPLC-DAD-MS profile showed no notable differences between the ΔencA mutant and its parental wild-type strain (data not shown). Therefore, we performed a large-scale extraction followed by extensive fractionation as described in Materials and Methods to purify the major of the three red metabolites. The crude extract was fractionated through an LH-20 column to afford 15 fractions (FY01A to FY01O). The major red metabolite was found to be highly concentrated in fractions FY01J and FY01K (see Fig. S3 in the supplemental material) and was obtained after further purification using HPLC. The structure of the red metabolite was elucidated as the known anthraquinone endocrocin (9,10-dihydro-1,6,8-trihydroxy-3-methyl-9,10-dioxo-2-anthracenecarboxylic acid) by NMR (1H NMR and HSQC), HRMS data, and comparison with previously reported data (4) (Fig. 2B; see Fig. S4 in the supplemental material). In addition, the purified compound was identical to an authentic endocrocin sample kindly provided by Clay Wang at the University of Southern California. This is the first report of this anthraquinone species in A. fumigatus.
Transcriptional and bioinformatic identification of the endocrocin biosynthetic genes.
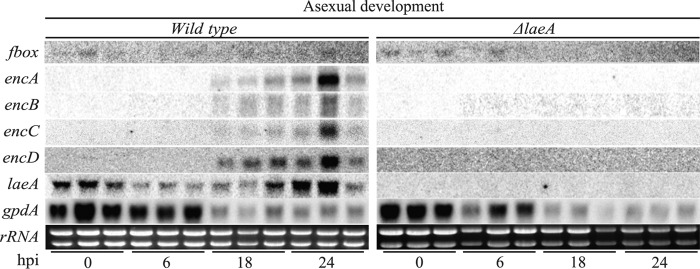
As previously found, the NR-PKS gene, encA, was downregulated in a ΔlaeA mutant of wild-type A. fumigatus AF293 (29). In agreement with the microarray data, endocrocin cannot be detected in SM extracts from the ΔlaeA mutant of A. fumigatus CEA17 ΔnkuB, which we used to create the enc mutants (Fig. 2C). We then examined gene expression of this region via Northern analysis in the wild type and the ΔlaeA mutant of the CEA17 ΔnkuB strain background. Based on the Northern analysis, four transcripts, termed encA-D, accumulated to high levels in the CEA17 ΔnkuB wild-type strain at 18 and 24 h postinduction (hpi) of asexual development (Fig. 3). A decrease in expression levels for all four enc genes was observed at 42 hpi, with low to no expression at 54 hpi (data not shown). However, an F-box domain-encoding gene situated beside encA was not regulated in this fashion. As we expected, transcripts for encA-D were undetectable at all time points in the CEA17 ΔnkuB ΔlaeA strain (Fig. 3).
Fig 3.
Northern analysis of the endocrocin gene cluster of the wild-type CEA17 ΔnkuB strain compared to that of strain TJHO3.1 (CEA17 ΔnkuB ΔlaeA). rRNA was used as a loading control. hpi, hours after induction of asexual development.
Identification of an anthrone oxidase encoded in the endocrocin gene cluster.
Not all of the Aspergillus databases showed the presence of encC. However, this ORF was annotated in the CADRE site as AFUA_4G00225 (28). As shown in Fig. 3, mRNA supported the existence of this ORF. Additionally, a PSI-BLAST search across the Aspergillus genomes using two iterations and the query protein sequence of AFUA_4G00225/encC gave a top hit with the recently characterized anthrone oxidase HypC of the aflatoxin biosynthetic cluster (13). Multiple-sequence alignment of amino acid sequences of EncC, HypC, and several other secondary metabolite monooxygenases involved in anthraquinone (or likely anthraquinone) metabolism revealed conservation of essential amino acid residues in the catalytic sites (Fig. 4). The flavin-dependent monooxygenase (FMO) MdpD of the monodictyphenone cluster was used as an outlier for the alignment. The alignment revealed that the essential tryptophan and tyrosine residues (hydrogen bond donor and receptor, respectively) responsible for substrate binding and enzyme activity (13, 33) are conserved in EncC and a HypC-like monooxygenase (XM_001911480) in Podospora anserina (Fig. 4) but not in MdpD, HypB (another monooxygenase with some sequence similarity to HypC but which has no known anthrone oxidase activity), AptC (discussed below), and AFUA_7G00150 (an A. fumigatus putative ortholog to AptC). In addition, an arginine in the motif RAL responsible for proton transfer was also conserved in EncC (Fig. 4). Between the two catalytic sites lay a DUF 1772 domain that has been speculated to play a role in protein-protein interactions (13).
Fig 4.
Clustal W alignment of EncC to the known anthrone oxidase HypC and other related monooxygenases. Gray denotes amino acid residues within the two catalytic sites, the DUF 1772 domain, and a highly conserved site of unknown function. Black denotes amino acid residues that are crucial for anthrone oxidase activity.
Genome-based deletion confirms cluster genes involved in endocrocin production.
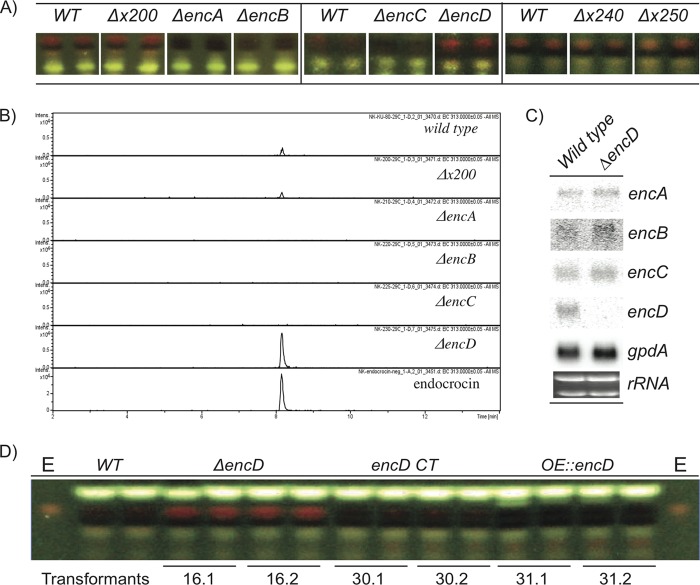
Based on SMURF prediction and Northern analysis, we sought to define which putative ORFs are required for endocrocin production. We created deletion mutants with mutations in 6 putative ORFs flanking encA, namely, AFUA_4G00200 (encoding an F-box domain protein), AFUA_4G00220/encB (encoding a metallo-β-lactamase domain protein), AFUA_4G00225/encC (encoding an anthrone oxidase), AFUA_4G00230/encD [encoding a 2-oxoglutarate-Fe(II) type oxidoreductase], AFUA_4G00240 (encoding an amino acid transporter), and AFUA_4G00250 (encoding a WD40 repeat protein) (Table 1; Fig. 1). Based on TLC analysis comparing total crude extracts of wild-type and mutant strains, endocrocin production was not affected by AFUA_4G00200, AFUA_4G00240, or AFUA_4G00250 loss but was abolished in the ΔencB mutant (Fig. 5A). However, we noticed minute amounts of endocrocin present in extracts of the ΔencC mutant (data not shown). Because anthrone species are relatively unstable and can slowly undergo spontaneous oxidation, we hypothesized that the small amounts of endocrocin in the crude extracts of the ΔencC mutant could be due to spontaneous oxidation of the endocrocin-9-anthrone to endocrocin (13). To examine this possibility, we repeated the above-described extraction process and ran TLC with the extracts immediately upon extraction and in vacuo dehydration to minimize postextraction oxidation of the unstable anthrone species. No endocrocin was detected in the ΔencC mutant, supporting the view that loss of EncC leads to unstable anthrone production (Fig. 5A).
Fig 5.
SM analysis of endocrocin gene cluster mutants. (A) TLC analysis of endocrocin cluster deletion mutants. The three panels represent TLC analyses from separate growth events. The wild-type crude extracts corresponding to each growth event are displayed in each panel. (B) Extracted ion chromatogram from HPLC/MS analysis of endocrocin levels, comparing the wild type to the ΔAFUA_4G00200, ΔencA, ΔencB, ΔencC, and ΔencD deletion mutants. (C) Northern analysis comparing the WT and the ΔencD mutant. rRNA was used as a loading control. (D) TLC of various EncD mutants grown at 29°C on solid GMM. Deletion of EncD resulted in a significant increase in endocrocin production, complementation of ΔencD restored the WT endocrocin production level, and overexpression of EncD (OE::encD) resulted in loss of endocrocin production. E, endocrocin standard; x, AFUA_4G00.
Unexpectedly, deletion of encD resulted in increased endocrocin production (Fig. 5A). To further support these TLC results, the extraction process was repeated using the wild-type strain and the ΔAFUA_4G00200, ΔencA, ΔencB, ΔencC, and ΔencD mutants. Endocrocin production was then quantitated via HPLC, in which we observed that the endocrocin level was wild type in the ΔAFUA_4G00200 mutant, eliminated in the ΔencA, ΔencB, and ΔencC mutants, and increased approximately 6-fold in the ΔencD mutant (Fig. 5B). To assess whether the increase in endocrocin production in the ΔencD mutant could be a result of higher expression of encA-C, we performed a Northern analysis and found that deletion of encD did not alter expression of encA-C (Fig. 5C). In addition, complementation of EncD in the ΔencD background (encD CT) restored WT levels of endocrocin production (Fig. 5D). Next we created an overexpression strain of encD (designated the OE::encD strain) and found that the strain did not produce endocrocin (Fig. 5D).
Overexpression of encA results in endocrocin overproduction.
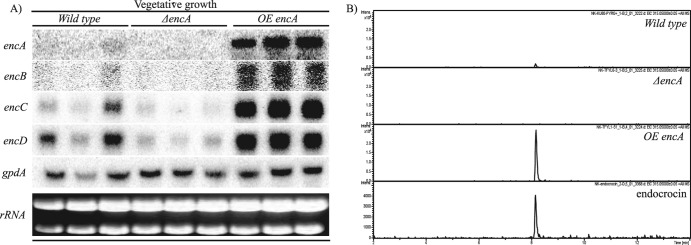
Having found no cluster-specific transcription factor within or in close proximity to the endocrocin cluster, we asked whether EncA itself could affect expression of other genes in the cluster and, if so, whether this would result in the overproduction of endocrocin. Regulation of cluster gene expression by other enzymatic genes has been observed in the gliotoxin gene cluster, in which the disruption of GliP, a multimodular nonribosomal peptide synthetase (NRPS), resulted in the inhibition of expression of other gli genes (10, 39). We found that the deletion of encA resulted in loss of expression of encB-D, while overexpression of encA resulted in increased expression of encB-D (Fig. 6A). There was also a significant increase in endocrocin production in the overexpression strain compared to the wild type (Fig. 6B).
Fig 6.
Overexpression of encA (OE::encA) results in overproduction of endocrocin. (A) Northern analysis comparing the wild-type, ΔencA, and OE::encA strains. rRNA was used as a loading control. (B) Extracted ion chromatogram from HPLC/MS analysis of total crude extracts from the wild-type, ΔencA, and OE::encA strains.
Comparison between the endocrocin and asperthecin gene clusters.
The organization of the asperthecin gene cluster in A. nidulans is very similar to that of the endocrocin cluster in A. fumigatus, in that they both consist of genes encoding a TE-less NR-PKS (AptA/EncA), an MβL-TE (AptB/EncB), and a monooxygenase (AptC/EncC). However, the endocrocin cluster contains a gene encoding an additional enzyme, EncD (2-oxoglurate-Fe-oxidoreductase), that is not present in the asperthecin cluster. For asperthecin synthesis, it was suggested that endocrocin could undergo spontaneous conversion to yield emodin, which is then catalyzed by AptC to yield asperthecin (9, 40). This led us to ask whether the endocrocin biosynthetic pathway could share precursors/end products with the asperthecin pathway. To address this, we performed LC-MS analysis on extracts of the A. fumigatus CEA17 ΔnkuB wild-type strain to identify the presence of either asperthecin or emodin; however, neither could be found (data not shown).
We then performed a stability assay to test whether endocrocin could undergo spontaneous conversion to yield emodin, a step suggested for formation of asperthecin by Szewczyk et al. (40). However, when incubated at 37°C for 15 days, endocrocin showed no signs of degradation or conversion to emodin or asperthecin (see Fig. S5 in the supplemental material). We also note that the amino acid residues crucial to anthrone oxidase activity in HypC are conserved in EncC but not in AptC (Fig. 4). This result suggests that AptC might catalyze a biosynthetic step distinct from that catalyzed by EncC and HypC.
DISCUSSION
Anthraquinones have many industrial, medicinal, and agricultural uses, which have generated much interest in anthraquinone production. Nonreducing type I iterative PKS (NR-PKS) mediate the syntheses of many anthraquinone species. Recently, a group of 18 unique NR-PKS that do not possess the canonical NR-PKS TE/CLC domain were identified in the aspergilli (9, 11, 40). Release of the nascent polyketide intermediate from these NR-PKS is instead achieved by activity of an MβL-TE domain protein (4, 9, 27, 40). Here we describe our characterization of an MβL-TE NR-PKS gene cluster in A. fumigatus and show that it is responsible for the production of endocrocin.
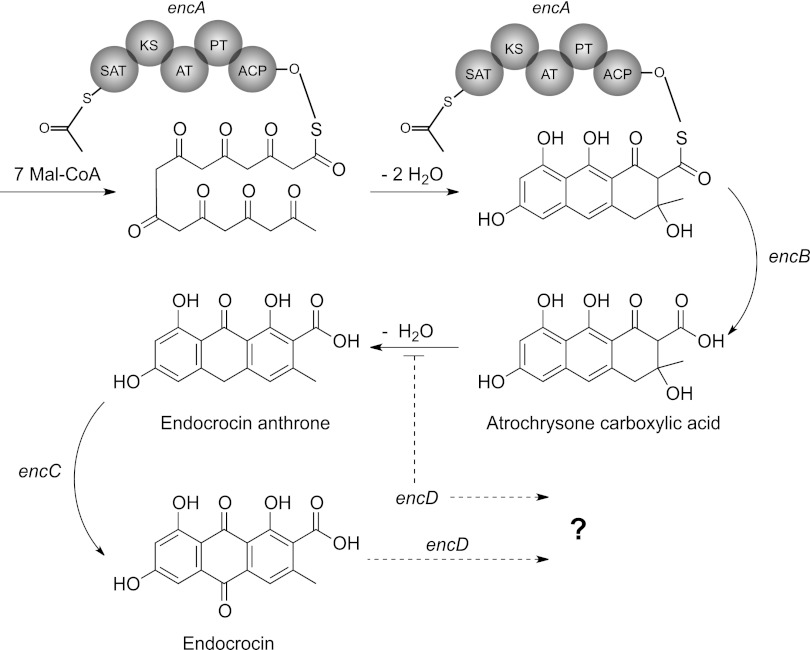
The endocrocin gene cluster is located in the subtelomeric region of the left arm of chromosome IV. Single-deletion mutants revealed that this cluster comprises four genes, of which three (encA, encB, and encC) are required for endocrocin production, while one (encD) negatively regulates the level of endocrocin production by a yet-uncharacterized mechanism. Based on the in silico predictions, genetic manipulations, and chemical assays presented in this study, we propose a model for the biosynthetic pathway of endocrocin (Fig. 7). Here the NR-PKS EncA couples with the MβL-TE, EncB, to form atrochrysone carboxylic acid (ACA) from 7-malonyl coenzyme A (7-malonyl-CoA). ACA then undergoes a spontaneous dehydration to form endocrocin-9-anthrone, which is then catalyzed by the predicted anthrone oxidase, EncC, to form endocrocin. Multiple-sequence alignment shows that EncC and HypC-like monooxygenases resemble bacterial anthrone oxidases, in that they do not require metal ions, prosthetic groups, or cofactors associated with oxygen activation (13, 14). A previous study showed that the spontaneous oxidation of anthrones could bypass enzymatic activity of the anthrone oxidases (13). Although spontaneous anthrone oxidation is likely in vitro, the efficiency of this phenomenon within the fungal cell is unclear. Our results suggested that EncC is crucial to endocrocin production, where deletion of encC resulted in loss of endocrocin production when extracted for SMs under minimal oxidation conditions. EncD, on the other hand, is not required for endocrocin biosynthesis, but rather loss of this protein enhances endocrocin production. Northern analysis demonstrated that the increase in endocrocin production in the ΔencD strain is not due to the insertion of the transformation cassette affecting the local genome architecture and expression level of encA-C. We note that the increase in encD transcript level in the encA overexpression strain did not appear to have a negative impact on increased endocrocin synthesis, suggesting that EncD could be a possible bottleneck in metabolite processing (Fig. 6). We propose two possible roles for EncD: (i) catalyzation of endocrocin to form an unknown product or (ii) inhibition of endocrocin synthesis through conversion of a precursor to an unknown product. The latter possibility is reminiscent of the monodictyphenone cluster in A. nidulans, where endocrocin is formed only when the decarboxylase (MdpH) is deleted from the cluster (9, 35). However, comparison of the MdpH sequence to the A. fumigatus genome revealed that its highest hit was to a gene in an uncharacterized TE-less NR-PKS cluster in A. fumigatus (AFUA_4G14470). It is likely that while MdpH and EncD elicit similar overall effects on the level of endocrocin production in vivo, they do so through different processes. EncD complementation of ΔencD restored WT endocrocin production levels, while overexpression of encD resulted in loss of endocrocin production. This observation suggests that EncD likely acts through the modification of endocrocin itself. Hence, we think that without the overall increase in enc cluster gene expression (as seen in the OE::encA strain), the increased EncD level in the OE::encD strain reduces the amount of endocrocin made and hence results in the loss of endocrocin production in this strain. Figure 5D hints at a possible accumulation of other metabolites in the OE::encD strain. One future goal of our lab is enzymatic characterization of EncD coupled with metabolite assessment of the OE::encD strain to further understand this pathway.
Fig 7.
Proposed biosynthetic model for endocrocin synthesis in A. fumigatus.
The gene organization of the endocrocin cluster shows high similarity to that of the asperthecin cluster in the sense that both do not contain a pathway-specific transcription factor but do contain a PKS, an MβL-TE, and a monooxygenase. However, our results showed that the biosynthetic pathways for these two clusters are distinct, as seen by the absence of an EncD-like protein in the asperthecin gene cluster and the different biosynthetic steps proposed to be catalyzed by the monooxygenases, EncC and AptC. Examination of the stability of purified endocrocin (see Fig. S5 in the supplemental material) as well as chemical analysis of wild-type and enc mutant strains suggests that endocrocin is not spontaneously converted to emodin as suggested for the asperthecin cluster by Szewczyk et al. (40). Indeed, a study in 1972 using labeled endocrocin did not find this compound to incorporate into emodin or other anthraquinones in the fungus Dermocybe sanguinea (36). These results suggest that although the asperthecin and endocrocin clusters share a high level of similarity in the organization of their biosynthetic genes, the distinct biosynthetic steps proposed to be catalyzed by EncC and AptC and the presence of EncD only in the endocrocin cluster contribute to the production of diverse metabolites from these two clusters. Furthermore, unlike for AptC, a flavin-dependent monooxygenase (FMO), multiple-sequence alignment showed that EncC resembles bacterial anthrone oxidases, in that they do not require metal ions, prosthetic groups, or cofactors associated with oxygen activation (13, 14, 27). The two FMO gene-containing clusters (apt and ada) known thus far belong to the SAT(GXSXG)-PT(V2)-MβL(V2) PKS group, while the anthrone oxidase gene-containing cluster described here (enc) belongs to the SAT(GXGXG)-PT(V1)-MβL(V1) PKS group (11, 25, 27). In all, our results lead us to propose that the TE-less NR-PKS fall into two distinct types, one containing the SAT(GXSXG)-PT(V2)-MβL(V2) FMO and the other containing SAT(GXGXG)-PT(V1)-MβL(V1) anthrone oxidase.
Simple anthraquinones are often precursors to a myriad of clinically or agriculturally important mycotoxins, pigments, and even small molecules signaling fungal protection and developmental processes such as conidiation or sexual development (17, 31, 43). The fact that endocrocin has been isolated from many Aspergillus spp., now including A. fumigatus, as well as from other genera and kingdoms raises the question of its role to its native producer. In other words, are simple anthraquinones such as endocrocin being made as a “by-product” of higher, more complex secondary metabolic pathways, or do they have an important yet undefined biological role for the endogenous organism? Interestingly, the enc cluster in A. fumigatus is subject to regulation by LaeA, a global regulator of SMs and morphological differentiation in fungi (3, 5–7, 24, 38). Future studies should elucidate any role of endocrocin and other SMs in fungal development, perhaps as mediated by LaeA regulation.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded in part by NIH grant 1 R01 Al065728-01 and NIH grant GM084077 to N.P.K. This research was supported in part by the National Research Foundation of Korea (NRF 2010-0012426 to I.L.). This study made use of the National Magnetic Resonance Facility at Madison, WI, which is supported by NIH grants P41RR02301 (BRTP/NCRR) and P41GM66326 (NIGMS). Additional equipment was purchased with funds from the University of Wisconsin, the NIH (RR02781 and RR08438), the NSF (DMB-8415048, OIA-9977486, and BIR-9214394), and the USDA.
We thank the Analytical Instrumentation Center at the University of Wisconsin—Madison School of Pharmacy for the facilities to acquire spectroscopic data, especially NMR and MS data.
Footnotes
Published ahead of print 6 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alexander NJ, Hohn TM, McCormick SP. 1998. The TRI11 gene of Fusarium sporotrichioides encodes a cytochrome P-450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl. Environ. Microbiol. 64:221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Amaike S, Keller N. 2009. Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot. Cell 8:1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Awakawa T, et al. 2009. Physically discrete beta-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase. Chem. Biol. 16:613–623 [DOI] [PubMed] [Google Scholar]
- 5. Bayram O, et al. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506 [DOI] [PubMed] [Google Scholar]
- 6. Bok J, et al. 2005. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 4:1574–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bok J, Keller N. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3:527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calvo A, Bok J, Brooks W, Keller N. 2004. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 70:4733–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiang Y, et al. 2010. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl. Environ. Microbiol. 76:2067–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cramer RA, Jr, et al. 2006. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot. Cell 5:972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crawford JM, Vagstad AL, Ehrlich KC, Townsend CA. 2008. Starter unit specificity directs genome mining of polyketide synthase pathways in fungi. Bioorg. Chem. 36:16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. da Silva Ferreira M, et al. 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5:207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ehrlich KC, Li P, Scharfenstein L, Chang P-K. 2010. HypC, the anthrone oxidase involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 76:3374–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fetzner SF. 2002. Oxygenases without requirement for cofactors or metal ions. Appl. Microbiol. Biotech. 60:243–257 [DOI] [PubMed] [Google Scholar]
- 15. Fujii I, Watanabe A, Sankawa U, Ebizuka Y. 2001. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem. Biol. 8:189–197 [DOI] [PubMed] [Google Scholar]
- 16. Gatenbeck S. 1959. The occurence of endocrocin in Penicillium islandicum. Acta Chem. Scand. 13:386–387 [Google Scholar]
- 17. Gautam R, Karkhile KV, Bhutani KK, Jachak SM. 2010. Anti-inflammatory, cyclooxygenase (COX)-2, COX-1 inhibitory, and free radical scavenging effects of Rumex nepalensis. Planta Med. 76:1564–1569 [DOI] [PubMed] [Google Scholar]
- 18. Gill M, Steglich W. 1987. Pigments of fungi (Macromycetes). Fortschr. Chem. Org. Naturst. 51:1–317 [DOI] [PubMed] [Google Scholar]
- 19. Hirayama T, et al. 1980. Anti-tumor activities of some lichen products and their degradation products. Yakugaku Zasshi 100:755–759 [DOI] [PubMed] [Google Scholar]
- 20. Hong SB, Go SJ, Shin HD, Frisvad JC, Samson RA. 2005. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 97:1316–1329 [DOI] [PubMed] [Google Scholar]
- 21. Khaldi N, et al. 2010. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 47:736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kikuchi N, et al. 2011. Endocrocin and its derivatives from the Japanese mealybug Planococcus kraunhiae. Biosci. Biotechnol. Biochem. 75:764–767 [DOI] [PubMed] [Google Scholar]
- 23. Korman TP, et al. 2010. Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 107:6246–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kosalkova K, et al. 2009. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 91:214–225 [DOI] [PubMed] [Google Scholar]
- 25. Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. U. S. A. 100:15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kurobane I, Vining LC, Mcinnes AG. 1979. Biosynthetic relationships among the secalonic acids—isolation of emodin, endocrocin and secalonic acids from Pyrenochaeta terrestris and Aspergillus aculeatus. J. Antibiot. 32:1256–1266 [DOI] [PubMed] [Google Scholar]
- 27. Li Y, Chooi YH, Sheng Y, Valentine JS, Tang Y. 2011. Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an alpha-hydroxylation-dependent Claisen-like cyclization catalyzed by a dimanganese thioesterase. J. Am. Chem. Soc. 133:15773–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mabey JE, et al. 2004. CADRE: the Central Aspergillus Data REpository. Nucleic Acids Res. 32:D401–D405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perrin R, et al. 2007. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3:508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raisanen R, Bjork H, Hynninen PH. 2000. Two-dimensional TLC separation and mass spectrometric identification of anthraquinones isolated from the fungus Dermocybe sanguinea. Z. Naturforsch. C 55:195–202 [DOI] [PubMed] [Google Scholar]
- 31. Rohlfs M, Albert M, Keller N, Kempken F. 2007. Secondary chemicals protect mould from fungivory. Biol. Lett. 3:523–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Sciara G, et al. 2003. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 22:205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shimizu K, Keller N. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steglich W. 1972. The biosynthesis of fungal quinones. Hoppe-Seylers Z. Physiol. Chem. 353:124–125 [PubMed] [Google Scholar]
- 36. Steglich W, Arnold R, Losel W, Reininger W. 1972. Biosynthesis of anthraquinone pigments in Dermocybe. J. Chem. Soc. Chem. Commun. 1972:102–103 [Google Scholar]
- 37. Steglich W, Reininger W. 1970. A synthesis of endocrocin, endocrocin-9-anthrone, and related compounds. J. Chem. Soc. Chem. Commun. 1970:178a [Google Scholar]
- 38. Sugui JA, et al. 2007. Role of LaeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot. Cell 6:1552–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugui JA, et al. 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 6:1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Szewczyk E, et al. 2008. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl. Environ. Microbiol. 74:7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Szewczyk E, et al. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120 [DOI] [PubMed] [Google Scholar]
- 42. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang C, Chiang Y, Kuo P, Chang J, Hsu Y. 2008. Norsolorinic acid from Aspergillus nidulans inhibits the proliferation of human breast adenocarcinoma MCF-7 cells via Fas-mediated pathway. Basic Clin. Pharmacol. Toxicol. 102:491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.