Abstract
Microbial consortia confer important benefits to animal and plant hosts, and model associations are necessary to examine these types of host/microbe interactions. The accessory nidamental gland (ANG) is a female reproductive organ found among cephalopod mollusks that contains a consortium of bacteria, the exact function of which is unknown. To begin to understand the role of this organ, the bacterial consortium was characterized in the Hawaiian bobtail squid, Euprymna scolopes, a well-studied model organism for symbiosis research. Transmission electron microscopy (TEM) analysis of the ANG revealed dense bacterial assemblages of rod- and coccus-shaped cells segregated by morphology into separate, epithelium-lined tubules. The host epithelium was morphologically heterogeneous, containing ciliated and nonciliated cells with various brush border thicknesses. Hemocytes of the host's innate immune system were also found in close proximity to the bacteria within the tubules. A census of 16S rRNA genes suggested that Rhodobacterales, Rhizobiales, and Verrucomicrobia bacteria were prevalent, with members of the genus Phaeobacter dominating the consortium. Analysis of 454-shotgun sequencing data confirmed the presence of members of these taxa and revealed members of a fourth, Flavobacteria of the Bacteroidetes phylum. 16S rRNA fluorescent in situ hybridization (FISH) revealed that many ANG tubules were dominated by members of specific taxa, namely, Rhodobacterales, Verrucomicrobia, or Cytophaga-Flavobacteria-Bacteroidetes, suggesting symbiont partitioning to specific host tubules. In addition, FISH revealed that bacteria, including Phaeobacter species from the ANG, are likely deposited into the jelly coat of freshly laid eggs. This report establishes the ANG of the invertebrate E. scolopes as a model to examine interactions between a bacterial consortium and its host.
INTRODUCTION
Many aquatic and marine invertebrates, including some cephalopods (squid, octopuses, and cuttlefish), lay their eggs in clutches or masses on benthic substrates, where they take weeks or even months to develop before hatching (2, 5, 12, 33). During this time, the developing embryos are unprotected, and prior observations suggest these egg clutches resist predation and/or fouling by microorganisms, although clear mechanisms for this resistance have yet to be described. Sexually mature females of some species have an accessory nidamental gland (ANG), a reproductive organ that houses a dense consortium of bacteria in pigmented epithelium-lined tubules and is attached to the nidamental gland (NG), the organ that secretes the jelly coat surrounding fertilized eggs (6). Culture-dependent and -independent methods have identified the dominant members of these microbial communities for some squid species (3, 6, 25, 39). All squid ANGs examined to date are dominated by alphaproteobacteria, usually members of the Roseobacter clade within the Rhodobacterales (6, 16, 39) with additional members belonging to the Gammaproteobacteria (vibrios, pseudoalteromonads, and pseudomonads) and the Bacteroidetes. Similar taxonomic groups were also found in the egg casings of the squid Loligo pealei, suggesting that the ANG serves to inoculate the egg clutches with a bacterial population (6). Although the exact role of these consortia has not been determined, those past studies suggest a symbiotic relationship between these bacteria and their hosts that should be investigated further.
In this study, we examined the accessory nidamental gland of the Hawaiian bobtail squid, Euprymna scolopes (Fig. 1). The symbiosis between E. scolopes and the bioluminescent bacterium Vibrio fischeri is used as a model system to study the effects of beneficial bacteria on the development of animal host tissues (26, 29, 30, 35). Adult E. scolopes squid can easily be collected and bred in the laboratory and are readily accessible to use as experimental animals to research host/microbe interactions. In addition, its responses, i.e., biochemical, cellular, genetic, and developmental, to bacterial colonization are the best characterized for any cephalopod species.
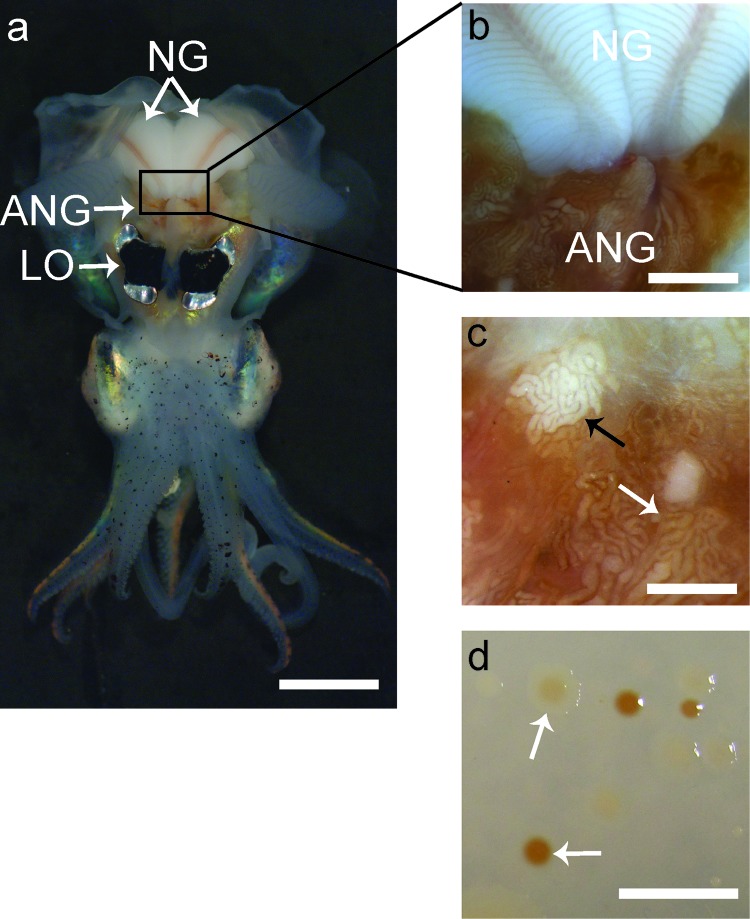
Fig 1.
Anatomy of a female Euprymna scolopes and morphology of ANG isolates. (a) Ventral dissection of E. scolopes, showing the accessory nidamental gland (ANG) located posterior to the light organ (LO) and in close proximity to the nidamental gland (NG). (b) Pigmented ducts in the NG converge at the ANG. (c) Magnification of the ANG reveals convoluted tubules, most of which are dark orange in pigmentation (white arrow), but others appear white (black arrow) or yellow (not shown). (d) Culturing of ANG symbionts results in many colonies with pigmentation similar to that of the ANG tubules (white arrows). Bars, 1 cm for panels a and d, 2.5 mm for panel b, and 1 mm for panel c.
In order to better understand the role of the ANG consortium in the biology of E. scolopes, the host and bacterial cell morphologies as well as the microbial diversity were characterized using transmission electron microscopy (TEM), 16S ribosomal sequence analysis, restriction fragment length polymorphism (RFLP) analysis, fluorescent in situ hybridization (FISH), and high-throughput 454 metagenomic sequencing. Here we report the initial characterization of the ANG microbiota for the model host, E. scolopes. This study is the first to use high-throughput sequencing to characterize the bacteria in any accessory nidamental gland. More importantly, it sets the foundation for exploration of a bacterial consortium in the same host as has already been used to research a well-studied monospecific symbiosis.
MATERIALS AND METHODS
Animal maintenance.
Adult animals were collected from shallow sand flats off Oahu, HI, by dip net and maintained in 42-liter recirculating aquaria at the University of Connecticut with artificial seawater (ASW; Instant Ocean) at 23°C and kept on an approximately 12 h light/12 h dark cycle (33).
Dissection and DNA extraction.
Female squid that had been maintained in the laboratory for between 24 h and 4 months were anesthetized in 2% ethanol in filter-sterilized ASW and ventrally dissected to remove the accessory nidamental gland. Once removed, the ANGs were flash frozen in liquid nitrogen and stored at −80°C until use. For 16S clone libraries, ANGs were homogenized in lysis buffer and total DNA was isolated using a DNeasy Tissue Prep kit (Qiagen, Hilden, Germany). To obtain DNA for 454 library construction, frozen ANGs were first thawed and then homogenized in squid Ringer's solution (530 mM NaCl, 25 mM MgCl2, 10 mM CaCl2, 20 mM HEPES, pH 7.5) using a ground glass homogenizer. The homogenate was spun for 10 min at 5,000 × g at 4°C. To remove solubilized host tissues, the supernatant was removed and the pellet was repeatedly washed (at least three times) with squid Ringer's solution until the protein concentration of the supernatant was sufficiently low (<0.5 mg/ml), as measured spectrophotometrically by A280 analysis. For 454 sequencing, total genomic DNA was extracted from the resulting pellet by the use of a DNA MasterPure kit (Epicentre, Madison, WI).
Culturing bacteria from the ANG.
Frozen ANGs from three sexually mature ANGs were homogenized in squid Ringer's solution, and the homogenate was serially diluted 10-fold and plated in triplicate onto R2A media (42) supplemented with 27 g of marine salts (Instant Ocean). Plates were incubated aerobically at 28°C for 3 days, and the resulting colonies were observed for pigmentation.
Microscopy.
Immediately after dissection, ANGs were cut in half and fixed at room temperature in 2.0% paraformaldehyde–2.5% glutaraldehyde in buffer A (0.1 M sodium cacodylate, 0.375 M NaCl, 1.5 mM CaCl2, and 1.5 mM MgCl2, pH 7.4). After an initial 15-min fixation period, the tissue samples were cut into smaller pieces (∼0.25 cm thick) and placed in fresh fixative for an additional 5 h at 4°C. Following fixation, tissue pieces were washed several times in cold buffer A and left at 4°C overnight. The following day, tissues were postfixed in a solution of 1% osmium tetroxide–0.8% potassium ferricyanide–0.1 M sodium cacodylate–0.375 M NaCl for 1.5 h at 4°C and then washed in distilled water, dehydrated through an ascending ethanol series, cleared in 100% acetone, and embedded in an epoxy mixture of Embed 812 (Electron Microscopy Sciences, Hatfield, PA) and Araldite 506 (Ernest Fulham Inc., Albany, NY). Semithin (2-μm) sections were obtained with a glass knife using an LKB Ultramicrotome V and stained with methylene blue and azure II followed by counterstaining with basic fuchsin. Stained sections were viewed on an Axiovert 200 M (Zeiss, Oberkochen, Germany) microscope. Thin (80-nm) sections were obtained using a diamond knife on a LKB Ultramicrotome V followed by staining with 2% uranyl acetate and Reynold's lead citrate (43) and viewed with an FEI Tecnai Biotwin G2 Spirit electron microscope (Hillsboro, OR) operated at 80 kV.
16S clone library construction and RFLP and sequencing analyses.
To examine the bacterial diversity in ANGs, total genomic DNA from the ANGs of five sexually mature females were used to make five separate 16S clone libraries. ANGI, ANGII, ANGIII, and ANGIV came from each of four females that were kept in our squid facility for 9, 14, 12, and 17 weeks, respectively. ANGV came from an individual that had been field caught and was maintained in our facility for 24 h. 16S genes were amplified using 25-μl GoTaq reaction mixtures (Promega, Madison, WI) with the 27F and 1406R eubacterial 16S primers (Table 1). PCR conditions were as follows: 95°C for 3 min, then 35 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 90 s, followed by a final elongation at 72°C for 10 min. PCR products were ligated and cloned using a PGEM-T Easy kit with JM109 cells (Promega, Madison WI). A total of 417 colonies were selected for restriction fragment length polymorphism (RFLP) analysis by incubating cloned genes with 10 U of MspI restriction enzyme (New England BioLabs, Ipswich, MA) at 37°C for 15 min. The resulting fragments were visualized on 1.5% agarose gels, and clones were grouped according to unique RFLP patterns. Representative clones from each group were sequenced using BigDye version 1.1 (Applied Biosystems, Carlsbad, CA) according to the manufacturer's specifications. Any clone that could not be grouped with an RFLP pattern was also sequenced. The 16S rRNA genes from 25, 45, and 27 clones from ANGI, ANGII, and ANGIII, respectively, were fully sequenced to confirm the accuracy of the restriction digest grouping. Sequences were analyzed with the Bellerophon chimera server (18), and 25 chimeric sequences were discarded, leaving 392 clones that were included in the analysis. The full-length sequences were used to search the Greengenes 16S rRNA gene database of named isolates by the use of BLAST (11). Operational taxonomic units (OTUs) were assigned to each sequence based on highest percent identity. Sequences from the Verrucomicrobia isolates had few quality alignments and were therefore characterized as representing a phylum.
Table 1.
Primers and FISH probes used in this study
| Primer, probe, or target organism category | Name | Sequence | Hybridization buffer % formamide (probes only) | Reference or source |
|---|---|---|---|---|
| Primer | ||||
| Eubacterial | 27F | AGAGTTTGATCCTGGCTCAG | Lane (23) | |
| 1406R | ACGGGCGGTGTGTRCAA | Lane (23) | ||
| FISH probes | ||||
| Eubacterial (universal) | Eub338I | GCTGCCTCCCGTAGGAGT | 30 | Amann (1) |
| Eub338III | GCTGCCACCCGTAGGTGT | 30 | Daims et al. (9) | |
| Roseobacter | G Rb | GTCAGTATCGAGCCAGTGAG | 30 | Giuliano et al. (13) |
| Bacteroidetes (Cytophaga-Flavobacteria) | CF319 | TGGTCCGTGTCTCAGTAT | 30 | Manz et al. (28) |
| Verrucomicrobia | Verruco_193 | CGCCATTACAAGCTTTAGTA | 20 | This study |
| Phaeobacter | Phaeo_126 | TGGCTATTTTAGAGAAGGGCA | 20 | This study |
| Alphaproteobacteria | Alph_968 | GGTAAGGTTCTGCGCGTT | 30 | Neef (34) |
| Eubacteria (negative control) | NonEub338 | ACTCCTACGGGAGGCAGC | 30 | Wallner et al. (49) |
454 metagenomic sequencing.
To identify other bacterial members isolated from the ANG that might not have been detected with 16S clone libraries and to increase our sequencing depth, we analyzed bacterial diversity using 454-metagenomic analyses. Bacterial DNA was extracted from 3 ANGs as described above. The samples were pooled, and 500 ng was used to construct a 454-shotgun metagenomic library using a Rapid Library kit (Roche Applied Science, Basel Switzerland). After the small-volume (SV) emulsion PCR (emPCR) titration was performed, the library was used in two 454 sequencing runs with FLX Titanium chemistry (Roche Applied Science, Basel, Switzerland). After removing 454 artifacts by the use of a 454 replicate filter (15), 622,987 sequences with an average length of 389.68 bases (total = 242.77 Mb) were analyzed. Roughly 1% of the reads (6,350) were eukaryotic in origin and not used in our analyses.
For 16S analysis of 454 data, reads were annotated using the MG-RAST server (32). Using the algorithm available from the Ribosomal Database Project (RDP), reads with at least a 200-bp alignment to a known 16S gene were extracted and used to search the NCBI nucleotide database with BLAST. OTUs were assigned as described above.
FISH.
To localize bacteria to the ANG, organs were dissected from six sexually mature female squid and prepared for fluorescent in situ hybridization (FISH). Two were freshly collected and dissected in Hawaii; the other four were kept in our animal facility for 8 to 14 weeks prior to dissection. Time in captivity did not affect results (data not shown). Three ANGs were fixed with Carnoy's solution (ethanol:chloroform:acetic acid [6:3:1]) overnight, and three were fixed in 1× PBS–4% paraformaldehyde for 4 h. Tissues were embedded in paraffin, and hybridization was performed as previously described (21). Three egg capsules were removed from freshly laid egg clutches, fixed in squid Ringer's solution–4% paraformaldehyde for 4 h, and embedded in paraffin as described above.
Based on our 16S data, several ribosomal probes were used at 50 pmol/ml each for hybridization (Table 1). Probes that corresponded to species of Eubacteria, Alphaproteobacteria, the Roseobacter clade, or Cytophaga-Flavobacteria-Bacteroidetes (CFB) were designed on the basis of published data. Novel 16S probes for Verrucomicrobia and Phaeobacter species were designed based on 16S sequence data, and specificity was confirmed with ProbeCheck (Table 1) (24) and fixed cultures of closely related members of genera of Rhodobacterales (e.g., Phaeobacter, Ruegeria, Tateyamaria, and Nautella for the Phaeobacter-specific probes; data not shown). All probes were synthesized by Eurofins MWG Operon (Huntsville, AL) and conjugated to fluorescein isothiocyanate (FITC), Cy3, or Cy5. After an overnight hybridization at room temperature in formamide hybridization buffer (0.9 M NaCl, 20 mM Tris [pH 8], 0.01% sodium dodecyl sulfate [SDS]) (Table 1), the tissue was washed in hybridization buffer and then counterstained with 300 nM DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen, Carlsbad, CA) in 1× PBS for 5 min. The following negative controls were performed: no probe, a nonsense probe (complementary to the eubacterial probe Eub338), and competition with nonlabeled probes. Tissue sections were imaged on a Leica SP2 confocal microscope (Wetzlar, Germany) or a Zeiss Axiovert 200 M epifluorescence microscope (Carl Zeiss, Germany) using DAPI, FITC, Cy3, and Cy5 filter sets.
Accession numbers.
16S clone library sequences were deposited in the European Nucleotide Archive (ENA) with accession numbers HE574851 to HE574928. Metagenomic reads were deposited in the NCBI Short Read Archive with accession numbers SRR329677.8 and SRR329678.5.
RESULTS
Morphological and EM observations.
The ANG of E. scolopes (Fig. 1a) contains many convoluted tubules that are highly pigmented (Fig. 1b). While most tubules have a dark orange pigmentation, some appear white (Fig. 1c) or, more rarely, yellow (not shown). As with other ANGs, the bacteria within the tubules likely synthesize these pigments, as colonies isolated from the organ also appeared similarly pigmented when grown in culture (Fig. 1c) (6).
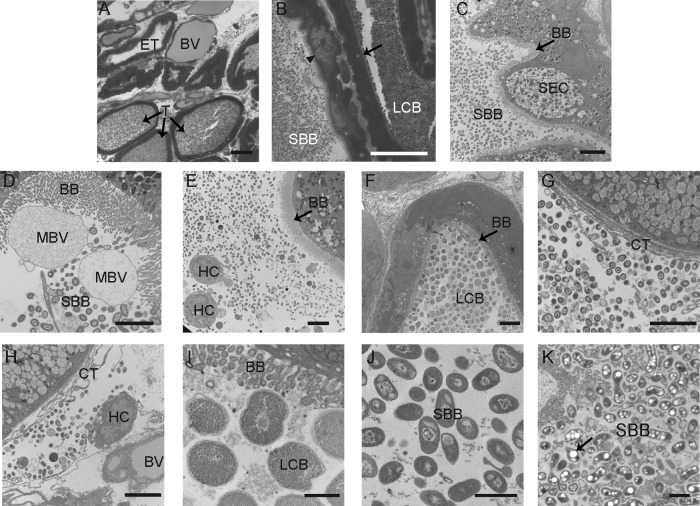
Light and electron microscopy of fixed sections of the ANG revealed that the organ is highly vascularized, with many blood vessels among tubules lined with ciliated epithelial cells and containing populations of bacteria. In some tubules, however, bacteria were not observed (Fig. 2A). Two morphologically distinct and segregated cell types were observed (Fig. 2B): a large coccoid bacterium (LCB) and a smaller bacillus bacterium (SBB). These two bacterial morphotypes appeared in separate tubules with strikingly different epithelia. One type of epithelium, associated with the SBB, appeared vacuole-rich (Fig. 2B and C), while the other associated with the LCB, had an electron-dense staining pattern lacking vacuoles (Fig. 2B and F). Within the tubules housing the bacteria were microvillar brush borders 1 to 5 μm in thickness along with membrane-bound vesicles that may be secreted or blebbed by the host (Fig. 2D). Some of the vacuole-rich epithelial cells had the distinct appearance of being secretory in nature, containing numerous large electron-light vacuoles and smaller electron-dense granules located at the apical surfaces of the epithelium (Fig. 2C). Hemocytes, the primary innate immune cells of E. scolopes, were observed in the lumina of the tubules; however, phagocytosed bacteria were not observed within these cells (Fig. 2E). Each tubule was dominated by one of the two morphologies: either the SBB (Fig. 2C and D) or the LCB (Fig. 2F). Mixtures of both LCB and SBB morphotypes were also observed outside the tubules within the connective tissue (Fig. 2G and H). The epithelial membranes appeared well-preserved, suggesting that these observations were not from a fixation artifact and that the bacteria can travel outside the ANG lumina. Hemocytes were also observed within the connective tissue (Fig. 2H), but as in the lumina of the tubules, no intracellular or phagocytosed bacteria were noted. Under higher magnification, the LCB cells appeared to be filled with many granules (Fig. 2I). This was in stark contrast to SBB, which were either mostly electron dense (Fig. 2J) or contained large electron-light storage vacuoles which resembled polyhydroxybutarate (PHB) (19, 27) (Fig. 2K).
Fig 2.
Light microscopy and TEM of fixed sections from the E. scolopes ANG. (A) Cross-section of ANG tissue, showing many tubules interspersed between blood vessels (BV). Most tubules contained dense bacterial populations (T); however, some were empty (ET). (B) Closer inspection revealed that tubules have two distinct morphotypes, comprising a large coccoid bacterium (LCB) and a smaller bacillus-shaped bacterium (SBB) that were observed in tubules with differing epithelial morphologies, either a vacuole-rich epithelium (arrowhead) or dense epithelium (black arrow). (C) Tubule with brush border (BB) dominated by SBB with an epithelial morphology suggesting secretory cells (SEC). (D) SBB inhabiting tubules with a thick microvillar brush border (BB) and membrane-bound vesicles (MBV). (E) Hemocytes (HC) in the lumen of a tubule with a population of bacteria. (F) Tubule dominated by LCB. (G) Bacteria were also seen in the connective tissue (CT) outside the tubules; note the absence of a brush border. (H) Hemocytes were also seen with bacteria among the connective tissue. (I) Closer inspection of the LCB revealed many storage granules. (J and K) One SBB morphotype showed a nucleoid structure, while the other (K) showed many polyhydroxybutyrate-like granules (black arrow). Bars, 30 μm (A and B), 5 μm (C to H), and 1 μm (I to K).
16S diversity.
In order to identify members of the microbial community of the E. scolopes ANG, we constructed five 16S clone libraries from five sexually mature adult female squid. Sequences of 417 clones were binned by RFLP analysis. Of these, 96 full-length 16S sequences were analyzed and 25 identified chimeric sequences were removed. Analysis of these data showed that most (302/392; Table 1) clones belonged to the Alphaproteobacteria and that Phaeobacter was the most commonly observed genus (221/392). Other Alphaproteobacteria species belonged to genera within the Rhodobacterales, primarily of the Roseobacter clade (for example, Ruegeria, Labrenzia, and Pseudoruegeria). Twelve were from the genus Kordiimonas, and eight sequences were from the Rhizobiales. The next most common group of sequences (89/392) had greatest similarity to the sequences corresponding to members of the phylum Verrucomicrobia. These Verrucomicrobia sequences displayed only ∼90% identity to those from the Greengenes database, most likely due to the lack of characterized verrucomicrobial isolates. Only one sequence belonged to the Gammaproteobacteria and corresponded to the genus Shewanella.
The overall bacterial populations of ANGs from separate animals were similar, with two OTUs conserved across all five clone libraries, Phaeobacter and Verrucomicrobia (Table 2). Members of four other genera of the alphaproteobacteria (Ruegeria, Kordiimonas, Cohaesibacter, and Nautella) were conserved among the same four ANG libraries. Length of time spent in the mariculture facility did not seem to influence the microbial communities found in the ANG, as animals maintained for either 1 day (library ANG5) or 4 months (library ANG4) were found to have similar bacterial taxa (Table 2).
Table 2.
Operational taxonomic units within five ANG 16S clone librariesa
| Phylotype | No. of OTUs |
||||
|---|---|---|---|---|---|
| ANG1 | ANG2 | ANG3 | ANG4 | ANG5 | |
| Alphaproteobacteria | |||||
| Rhodobacterales | |||||
| Phaeobacter | 20 | 49 | 78 | 40 | 26 |
| Ruegeria | 9 | 6 | 0 | 6 | 4 |
| Nautella | 2 | 4 | 0 | 5 | 1 |
| Labrenzia | 0 | 6 | 0 | 1 | 0 |
| Pseudoruegeria | 0 | 1 | 0 | 0 | 0 |
| Oceanicola | 0 | 1 | 0 | 0 | 0 |
| Marinovum | 0 | 0 | 2 | 0 | 0 |
| Salipiger | 0 | 0 | 4 | 0 | 0 |
| Rhizobiales | |||||
| Cohaesibacter | 0 | 1 | 0 | 2 | 3 |
| Mesorhizobium | 0 | 0 | 2 | 0 | 0 |
| Kordiimonadales | |||||
| Kordiimonas | 1 | 4 | 0 | 3 | 4 |
| Verrucomicrobia | 37 | 12 | 3 | 25 | 29 |
| Gammaproteobacteria | |||||
| Shewanella | 0 | 1 | 0 | 0 | 0 |
| Total no. of clones | 69 | 85 | 89 | 82 | 67 |
A total of 392 clones were binned into taxonomic groups by RFLP analysis. The full-length 16S rRNA gene of 71 clones from ANG1, ANG2, and ANG3 was analyzed using the Greengenes database (11) after chimeric sequence removal (Materials and Methods).
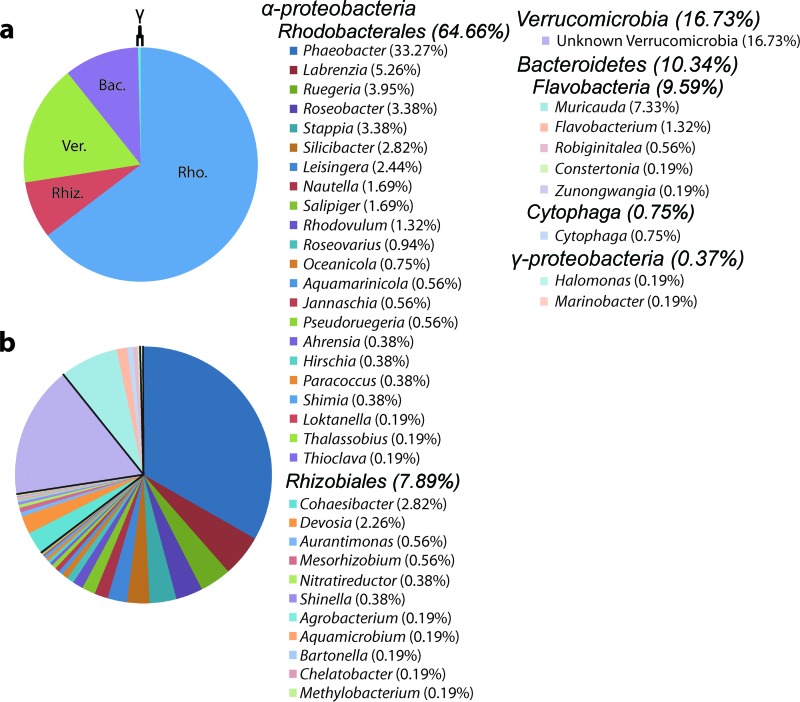
In addition to the RFLP and sequence data from the five clone libraries, 16S gene fragments from the 454 metagenome were also analyzed. A total of 532 genomic fragments with at least a 200-bp alignment to a reference 16S sequence in the RDP database were used for this analysis. The taxonomies of these 16S sequences were similar to those identified in the 16S libraries; species of Rhodobacterales, Rhizobiales, and Verrucomicrobia were dominant (Fig. 3). 72.55% of the 16S sequences belonged to the Alphaproteobacteria, and the most common genus was Phaeobacter (177/532). Members of the order Rhodobacterales was the most common, with Verrucomicrobia being the second largest taxonomic contingent overall (89/532). Members of the Rhizobiales and the phylum Bacteroidetes, which were not seen in our 16S library, accounted for less than 20% of the 16S sequences. Only 2 of the 532 16S sequences were from the Gammaproteobacteria.
Fig 3.
Taxonomic analysis of 454 metagenomic data. Higher (a) and lower (b) taxonomy of 532 16S gene fragments found within the ANG metagenome. Members of the Alphaproteobacteria, comprising mostly Rhodobacterales (Rho.), accounted for the vast majority of sequences (72.55%). Members of the Verrucomicrobia (Ver.) were the next largest contingent (16.73%), and members of the Bacteroidetes (Bac.), mostly from the Flavobacteria, made up a smaller fraction (10.34%). Less than 1% of 16S fragments belonged to the Gammaproteobacteria. The presence of a single 16S sequence is represented by “0.19%.” Rhiz., Rhizobiales.
FISH.
Observations from electron microscopy suggested that different morphotypes (SBB or LCB) dominated individual tubules within the ANG (Fig. 2). To test whether this could have been due to different phylogenetic groups occupying separate tubules, fluorescent in situ hybridization (FISH) was used to visualize dominant bacterial taxa within the ANG. Ribosomal FISH revealed that most tubules within the ANG contained a specific bacterial group (Fig. 4). Staining with an FITC-labeled Roseobacter clade-specific probe and a cocktail of the eubacterial probes Eub338 and Eub338III (Table 1) showed that the Roseobacter clade probe hybridized to the majority of the bacteria of one tubule (Fig. 4a). Similarly, using both the Verrucomicrobia- and Alphaproteobacteria-specific probes, tubules were dominated by only one of the two fluorescent signals (Fig. 4b), suggesting bacterial partitioning among the ANG tubules. The presence of members of the Cytophaga-Flavobacteria-Bacteroidetes (CFB) that were identified in the 454 metagenomic sequencing analysis was also confirmed with this technique (Fig. 4c).
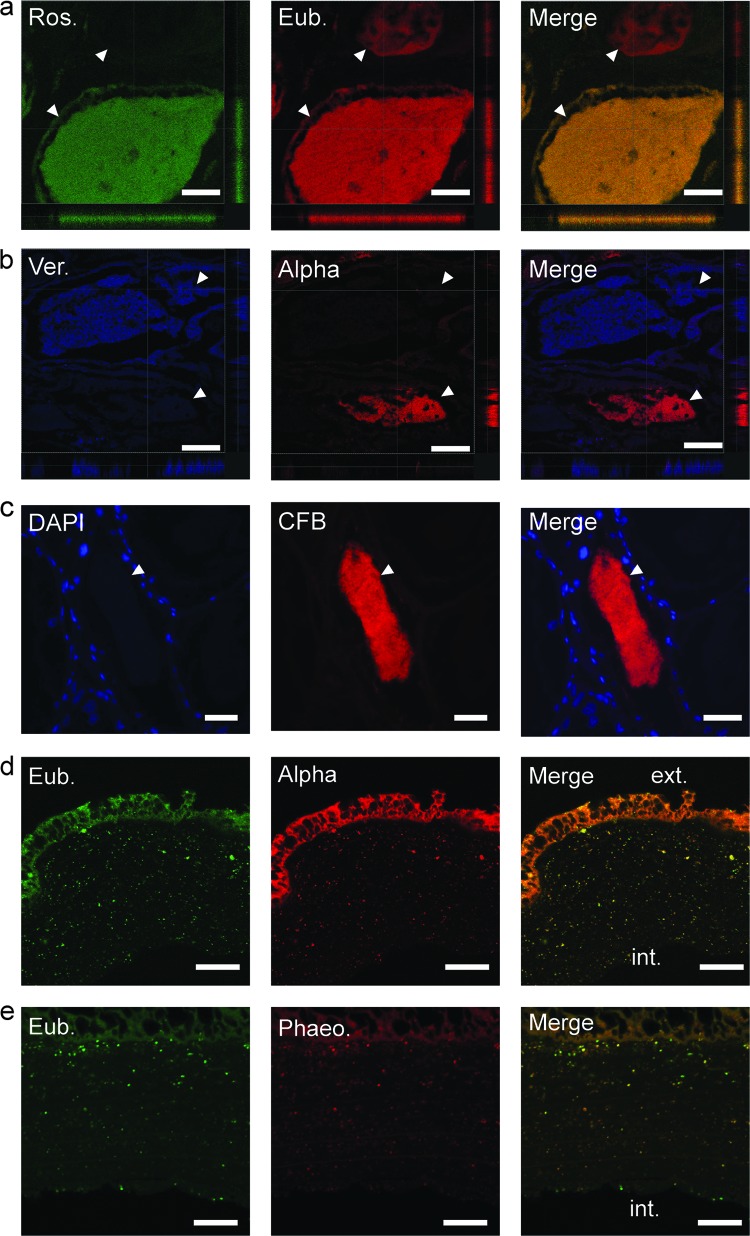
Fig 4.
Fluorescent in situ hybridization of fixed ANG paraffin-embedded sections. (a) 16S FISH with FITC-conjugated Roseobacter (Ros.) probe (green) and CY3-conjugated eubacterial (Eub.) cocktail (red). (b) Cy3-conjugated Alphaproteobacteria (Alpha; red) and Cy5-conjugated Verrucomicrobia (Ver.; blue) probes were observed dominating separate tubules, suggesting specificity and/or segregation of the bacterial populations. Axes denote positions of “slices” through confocal sectioning. (c) 16S FISH with Cy3-conjugated Cytophaga-Flavobacteria-Bacteroidetes probe (CFB; red) and DAPI staining (blue). (d) Hybridization with an FITC-conjugated eubacterial cocktail (green) and a Cy3-conjugated Alphaproteobacteria probe (red) revealed a population of bacteria within the jelly capsule of a freshly laid egg. (e) Hybridization with a Cy3-conjugated Phaeobacter (Phaeo.) probe confirmed that many of the bacteria in the jelly capsule were Phaeobacter species. The exterior (ext.) and interior (int.) of the capsule are labeled. White arrowheads indicate tubules. Bars, 30 μm (a), 50 μm (b), and 20 μm (d and e).
Applying FISH to the jelly capsule of freshly laid squid eggs also revealed a mixture of bacteria within the capsule, with an abundance of Alphaproteobacteria present, including Phaeobacter sp. (Fig. 4d and e). No bacterial cells were observed in direct contact with the developing embryo (not shown). These data suggest that bacteria from the ANG are deposited directly into host egg capsules.
DISCUSSION
We used a variety of microscopy and molecular methods to characterize the bacterial population of the ANG of E. scolopes, ultimately to understand its role in host reproduction. These analyses show that Alphaproteobacteria species from the Roseobacter clade within the Rhodobacterales are prevalent and that members of the genus Phaeobacter dominate the consortium, while other major constituents are members of the Rhizobiales, Verrucomicrobia, and Flavobacteria (Table 2 and Fig. 3). This bacterial consortium is contained within heteromorphic epithelium-lined tubules that are infiltrated by host hemocytes. Moreover, FISH analyses confirmed that many tubules of the ANG are dominated by single taxonomic groups, suggesting niche specificity in this association (Fig. 4).
The dominance of Roseobacter clade members in the ANG of Euprymna scolopes is similar to what has been described for other cephalopod ANGs, including those of other squid (3, 39) and cuttlefish (16), by the use of 16S clone sequencing. Like that of Loligo pealei, the ANG of Euprymna has a large Alphaproteobacteria contingent, comprising Roseobacter clade members as well as members of the marine Rhizobiales. The E. scolopes ANG also has a Flavobacteria contingent, similar to observations made using the egg casings of L. pealei (3).
The presence of Verrucomicrobia and the lack of Gammaproteobacteria make the consortium in E. scolopes strikingly different from the ANG consortia previously described for other cephalopods. Members of the Verrucomicrobia have been detected in relatively few host/microbe associations (38, 44, 47, 51), and the major presence of this group in the squid ANG represents a potentially novel symbiotic role for this phylum. Less than 1% of 16S genes from our clone libraries and from the 454 metagenome belonged to the Gammaproteobacteria. This is surprising for a number of reasons. In L. peali, it has been estimated that 5% of the bacterial population of the ANG is made up of this group (3). Furthermore, E. scolopes has a binary association with the bioluminescent Gammaproteobacterium Vibrio fischeri (29, 30, 35). Given that the host expels 106 to 109 symbionts from its light organ as part of a daily rhythm (7, 36), the close proximity of the two organs, and that the ANG consortium is likely environmentally transmitted (see below), it is surprising that V. fischeri was not detected in our analyses.
Previous work has shown that the bacterial consortia within cephalopod ANGs are likely established by horizontal/environmental transmission (20). In that work, Kaufman et al. examined development of the ANG in Loligo opalescens and found that the organ develops 11 weeks after hatching and that colonization is likely due to horizontal/environmental transmission. This conclusion is also supported by the observation that the nearest relatives of ANG isolates from L. pealei are environmental strains (3). The ANG of E. scolopes is absent at hatching, and females tend to reach sexual maturity within 60 days (17). Therefore, horizontal transmission of the E. scolopes ANG consortium is also probable. Field-caught animals at different stages of development of the ANG symbiosis will be used for future analyses of both the organ and the microbial community. Current efforts are also under way to rear animals to sexual maturity in our laboratory.
The data presented here suggest that establishment and maintenance of the bacterial consortium may be an intricate process, as both electron microscopy and FISH analyses showed bacterial partitioning among the ANG tubules (Fig. 2 and 4). Electron microscopy revealed distinct morphotypes (LCB and SBB) prevalent in each tubule, and FISH revealed that members of the Rhodobacterales, Cytophaga-Flavobacteria-Bacteroidetes, and Verrucomicrobia dominated separate tubules (Fig. 2 and 4). Other studies have noted dominant morphotypes (coccus and bacillus) within the lumina of cephalopod ANGs (3, 6). Electron micrographs from the cuttlefish Sepia officinalis show a coccoid bacterium with a morphology very similar to that of the granular, coccoid cells observed in the E. scolopes ANG (48). The data from this study suggest that the different bacterial morphotypes are different taxa occupying separate tubules. Bacterial morphology by itself is not a reliable taxonomic identifier; however, studies of the marine verrucomicrobium Coraliomargarita akajimensis revealed a morphology similar to the LCB morphotype observed in this study (52). The two epithelial morphologies of the ANG tubules are very distinct from one another, suggesting that each tubule fosters a unique microenvironment optimized to contain a specific bacterial taxon or that specific bacteria influence development of different epithelia. The mechanism(s) for establishing and maintaining bacterial tubule dominance is not yet known, but these bacterial groups may be adapted to specific niches or microenvironments within the ANG. Alternatively different taxa may dominate specific tubules during colonization due to a founder effect.
Just as carbon and energy sources influence bacterial diversity in digestive tracts, nutrition may play a role in the segregation of the bacteria within the ANG. While members of the Verrucomicrobia have not been thoroughly described, many have been shown to degrade polysaccharides such as mucin in the human gut (10) or fucoidan in the gut of a sea cucumber (45). The genome of Phaeobacter gallaeciensis ANG1, a dominant member of the ANG consortium in E. scolopes, reveals that it has many pathways for energy and carbon assimilation; however, it lacks enzymes to degrade polysaccharides, including chitinases, amylases, agarases, and α-l-fucosidase (8). Therefore, the host may provide different nutrients in different tubules, thereby enriching for certain bacteria. The presence of PHB-like granules in some cells (Fig. 2K) could be explained by nutrient restriction, as PHB improves survivability during starvation and/or stress tolerance in other systems (27, 41).
There are other clues that can be gathered from the host as to how a microenvironment can be created to foster dominance of specific bacteria. The light organ symbiosis between E. scolopes and V. fischeri has been studied in detail for more than 20 years, and previous studies have shown that the host and symbiont work in concert to create a microenvironment that selects for V. fischeri to the exclusion of nonsymbiotic bacteria (30, 35, 50). The stark differences in epithelial tissues in the ANG suggest that unique microenvironments exist between tubules. In the light organ, hemocytes, representing the sole cellular component of the host's innate immune system, have been implicated in establishing and maintaining specificity (22, 31, 36, 37). Hemocytes were also observed to infiltrate the lumina of the ANG tubules and were found to come in direct contact with the bacterial consortium (Fig. 2). Whether these hemocytes contribute to specificity in the ANG association remains to be determined, but we have isolated several ANG bacterial strains that are available to use in adhesion and phagocytosis assays. Future research should examine how components of the innate immune system as well as other host and symbiont factors may influence the development and maintenance of this association.
Despite numerous studies that have characterized the bacterial communities within cephalopod ANGs, its function is still unknown. The ANG may provide antimicrobial or antifouling compounds that protect the squid's eggs throughout their development (4, 14). The genome of Phaeobacter gallaeciensis ANG1 revealed no classical antibiotic synthesis pathways (8), but future analyses should take into account the genomes of the other ANG members and possible uncharacterized pathways of novel antimicrobial compounds. Two other P. gallaeciensis strains have shown the ability to inhibit fungal, bacterial, and/or algal growth (40, 46), and future studies should test whether removing the ANG consortium from female squid and/or their egg clutches influences fecundity and egg development. Currently, we have 14 other E. scolopes ANG rhodobacterial isolates in culture, and characterizing these strains in greater detail should shed light on the function of the ANG.
The female members of many squid species found worldwide harbor a consortium of bacteria within their ANG. Surprisingly, much of the composition of the microbial communities is the same (e.g., the dominance of Rhodobacterales), even though these hosts are found in very different environments with different physical and biological parameters (e.g., salinity, temperature, predators, and life histories). This trend suggests that these microbial consortia play similar roles in their squid hosts. Future experiments can utilize high-throughput sequencing techniques to reveal gene expression of the bacteria in the ANG and within the egg capsule. The results of this study lay the foundation for the development of E. scolopes as a model for studying a consortial symbiosis (ANG) and a binary symbiosis (light organ) in the same host.
ACKNOWLEDGMENTS
We thank Mark Martindale and the entire staff of the Kewalo Marine Laboratory and Ruth Gates and the Hawaii Institute of Marine Biology at the University of Hawaii for laboratory and aquarium space and assistance with animal collections. We thank Stephen Daniels and Marie Cantino of the Electron Microscopy Laboratory of the University of Connecticut for help with electron microscopy, Craig Obergfell and Rachel O'Neil of the Center for Applied Genetics and Technology for help with 454 sequencing, Pascal LaPierre of the Bioinformatics Facility for bioinformatics advice, and Carol Norris of the Cytometry and Confocal Microscopy Facility for assistance with confocal microscopy. We also thank Bethany Rader, Tyler Schleicher, and Corey Bunce for helpful comments on the manuscript.
This research was funded by NSF IOS-0958006 and the University of Connecticut Research Foundation to S.V.N., the Romano Graduate Student Fellowship to A.J.C., and NSF-NEAGEP to A.J.C. and J.S.
Footnotes
Published ahead of print 13 April 2012
REFERENCES
- 1. Amann RI, et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnold JM, Singley CT, Williams-Arnold LD. 1972. Embryonic development and post-hatching survival of the sepiolid squid Euprymna scolopes under laboratory conditions. Veliger 14:361–375 [Google Scholar]
- 3. Barbieri E, et al. 2001. Phylogenetic characterization of epibiotic bacteria in the accessory nidamental gland and egg capsules of the squid Loligo pealei (Cephalopoda: Loligindae). Environ. Microbiol. 3:151–167 [DOI] [PubMed] [Google Scholar]
- 4. Barbieri E, Barry K, Child A, Wainwright N. 1997. Antimicrobial activity in the microbial community of the accessory nidamental gland and egg cases of Loligo pealei (Cephalopoda: Loliginidae). Biol. Bull. 193:275–276 [DOI] [PubMed] [Google Scholar]
- 5. Benkenorff K, Davis AR, Bremner JB. 2001. Chemical defense in the egg masses of benthic invertebrates: an assessment of antibacterial activity in 39 mollusks and 4 polychaetes. J. Invertebr. Pathol. 78:109–118 [DOI] [PubMed] [Google Scholar]
- 6. Bloodgood RA. 1977. The squid accessory nidamental glad; ultrastructure and association with bacteria. Tissue Cell 9:197–208 [DOI] [PubMed] [Google Scholar]
- 7. Boettcher KJ, Ruby EG, McFall-Ngai MJ. 1996. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J. Comp. Physiol. 179:65–73 [Google Scholar]
- 8. Collins AJ, Nyholm SV. 2011. Draft genome of Phaeobacter gallaeciensis ANG1; a dominant member of the accessory nidamental gland of Euprymna scolopes. J. Bacteriol. 193:3397–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daims G, Brühl A, Amann R, Scheifer KH, Wagner M. 1999. The domain-specific probe Eub338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 10. Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54(Pt. 5):1469–1476 [DOI] [PubMed] [Google Scholar]
- 11. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fields WG. 1965. The structure, development, food relations, reproduction, and life history of squid Loligo opalescens Berry. Fish Bulletin 131. State of California Department of Fish and Game, Sacramento, CA [Google Scholar]
- 13. Giuliano L, De Domenico M, De Domenico E, Hofle MG, Yakimov MM. 1999. Identification of culturable oligotrophic bacteria within naturally occurring bacterioplankton communities of the Ligurian Sea by 16S rRNA sequencing. Microb. Ecol. 37:77–85 [DOI] [PubMed] [Google Scholar]
- 14. Gomathi P, Nair JR, Sherief PM. 2010. Antibacterial activity in the accessory nidamental gland extracts of the Indian squid, Loligo duvauceli Orbigny. Indian J. Mar. Sci. 39:100–104 [Google Scholar]
- 15. Gomez-Alvarez V, Teal TK, Schmidt TM. 2009. Systemic artifacts in metagenomes from complex microbial communities. ISME J. 3:1314–1317 [DOI] [PubMed] [Google Scholar]
- 16. Grigioni S, Boucher-Rodoni R, Demarta A, Tonolla M, Peduzzi R. 2000. Phylogenetic characterization of bacterial symbionts in the accessory nidamental glands of the sepioid Sepia officinalis (Cephalopoda: Decapoda). Mar. Biol. 136:217–222 [Google Scholar]
- 17. Hanlon RT, Claes MF, Ashcraft SE, Dunlap PV. 1997. Laboratory culture of the sepiolid squid Euprymna scolopes: a model system for bacteria-animal symbiosis. Biol. Bull. 192:364–374 [DOI] [PubMed] [Google Scholar]
- 18. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 19. Jensen TE, Sicko LM. 1971. Fine structure of poly-β-hydroxybutryic acid granules in blue-green alga, Chlorogloea fritschii. J. Bacteriol. 106:683–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaufman MR, Ikeda Y, Patton C, Van Dykhuizen G, Epel D. 1998. Bacterial symbionts colonize the accessory nidamental gland of the squid Loligo oplaescens via horizontal transmission. Biol. Bull. 194:36–43 [DOI] [PubMed] [Google Scholar]
- 21. Kikuchi Y, Graf J. 2007. Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl. Environ. Microbiol. 73:1984–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koropatnick TA, Kimbell JR, McFall-Ngai MJ. 2007. Responses of host hemocytes during the initiation of the squid-Vibrio symbiosis. Biol. Bull. 212:29–39 [DOI] [PubMed] [Google Scholar]
- 23. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrant E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY [Google Scholar]
- 24. Loy A, et al. 2008. probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity. Environ. Microbiol. 10:2894–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lum-Kong A, Hastings TS. 1992. The accessory nidamental glands of Loligo forbesi (Cephalopoda: Loliginidae): characterization of symbiotic bacteria and preliminary experiments to investigate factors controlling sexual maturation. J. Zool. 228:395–403 [Google Scholar]
- 26. Mandel MJ. 2010. Models and approaches to dissect host-symbiont specificity. Trends Microbiol. 18:504–511 [DOI] [PubMed] [Google Scholar]
- 27. Mandon K, et al. 1998. Poly-β-hydroxybutyrate turnover in Azorhizobium caulinodans is required for growth and affects nifA expression. J. Bacteriol. 180:5070–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manz W, Amann R, Ludwig W, Vancannyet M, Schleifer KH. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the cytophaga-flavobacter-bacteroidetes in the natural environment. Microbiology 142(Pt. 5):1097–1106 [DOI] [PubMed] [Google Scholar]
- 29. McFall-Ngai MJ. 2002. Unseen forces: the influence of bacteria on animal development. Dev. Biol. 242:1–14 [DOI] [PubMed] [Google Scholar]
- 30. McFall-Ngai MJ, Ruby EG. 1991. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254:1491–1494 [DOI] [PubMed] [Google Scholar]
- 31. McFall-Ngai MJ, Nyholm SV, Castillo MG. 2010. The role of the immune system in the initiation and persistence of the Euprymna scolopes-Vibrio fischeri symbiosis. Semin. Immunol. 22:48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meyer F, et al. 2008. The metagenomic RAST server—a public resource for automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Montgomery MK, McFall-Ngai MJ. 1993. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Bio. Bull. 184:296–308 [DOI] [PubMed] [Google Scholar]
- 34. Neef A. 1997. Anwendung der in situ Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Ph.D. thesis Technical University, Munich, Germany [Google Scholar]
- 35. Nyholm SV, McFall-Ngai MJ. 2004. The winnowing: establishing the squid-Vibrio symbiosis. Nat. Rev. Microbiol. 2:632–642 [DOI] [PubMed] [Google Scholar]
- 36. Nyholm SV, McFall-Ngai MJ. 1998. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol. Bull. 195:89–97 [DOI] [PubMed] [Google Scholar]
- 37. Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. 2009. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ. Microbiol. 11:483–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petroni G, Spring S, Scheifer KH, Verni F, Rosati G. 2000. Defensive extrusive ectosymbionts of Euplotidium (Ciliophora) that contain microtubule-like structures are bacteria related to Verrucomicrobia. Proc. Natl. Acad. Sci. U. S. A. 97:1813–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pichon D, Gaia V, Norman MD, Boucher-Rodoni R. 2005. Phylogenetic diversity of epibiotic bacteria in the accessory nidamental glands of squids (Cephalopoda: Loliginidae and Idosepiidae). Mar. Biol. 147:1323–1332 [Google Scholar]
- 40. Rao D, Webb JS, Holmström C, Case R, Low A, Steinberg P, Kellenberg S. 2007. Low densities of epiphytic bacteria from marine alga Ulva australis inhibit settlement of fouling organisms. Appl. Environ. Microbiol. 73:7844–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ratcliff WC, Kadam SV, Denison RF. 2008. Poly-3-hydroxybutyrate (PHB) supports survival and reproduction in starving rhizobia. FEMS Microbiol. Ecol. 65:391–399 [DOI] [PubMed] [Google Scholar]
- 42. Reasoner DJ, Blannon JC, Geldreich EE. 1979. Rapid seven-hour fecal coliform test. Appl. Environ. Microbiol. 38:229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reynolds ES. 1963. The use of lead citrate at high H as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Romero-Pérez GA, Ominiski KH, McAllister TA, Krause DO. 2011. Effect of environmental factors and influence on bacterial communities in steers. Appl. Environ. Microbiol. 77:258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sakai T, Ishizuka K, Kato I. 2003. Isolation and characterization of a fucoidan-degrading marine bacterium. Mar. Biotechnol. (NY) 5:409–416 [DOI] [PubMed] [Google Scholar]
- 46. Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. 2011. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3:331–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vandekerckhove TT, Willems A, Gillis M, Coomans A. 2000. Occurrence of novel verrucomicrobial species, endosymbiotic and associated with parthenogenesis in Xiphinema americanum-group species (Nematoda, Longidoridae). Int. J. Syst. Evol. Microbiol. 50:2197–2205 [DOI] [PubMed] [Google Scholar]
- 48. Van den Branden C, Richard A, LeMaire J, Decleir W. 1978. La glande nidamentaire accessoire de Sepia officinalis L.: analyses biochimiques des pigments des bacteries symbiotiques. Ann. Soc. Zool. Belg. 108:123–139 [Google Scholar]
- 49. Wallner G, Amann R, Beisker W. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136–143 [DOI] [PubMed] [Google Scholar]
- 50. Wier AM, et al. 2010. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc. Natl. Acad. Sci. U. S. A. 107:2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yildirim S, et al. 2010. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One 5:e13963 doi:10.1371/journal.pone.0013963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoon J, et al. 2007. Coraliomargarita akajimensis gen. nov., sp. nov., a novel member of the phylum ‘Verrucomicrobia’ isolated from seawater in Japan. Int. J. Syst. Evol. Microbiol. 57(Pt. 5):959–963 [DOI] [PubMed] [Google Scholar]