Abstract
A total of 442 Listeria isolates, including 234 Listeria seeligeri, 80 L. monocytogenes, 74 L. welshimeri, 50 L. innocua, and 4 L. marthii isolates, were obtained from 1,805 soil, water, and other environmental samples collected over 2 years from four urban areas and four areas representing natural environments. Listeria spp. showed similar prevalences in samples from natural (23.4%) and urban (22.3%) environments. While L. seeligeri and L. welshimeri were significantly associated with natural environments (P ≤ 0.0001), L. innocua and L. monocytogenes were significantly associated with urban environments (P ≤ 0.0001). Sequencing of sigB for all isolates revealed 67 allelic types with a higher level of allelic diversity among isolates from urban environments. Some Listeria spp. and sigB allelic types showed significant associations with specific urban and natural areas. Nearest-neighbor analyses also showed that certain Listeria spp. and sigB allelic types were spatially clustered within both natural and urban environments, and there was evidence that these species and allelic types persisted over time in specific areas. Our data show that members of the genus Listeria not only are common in urban and natural environments but also show species- and subtype-specific associations with different environments and areas. This indicates that Listeria species and subtypes within these species may show distinct ecological preferences, which suggests (i) that molecular source-tracking approaches can be developed for Listeria and (ii) that detection of some Listeria species may not be a good indicator for L. monocytogenes.
INTRODUCTION
Members of the genus Listeria have traditionally been classified into three typically hemolytic species (Listeria monocytogenes, L. ivanovii, and L. seeligeri) and two typically nonhemolytic species (L. innocua and L. welshimeri) (59). While L. seeligeri is considered nonpathogenic, it includes both hemolytic and nonhemolytic isolates (15, 67), with hemolytic isolates of this species containing a homologue of the main virulence gene cluster (i.e., the prfA cluster), which carries key virulence genes in L. monocytogenes and L. ivanovii (21). An additional nonhemolytic species, Listeria grayi, has not been formally excluded as a member of the genus Listeria but has been shown to be very different from the other Listeria spp. (4, 59, 65). Although proposed at one time to represent a new genus, Murraya (61, 62), L. grayi is currently considered a Listeria species (11). Two new nonhemolytic Listeria species (i.e., Listeria rocourtii and L. marthii) were reported in 2010 (24, 38). Due to the importance of L. monocytogenes as a human food-borne and animal pathogen, there has been considerable effort to understand the epidemiology and distribution of L. monocytogenes and other Listeria spp. in human and animal disease, foods, and food-processing plants. Only limited information is available, though, on the occurrence of different Listeria spp. outside food processing plants and in natural environments. Many have demonstrated that Listeria spp. can be isolated from various different environments, including soil, vegetation, surface water, sewage, animal feeds, farm environments, and food-processing environments (58). Most studies on Listeria prevalence in the natural environment have focused on farm environments and associated croplands (8, 16, 30, 35, 43, 45), and only limited data on Listeria prevalence in nonagricultural environments (usually urban and suburban environments) are available (28, 29, 40, 47, 48, 71). A number of reports indicate a fairly high prevalence of Listeria spp. (often >20%) in various environments (58). For example, in 1975 Weis and Seeliger (69) reported Listeria species prevalences in vegetation samples ranging from 9.7 to 44% for samples from agricultural areas and from 21.3 to 23.1% for samples from nonagricultural areas. The same study reported even higher prevalences of Listeria spp. in soil samples, ranging from 8.7 to 51.4% for agricultural sites and from 15.2 to 43.2% for nonagricultural sites. Primarily due to subsequent changes in the taxonomy of Listeria (51), many of the earlier studies (e.g., reference 69) did not include reliable information on the diversity of Listeria spp. present in different environments. Some smaller, more-recent studies indicate considerable Listeria species diversity in samples collected from various environments. For example, a survey of urban environments in the United Kingdom showed that L. ivanovii and L. seeligeri represented the most-common Listeria spp. isolated from soil samples (40). Studies of surface water samples in different countries have identified L seeligeri, L. innocua, L. welshimeri, L. ivanovii, L. monocytogenes, and L. grayi (2, 19), with L. seeligeri (19) and L. monocytogenes reported as the most-prevalent species. In contrast, most studies on foods and food-processing environments seem to find L. monocytogenes and L. innocua as the most prevalent Listeria spp. (40).
Even though different studies have provided evidence that Listeria spp. are broadly distributed through the natural environment, our understanding of the ecology and reservoirs of Listeria species and L. monocytogenes is fairly limited. Even though molecular subtyping and characterization methods are now commonly used to characterize Listeria isolates (58), studies that include subtyping methods to characterize the ecology of Listeria spp. outside farm and food-processing environments are limited. Many studies on the distribution of Listeria in natural environments were actually conducted before molecular subtyping methods were available. Serotyping was most often used for strain discrimination in these studies but has been shown to have low discriminatory power and does not provide for reliable species-level identification (25). This study was designed to provide a better understanding of the occurrence of the genus Listeria outside food-processing and production and farm systems. A better understanding of the ecology of this genus not only will provide the basis for understanding the population genetics and natural history of closely related pathogenic and nonpathogenic Listeria species but is also needed to critically evaluate the validity of using the presence of any Listeria species as an indicator for L. monocytogenes contamination.
MATERIALS AND METHODS
Sample collection.
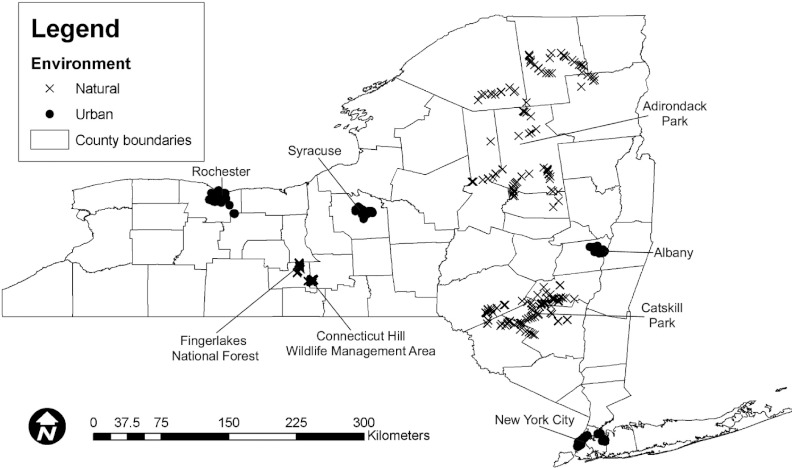
Samples were collected over 2 years (2001 and 2002) from multiple locations in four urban areas (Albany, New York City, Syracuse, and Rochester, NY) (Fig. 1) and four areas representing natural environments (Adirondack Park, Catskill Park, Connecticut Hill Wildlife Management Area [CT Hill], and Finger Lakes National Forest [FLNF]) (Fig. 1). The overall sampling scheme thus included two types of environment (which we designated “urban” and “natural”), four different areas for each type of environment, and multiple sampling locations within each area. While some samples collected in urban areas could also be considered “natural,” we used this designation, as the rural natural environments sampled represented undeveloped areas that are relatively undisturbed by human activity (i.e., state parks and wildlife management areas).
Fig 1.
Distribution of sample locations across New York State. The geographic distribution of all locations sampled in four urban and four natural areas across New York State in 2001 and 2002 is shown. Northerly direction and distance scale (km) as indicated.
In 2001, approximately even numbers of samples from urban (n = 295) and natural (n = 304) environments were obtained through 2 or 3 visits per area throughout spring, summer, and autumn. In 2002, every area was sampled once in each season (spring, summer, and autumn, yielding 594 and 612 samples from urban and natural areas, respectively). Samples were collected from different locations, and no exact location was sampled twice.
For samples collected in 2001, geographic location data were retrospectively obtained by plotting the sample locations in TopoUSA 4.0 (Delorme, Yarmouth, ME). In 2002, geographic location data were collected at the time of sampling using a Garmin Emap (Garmin International, Inc., Olathe, KS) handheld global positioning system (GPS) receiver and imported electronically into GPS Utility 4.04 (http://www.gpsu.co.uk). Geographic location data for each sample site were checked for accuracy by importing latitude and longitude coordinates into TopoUSA 4.0 and by comparing site locations with coordinate designations.
For areas representing natural environments, similar numbers of soil, vegetation, and surface water samples were collected. Soil and vegetation samples were collected from fields or forests; surface water samples were collected from standing water (pond/lake, swamp, or puddle) or flowing water (river/stream or runoff). Sample types collected in urban areas included soil and vegetation, surface water (standing or flowing), and sponge swipes of floors, sidewalks, and human contact surfaces (e.g., automated teller machines, benches, door handles, trash cans, mailboxes, parking meters, public telephones, picnic tables, railings, and vending machines). Soil and vegetation samples were taken from parks and playgrounds, surface water samples were taken from all available sources, and environmental sponges were taken from downtown areas and shopping malls. For soil and vegetation samples, approximately 50 to 100 g of material was collected, and for water samples, approximately 600 ml was collected. Sponges were rehydrated with 10 ml of sterile neutralizing buffer (Hardy Diagnostics, Santa Maria, CA) before the sampling. Samples were aseptically collected into sterile Whirl-Pak bags (Nasco, Fort Atkinson, WI), using sterile gloves and/or presterilized disposable plastic spatulas or scoops, and held on wet ice up to 24 h before culture.
Isolation of Listeria.
Listeria spp. and L. monocytogenes were isolated using selective enrichment in Listeria enrichment broth (LEB; Difco, Sparks, MD), followed by plating on Oxford medium (OX; Difco). Oxford agar was chosen because it is one of the selective agars specified by the U.S. FDA for the isolation of Listeria from foods. A 25-g aliquot of soil or vegetation was added to 225 ml of LEB in a Stomacher bag (Seward, Ltd., Norfolk, United Kingdom) and either mixed well by hand or mechanically stomached (model 400 Seward stomacher; Seward, Ltd.) for 2 min. For water samples, approximately 500 ml of each sample was filtered through at least three 150-ml Nalgene analytical filter (0.45-μm) units (Nalgene, Rochester, NY); some samples with high solid-content levels required more than three filters to achieve a filtered volume of 500 ml. Filters were subsequently placed into 100 ml of LEB and stomached for 2 min. For environmental sponges, 90 ml of LEB was added to each sponge, followed by manual mixing. After 24 and 48 h of incubation at 30°C, 100 μl of LEB was streaked onto OX as described previously (64). After incubation for 48 h at 30°C, up to four Listeria-like colonies (black, greenish, or gray colonies with esculin hydrolysis) were subcultured from OX to L. monocytogenes plating medium (LMPM; Biosynth International, Inc., Naperville, IL), a plating medium that differentiates L. monocytogenes and L. ivanovii (which appear as blue colonies due to phosphatidylinositol-specific phospholipase C activity) from other Listeria spp. (white colonies) (50). LMPM plates were incubated for up to 48 h at 37°C. Individual colonies on LMPM were classified as putative L. monocytogenes/L. ivanovii (blue), putative other Listeria spp. (white), or non-Listeria (atypical morphology and/or color).
Screening, confirmation, and phenotypic characterization.
Putative L. monocytogenes/L. ivanovii isolates were confirmed as L. monocytogenes using an L. monocytogenes-specific PCR assay targeting hly (72); this assay does not amplify L. seeligeri or L. ivanovii hly. In 2001, all other putative Listeria isolates (i.e., white colonies on LMPM) were identified to the species level using Gram-stain, motility at 25°C, oxidase, catalase, and hemolysis production and API-Listeria test strips (3). Since analysis of the putative Listeria isolates from 2001 showed that none of the nonmotile isolates represented Listeria, we concluded that the motility test is an appropriate screening tool for eliminating isolates that are not Listeria. Putative Listeria isolates from 2002 were thus screened using the motility test only and subsequently identified to the species level and subtyped using sigB sequencing as described below.
Gene sequencing and phylogenetic analysis.
All Listeria isolates were characterized by PCR amplification and sequencing of the partial open reading frame (ORF) of the stress response gene sigB. In addition, all isolates collected in 2001 were characterized by PCR amplification and sequencing of the housekeeping gene gap. PCR amplification and sequencing of sigB and gap were performed as described previously (46); for both genes, PCR products were directly sequenced in both directions. After sequences were assembled and proofread using Seqman (DNAStar, Madison, WI), they were aligned in Megalign (DNAStar) using the ClustalW algorithm and trimmed to consistent length for analysis. Allelic types (ATs) were assigned using DNAsp 4.06 (http://www.ub.es/dnasp); two gene sequences were classified as different allelic types if they differed by at least 1 nucleotide.
Phylogenetic analysis for sigB and gap allelic types was performed using a single isolate representing each unique allelic type. Phylogenetic trees were created in PAUP* (63) using the neighbor-joining (NJ) method (17). NJ trees were rooted with homologous gene sequences from Bacillus subtilis (http://genolist.pasteur.fr/SubtiList/) and bootstrapped for 2,000 replicates.
Simpson's index of discrimination.
Simpson's index of diversity and 95% confidence intervals were calculated as previously described (27, 32).
Categorical analysis.
Chi-square tests or a Fisher exact tests (if expected values were less than 5) were used to evaluate associations between Listeria spp. or sigB allelic types (representing subtypes within the different Listeria species) and (i) environmental source (urban or natural), (ii) individual areas (Albany, New York City, Syracuse, Rochester, Adirondacks, Catskills, CT Hill, or FLNF), (iii) specific sample sources (soil, vegetation, or surface water), (iv) and/or season (spring, summer, or fall). Since these tests determine whether the prevalences of different Listeria spp. differed among environments or sites, we used the total number of samples as a denominator (rather than the total number of isolates). Since one L. monocytogenes isolate and one other Listeria species isolate were identified in 30 samples, the overall chi-square analysis included two observations for each of these 30 samples. Thus, our denominator for each overall contingency table was 1,835 observations, while for individual 2-by-2 tables our denominator was the actual number of samples (n = 1,805).
All categorical analyses were performed using SAS 9.1 (SAS Institute, Inc., Cary, NC). P values of ≤0.05 were considered statistically significant and were not adjusted for the fact that multiple comparisons were made. Due to the large number of associations that were tested, it could be contended that the probability of a type 1 error was inflated and that the significance threshold should be adjusted. While we provide observed P values to avoid missing possible associations (caused by a very conservative P value), readers can evaluate the significance levels according to their preferred criteria (53).
Spatial analysis.
GPS data for all samples and isolates were imported into ArcGIS 9 (ESRI, Redland, CA) for spatial analyses. Nearest-neighbor analysis (68) as implemented in CrimeStat 2.0 (http://www.nedlevine.com/nedlevine17.htm) was used to test for spatial clustering of Listeria spp. and sigB allelic types that were significantly associated with specific sampling areas. Nearest-neighbor analysis provides an approximation as to whether points are more clustered or dispersed than would be expected by chance (68). Typically, this type of analysis compares the average observed distance of the nearest neighbor to a random distance by calculating the nearest-neighbor index (NNI), which is obtained by dividing the average observed nearest-neighbor distance by the expected random distance (39). To perform spatial clustering analysis, site-specific maximal dispersal boundaries were used to standardize comparisons to the same geographic area around each site. For each of the 8 sampling areas, a rectangular border was drawn around the maximum point dispersion in each geographic direction to approximate the size of a given area. Spatial coordinates for the sample locations that yielded the species or allelic type of interest in a specific site were subjected to nearest-neighbor analysis. In addition, equal numbers of randomly selected sample locations (chosen from all sample locations within a given site using SPSS 13.0 [SPSS, Inc., Chicago, IL]) were also subjected to the same nearest-neighbor analysis. Selection and nearest-neighbor analysis of random points were repeated 10 times, and the average mean nearest-neighbor distance (MNND) and its standard deviation were calculated. This random MNND was then compared using a 1-sample t test to the actual mean nearest-neighbor distance; P values of ≤0.05 were considered significant. If the observed MNND was significantly lower than the MNND for the randomly sampled locations, we concluded that there was evidence for clustering of the species or allelic types of interest.
Isolate and data curation.
All Listeria isolates were frozen at −80°C in brain heart infusion (BHI; Difco) broth containing 15% glycerol; all isolate characterization, including subtyping, was performed within <2 years after isolation. Isolate source information and subtyping data from this study are freely available through the Pathogen Tracker 2.0 database (http://www.pathogentracker.net).
RESULTS
Isolation and identification of Listeria spp. and L. monocytogenes.
Putative Listeria isolates were obtained from 525 (29%) of the 1,805 samples collected in 2001 and 2002. These samples yielded a total of 563 putative Listeria isolates, including 485 isolates that were white on LMPM and 78 isolates that were blue on LMPM. All 78 isolates that were blue on LMPM were confirmed as L. monocytogenes by hly PCR (72). Among the 485 isolates that were initially white on LMPM (indicating Listeria spp. other than L. monocytogenes or L. ivanovii), 370 were motile at 25°C. The Listeria-specific sigB PCR assay (46) yielded PCR products for 362 of these isolates, all of which were confirmed as Listeria spp. by sequencing of the sigB PCR product. Two of the putative Listeria species isolates from 2001 were identified as L. monocytogenes. In summary, we isolated and confirmed a total of 80 L. monocytogenes isolates (78 that were initially blue on LMPM and 2 that were initially white) and 362 isolates representing other Listeria spp.
Species-level identification and sigB allelic-type characterization of Listeria isolates.
In order to rapidly identify isolates to the species level and subtype them, we characterized all 119 Listeria isolates collected in 2001 (including 18 L. monocytogenes) by sequencing of sigB and gap. Preliminary phylogenetic analysis of the 119 gap sequences revealed clear clustering of all L. seeligeri (8 allelic types) and L. welshimeri (2 allelic types) gap sequences (with 31 phylogenetically informative sites). L. innocua, L. monocytogenes, and L. marthii did not form monophyletic clusters, probably due to the low number of phylogenetically informative sites (n = 9) among the gap sequences for these species. sigB sequencing provided more-discriminatory subtyping (35 sigB versus 20 gap allelic types) (Table 1) and more-reliable species identification. We thus chose sigB sequencing as a molecular species-level identification and subtyping method (allowing for differentiation within species) for the remainder of this study. sigB sequencing has also recently been validated, in a multilocus sequence typing (MLST) study, as a reliable method for identification of Listeria isolates to the species level (14).
Table 1.
sigB and gap allelic-type diversity among a subset of 119 isolates used to validate a rapid method for identifying isolates of Listeriaa
| Species (no. of isolates) | No. of allelic types based on: |
|
|---|---|---|
| sigB | gap | |
| L. marthii (1) | 1 | 1 |
| L. innocua (9) | 8 | 2 |
| L. monocytogenes (18) | 3 | 7 |
| L. welshimeri (20) | 11 | 2 |
| L. seeligeri (71) | 12 | 8 |
| Total (119) | 35 | 20 |
The method used morphology on Listeria monocytogenes plating medium (LMPM), motility at 25°C, and sigB sequencing.
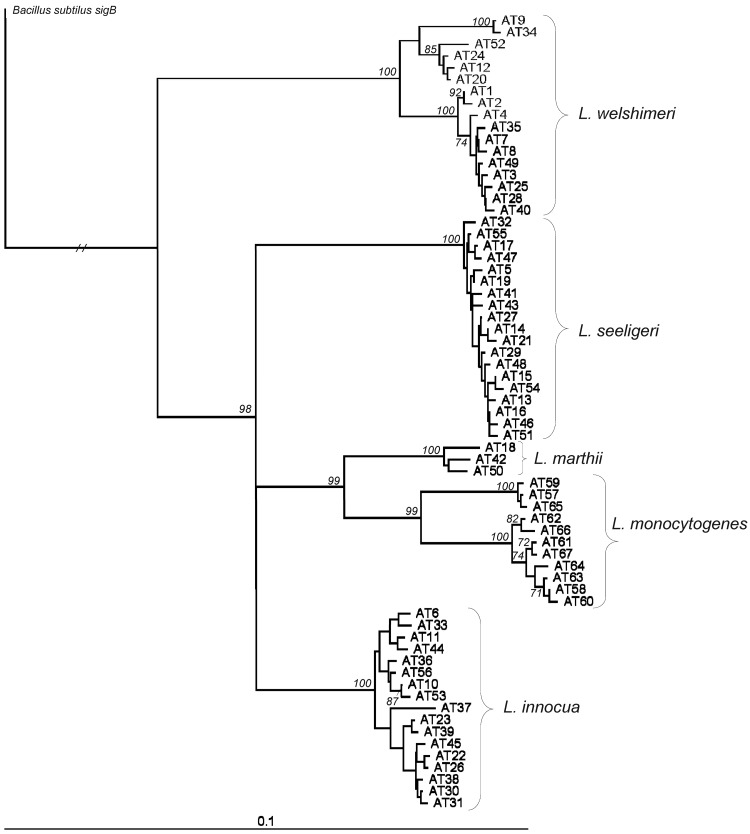
A neighbor-joining tree based on a 660-bp sigB sequence alignment, which represented all allelic types found among the 442 Listeria isolates characterized, revealed five well-supported sigB clusters (Fig. 2). Clustering of sigB sequences of the respective isolates was used to confirm and assign isolates to genospecies, including 234 L. seeligeri, 80 L. monocytogenes, 74 L. welshimeri, 50 L. innocua, and 4 L. marthii isolates (24). The 442 isolates represented 67 sigB allelic types (Table 2), including 19, 17, 17, 11, and 3 allelic types for L. welshimeri, L. seeligeri, L. innocua, L. monocytogenes, and L. marthii, respectively.
Fig 2.
Phylogenetic tree of unique sigB allelic types. Neighbor-joining tree rooted with Bacillus subtilis; branch length for B. subtilis collapsed (//) by a factor of 100 for display purposes. Bootstrap values were obtained from 2,000 replicates; bootstrap values of >70 are shown.
Table 2.
Distribution of sigB allelic types among 442 Listeria isolates from 2001 and 2002 by species, environment, and sampling area
| Species | sigB allelic type | No. of Listeria isolates from indicated sitea |
Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Natural |
Urban |
|||||||||||
| ADK | Catskills | CT Hill | FLNF | Subtotal | Albany | NYC | Rochester | Syracuse | Subtotal | |||
| L. marthii | 18 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| 42 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| 50 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Subtotal | 0 | 0 | 3 (+)* | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | |
| L. innocua | 6 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 2 | 7 | 7 |
| 10 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 2 | |
| 11 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 4 | 4 | |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 4 | 4 | |
| 23 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 1 | 6 | 6 | |
| 26 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 3 | 4 | |
| 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | |
| 31 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 2 | |
| 33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | |
| 36 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 0 | 5 | 5 | |
| 37 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 1 | 0 | 5 | 5 | |
| 38 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| 39 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 2 | |
| 44 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | |
| 45 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 3 | |
| 53 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| 56 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | |
| Subtotal | 0 | 0 | 0 | 1 | 1 | 14 | 19 (+)** | 10 | 6 (−)*** | 49 (+)*** | 50 | |
| L. monocytogenes | 57 | 4 | 4 | 1 | 2 | 11 | 9 | 6 | 4 | 11 | 30 (+)** | 41 |
| 58 | 0 | 0 | 1 | 0 | 1 | 11 (+)*** | 1 (−)* | 3 | 3 | 18 (+)*** | 19 | |
| 59 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 3 | |
| 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | |
| 61 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 3 | 3 | |
| 62 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | |
| 63 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| 64 | 0 | 0 | 0 | 0 | 0 | 5 | 2 | 0 | 0 | 7 | 7 | |
| 65 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| 66 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| 67 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 2 | |
| Subtotal | 4 | 5 | 2 | 2 | 13 | 27 (+)** | 15 | 7 (−)* | 18 | 67 (+)*** | 80 | |
| L. seeligeri | 1 | 8 | 6 (−)** | 35 (+)*** | 15 | 64 (+)*** | 5 | 1 | 4 | 4 | 14 | 78 |
| 2 | 0 (−)* | 1 (−)* | 4 | 11 (+)*** | 16 (+)** | 0 | 0 | 0 | 3 (+)* | 3 | 19 | |
| 3 | 3 | 11 | 6 | 8 | 28 (+)*** | 1 | 2 | 1 | 3 | 7 | 35 | |
| 4 | 0 | 2 | 2 | 4 | 8 | 0 | 0 | 2 | 2 | 4 | 12 | |
| 7 | 0 | 2 | 0 | 0 | 2 | 3 | 3 | 2 | 5 | 13 (+)** | 15 | |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 3 | |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 3 | 3 | |
| 12 | 3 | 4 | 1 (−)* | 10 (+)* | 18 | 8 (+)* | 2 | 2 | 3 | 15 | 33 | |
| 20 | 0 | 0 | 0 | 1 | 1 | 3 | 0 | 2 | 1 | 6 | 7 | |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 1 | 7 | 7 | |
| 25 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 3 | 3 | |
| 28 | 1 | 1 | 1 | 1 | 4 | 3 | 2 | 1 | 1 | 7 | 11 | |
| 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | |
| 35 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 3 | 3 | |
| 40 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| 49 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | |
| 52 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| Subtotal | 15 (−)** | 28 (−)* | 50 (+)*** | 51 (+)* | 144 (+)*** | 24 | 18 | 19 | 29 | 90 | 234 | |
| L. welshimeri | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | |
| 14 | 3 | 8 (+)** | 1 | 0 (−)* | 12 (+)*** | 0 | 0 | 0 | 0 | 0 | 12 | |
| 15 | 2 | 3 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 5 | |
| 16 | 2 | 5 | 3 | 0 (−)* | 10 | 1 | 1 | 1 | 0 | 3 | 13 | |
| 17 | 0 | 2 | 1 | 2 | 5 | 0 | 0 | 0 | 0 | 0 | 5 | |
| 19 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | |
| 21 | 0 | 8 (+)*** | 0 | 1 | 9 (+)* | 0 | 0 | 0 | 2 | 2 | 11 | |
| 27 | 1 | 1 | 2 | 0 | 4 | 3 | 1 | 1 | 0 | 5 | 9 | |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | |
| 32 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | |
| 41 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 2 | |
| 43 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 2 | |
| 46 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | |
| 47 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | |
| 48 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | |
| 51 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| 54 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| 55 | 0 | 3 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 1 | 4 | |
| Subtotal | 9 | 32 (+)*** | 7 | 4 (−)** | 52 (+)*** | 9 | 2 | 5 | 6 | 22 | 74 | |
| Total | 28 | 65 | 62 | 59 | 214 | 74 | 54 | 41 | 59 | 228 | 442 | |
Listeria species or sigB allelic-type prevalences that were significantly higher (+) or lower (−) for a specific environment (urban/pristine) or site as determined by categorical analyses were marked “*” (P ≤ 0.05), “**” (P ≤ 0.005), or “***” (P ≤ 0.0005). ADK, Adirondacks; CTHill, Connecticut Hill Wildlife Management Area; FLNF, Finger Lakes National Forest; and NYC, New York City.
Prevalence of Listeria species in natural and urban environments.
Among the 907 and 898 samples collected, over 2 years, from urban and natural environments, 23.4% and 22.3% were positive for Listeria, respectively (Table 3; see also Tables S1 and S2 in the supplemental material). Among the 412 Listeria-positive samples, 30 samples (2 and 28 from natural and urban areas, respectively) allowed for isolation of L. monocytogenes as well as another Listeria species; the 412 positive samples thus yielded 442 Listeria isolates (Tables 2 and 3). An initial overall 2-by-6 chi-square test clearly showed that the different Listeria spp. were not randomly distributed among urban and natural environments (P ≤ 0.0001) (Table 2). Subsequent 2-by-2 chi-square tests showed that L. seeligeri and L. welshimeri were significantly more common (P ≤ 0.0001) in natural environments, while L. monocytogenes and L. innocua were significantly more common (P ≤ 0.0001) in urban environments (Table 2).
Table 3.
Distribution of Listeria-positive samples by species, environment, and sampling area among 1,805 environmental samples obtained in 2001 and 2002
| Environment and area | Total no. of samples | No. of negative samples | No. of samples positive for indicated Listeria speciesa |
Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Only 1 species |
>1 species |
|||||||||||||
| LMa | LI | LM | LS | LW | Subtotal | LM+LMa | LM+LI | LM+LS | LM+LW | Subtotal | ||||
| Natural | ||||||||||||||
| Adirondacks | 187 | 159 | 0 | 0 | 4 | 15 | 9 | 28 | 0 | 0 | 0 | 0 | 0 | 28 |
| Catskills | 253 | 189 | 0 | 0 | 4 | 28 | 31 | 63 | 0 | 0 | 0 | 1 | 1 | 64 |
| CT Hill | 208 | 147 | 3 | 0 | 1 | 49 | 7 | 60 | 0 | 0 | 1 | 0 | 1 | 61 |
| Finger Lakes | 259 | 200 | 1 | 1 | 2 | 51 | 4 | 59 | 0 | 0 | 0 | 0 | 0 | 59 |
| Subtotal | 907 | 695 | 4 | 1 | 11 | 143 | 51 | 210 | 0 | 0 | 1 | 1 | 2 | 212 |
| Urban | ||||||||||||||
| Albany | 214 | 150 | 0 | 13 | 17 | 17 | 7 | 54 | 0 | 1 | 7 | 2 | 10 | 64 |
| New York City | 204 | 157 | 0 | 14 | 8 | 17 | 1 | 40 | 0 | 5 | 1 | 1 | 7 | 47 |
| Rochester | 207 | 171 | 0 | 10 | 2 | 15 | 4 | 31 | 0 | 0 | 4 | 1 | 5 | 36 |
| Syracuse | 273 | 220 | 0 | 4 | 12 | 25 | 6 | 47 | 0 | 2 | 4 | 0 | 6 | 53 |
| Subtotal | 898 | 698 | 0 | 41 | 39 | 74 | 18 | 172 | 0 | 8 | 16 | 4 | 28 | 200 |
| Total | 1,805 | 1,393 | 4 | 42 | 50 | 217 | 69 | 382 | 0 | 8 | 17 | 5 | 30 | 412 |
LMa, L. marthii; LI, L. innocua; LM, L. monocytogenes; LS, L. seeligeri; and LW, L. welshimeri. Detection of L. monocytogenes as well as another Listeria species was possible, as L. monocytogenes can be differentiated from other Listeria spp. on LMPM; the presence of two non-L. monocytogenes Listeria spp. in the same sample would not be detected with the approach used here, as only a single non-L. monocytogenes Listeria species colony was further characterized for each sample.
Listeria species prevalence by sample type (soil, plant, and vegetation) and season.
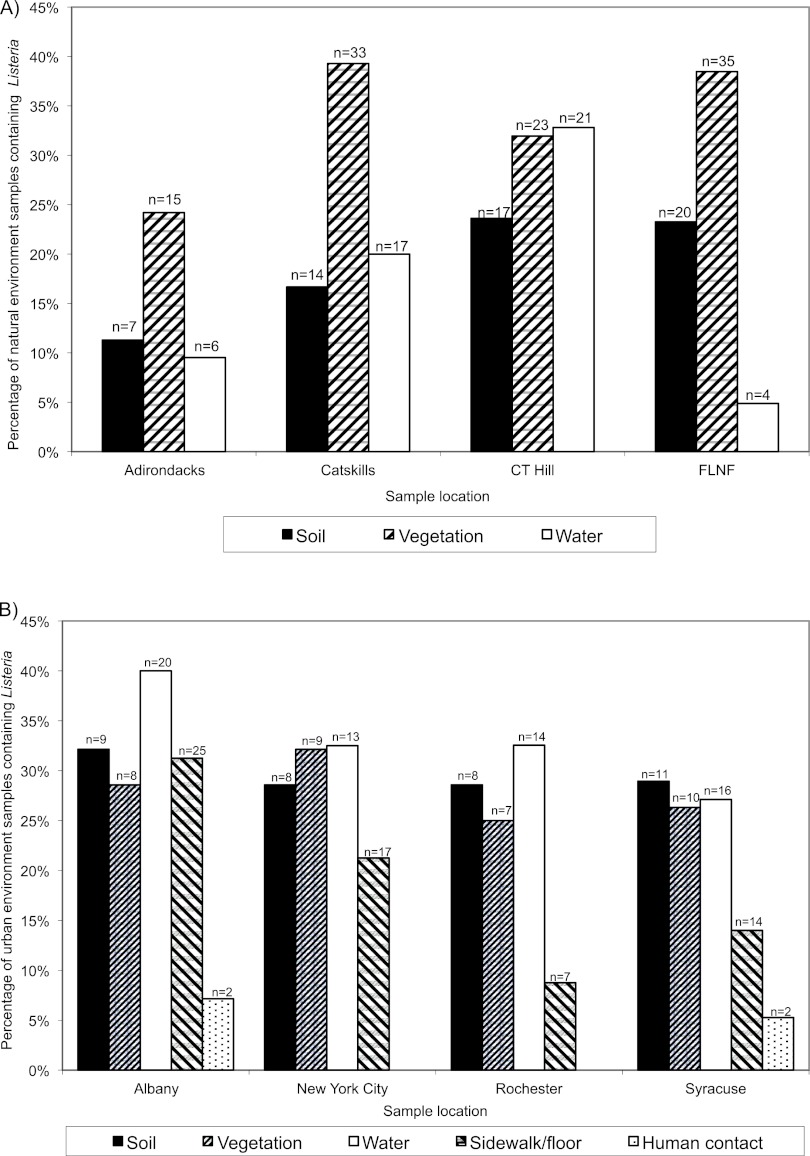
Overall Listeria prevalences in samples from natural environments ranged from 16% for surface water to 19% for soil and 34% for vegetation. Among samples from urban areas, Listeria prevalences were 33% for surface water, 30% for soil, 28% for vegetation, 19% for sidewalks/floors, and 3% for human contact surfaces. Listeria prevalences also differed considerably between sampling areas. For example, among natural areas, prevalences ranged from 5% in water from FLNF to 39% in vegetation from the Catskills, while among urban areas prevalences ranged from 0% (human contact surfaces in New York City and Rochester) to 40% (surface water in Albany) (Fig. 3).
Fig 3.
Listeria species isolation frequency by sample type. Prevalence of Listeria spp. (percentage of positive samples) in different sample types (soil, vegetation, water, sidewalk/floors, and human contact surfaces) collected from (A) natural and (B) urban environments (2001 and 2002).
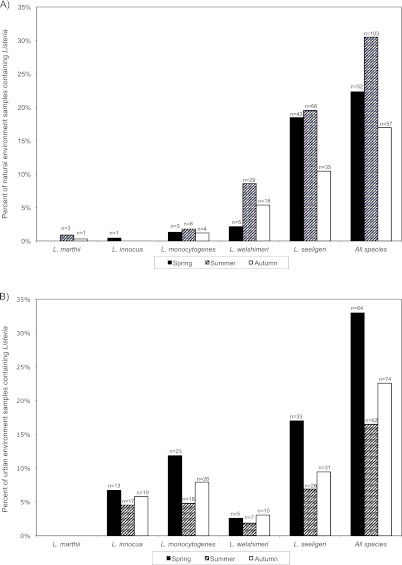
The seasonal Listeria prevalences ranged from 16% (urban environments, spring) to 33% (urban environments, summer) (Fig. 4). Overall Listeria prevalences differed significantly by season among samples from urban and natural environments (P = 0.000042 and P = 0.00017, respectively). In natural environments, prevalences for overall Listeria as well as for each individual Listeria species were highest in the summer, while prevalences for all Listeria species as well as for individual Listeria species in urban areas were lowest in the summer (Fig. 4).
Fig 4.
Listeria species isolation frequency by season. Seasonal prevalence (average of 2001 and 2002 data) of each Listeria species expressed as the percentage of positive samples from natural (A) and urban (B) environments with the corresponding number of positives samples indicated. A total of 233, 338, and 336 samples from natural environments and 194, 376, and 328 samples from urban environments were collected during spring, summer, and autumn, respectively.
Distribution of Listeria spp. by sample environment.
Separate 4-by-6 chi-square analyses for all urban and all natural environments showed that Listeria spp. were not independently distributed among individual natural (P ≤ 0.0001) and urban (P = 0.0015) sampling areas. Subsequent 2-by-2 chi-square analyses showed significant associations between specific Listeria spp. and certain sampling areas, including seven significant associations between natural sampling areas and specific Listeria spp. and four significant associations between urban areas and specific Listeria spp. (Table 2). For example, L. seeligeri was positively associated with CT Hill (P ≤ 0.0005) and FLNF (P ≤ 0.05), while L. welshimeri was positively associated with the Catskills (P ≤ 0.0005).
Spatial clustering of Listeria.
Listeria spp. that were associated with a specific sample site were also analyzed for evidence of spatial clustering using nearest-neighbor analysis (Table 4). Among the natural areas, we identified three instances of spatial clustering, including clustering of (i) L. welshimeri in the Catskills (Fig. 5), (ii) L. seeligeri in CT Hill, and (iii) L. seeligeri in FLNF (Table 4). Among the urban areas, we only identified evidence for spatial clustering of L. innocua in New York City (Table 4). All spatial clusters included isolation of the same species on at least four separate sampling dates, often several months apart. L. welshimeri was isolated from the Catskills on 5 out of 5 sampling dates, L. seeligeri was isolated from each CT Hill and FLNF on 5 out of 5 sampling dates, and L. innocua was isolated from New York City on 4 out of 6 sampling dates.
Table 4.
Spatial clustering, using nearest-neighbor analysis, of Listeria spp. and sigB allelic types associated with a specific area among isolates from 2001 and 2002
| Environment | Area | Size of area (km2) | No. of sample locations | Species (allelic type) | No. of positive samples | Actual MNNDa (km) | Random MNND (km) | Pb | Clustering |
|---|---|---|---|---|---|---|---|---|---|
| Natural | Catskills | 5,743 | 253 | L. welshimeri | 32 | 1.967 | 2.853 | 0.001 | Yes |
| L. welshimeri (AT14) | 8 | 5.787 | 8.887 | 0.007 | Yes | ||||
| L. welshimeri (AT21) | 8 | 3.185 | 7.448 | 0.003 | Yes | ||||
| CT Hill | 40 | 208 | L. marthii | 3 | 1.473 | 1.516 | NS | No | |
| L. seeligeri | 50 | 0.1357 | 0.1794 | 0.001 | Yes | ||||
| L. seeligeri (AT1) | 35 | 0.1822 | 0.2123 | 0.013 | Yes | ||||
| FLNF | 59 | 259 | L. seeligeri | 51 | 0.0711 | 0.1191 | ≤0.0001 | Yes | |
| L. seeligeri (AT2) | 11 | 0.4559 | 0.4963 | NS | No | ||||
| L. seeligeri (AT12) | 10 | 0.6856 | 0.5644 | NS | No | ||||
| Urban | Albany | 151 | 214 | L. monocytogenes | 27 | 0.4579 | 0.4675 | NS | No |
| L. monocytogenes (AT58) | 11 | 0.4863 | 0.749 | ≤0.0001 | Yes | ||||
| L. seeligeri (AT12) | 8 | 0.959 | 1.169 | NS | No | ||||
| NYC | 511 | 204 | L. innocua | 19 | 0.4868 | 1.013 | 0.003 | Yes | |
| Syracuse | 274 | 273 | L. seeligeri (AT2) | 3 | 1.968 | 3.087 | NS | No |
MNND, mean nearest-neighbor distance.
Value for a 1-sample t test comparing actual and random mean nearest-neighbor distances. NS, not statistically significant (P > 0.05).
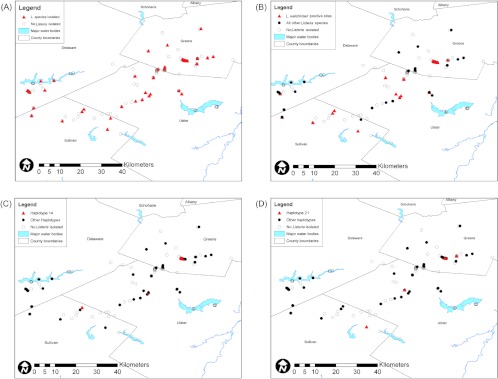
Fig 5.
Spatial clustering of L. welshimeri and L. welshimeri allelic types AT14 and AT21 in the Catskill Mountains. Spatial distribution is shown for all sampling locations and all sample locations positive for Listeria spp. (A), sample locations positive for L. welshimeri and other Listeria spp. (B), sample locations positive for L. welshimeri allelic type 14 (“haplotype 14”) and sample locations positive for other allelic types (regardless of Listeria species) (C), and sample locations positive for L. welshimeri allelic type 21 (“haplotype 21”) and sample locations positive for other allelic types (D).
Distribution of Listeria sigB allelic types by sample environment (natural and urban).
For categorical analysis of associations between Listeria sigB allelic types and urban and natural environments, individual allelic types with fewer than 10 occurrences were combined into a single category to avoid multiple categories with expected values less than 5. An initial overall 2-by-14 chi-square analysis showed that allelic types were not independently distributed between urban and natural environments (P ≤ 0.0001). Subsequent 2-by-2 chi-square analyses showed that three and five allelic types were significantly more common among urban and natural environments, respectively (Table 2).
Distribution of Listeria sigB allelic types by sampling environment.
Overall 4-by-14 chi-square tests showed that individual sigB allelic types were not independently distributed among either natural (P ≤ 0.0001) or urban (P ≤ 0.0001) sampling areas. Subsequent 2-by-2 chi-square analyses showed a significant positive association of five allelic types with specific natural areas (Table 2). For example, L. seeligeri allelic type 1 was significantly more likely to be isolated from samples collected in CT Hill (P ≤ 0.0005). In addition, three allelic types showed a significant positive association with specific urban areas (Table 2).
Spatial clustering of individual Listeria sigB allelic types.
Listeria sigB allelic types (ATs) that were specifically associated with individual sample areas were also analyzed for evidence of spatial clustering (Table 4). Among the natural areas, three instances of spatial clustering of sigB ATs were observed, including clustering of L. welshimeri ATs 14 and 21 in the Catskills (Table 4 and Fig. 5) and clustering of L. seeligeri AT 1 in CT Hill (Table 4). Among the urban areas, only one instance of spatial clustering was observed (allelic type 58 in Albany) (Table 4). All spatial clusters included isolation of the same AT on at least two separate sampling dates. ATs 14 and 21 were isolated from the Catskills on 3 out of 5 and 2 out of 5 sampling dates, respectively, AT 1 was isolated from CT Hill on 5 out of 5 sampling dates, and AT 58 was isolated from Albany on 3 out of 4 sampling dates.
Listeria sigB allelic-type diversity.
sigB AT data were used to evaluate overall subtype diversity as well as subtype diversity stratified by species and environment (urban or natural) (Table 5). The overall Simpson index of diversity (D), based on sigB ATs, was 0.939; D was higher for isolates from urban environments (0.958) than for isolates from natural environments (0.871). For L. monocytogenes, L. seeligeri, and L. welshimeri, the overall D was also higher for isolates from urban environments than for isolates from natural environments; these differences were significant for L. seeligeri and L. monocytogenes (as demonstrated by nonoverlapping 95% confidence intervals).
Table 5.
sigB allelic-type diversity stratified by Listeria species and sample environment among isolates from 2001 and 2002
| Species | Simpson index of diversity for isolates from indicated environment (no. of isolates)a |
||
|---|---|---|---|
| Natural | Urban | All | |
| L. monocytogenes | 0.295 (13)† | 0.722 (67)† | 0.678 (80) |
| L. seeligeri | 0.737 (144)† | 0.907 (99)† | 0.832 (234) |
| L. marthii | 0.833 (4) | NA (0) | 0.833 (4) |
| L. welshimeri | 0.867 (52) | 0.931 (22) | 0.902 (74) |
| L. innocua | NA (1) | 0.935 (49) | 0.935 (50) |
| Total | 0.871 (214)† | 0.958 (228)† | 0.939 (442) |
“†” marks Simpson indices of discrimination for urban and natural areas that have nonoverlapping 95% confidence intervals. NA, not applicable (Simpson's index of discrimination cannot be determined for fewer than two isolates).
DISCUSSION
Testing of more than 1,800 samples collected from urban and natural environments, in conjunction with phenotypic and molecular characterization of isolates, allowed us to assess the diversity and distribution of the genus Listeria in a well-defined geographic region (New York State) in areas other than farm environments and food-processing and production facilities. Our data show that even though Listeria represents a broadly disseminated genus, Listeria spp. as well as some specific subtypes within the species appear to show distinct ecological preferences. In addition, most Listeria spp. appear to include a proportion of subtypes that can establish persistent yet geographically dispersed populations outside food-processing and farm-associated environments, a trait likely to contribute to the wide geographical distribution of the genus Listeria.
PCR amplification and sequencing of sigB allow for rapid and reliable species-level identification and subtyping of Listeria isolates.
In addition to an MLST study that indicated that sigB sequence data allow for reliable species classification of Listeria isolates (14), another previous study (44), showed a good discriminatory power of sigB allelic typing based on 157 L. monocytogenes isolates. The data reported here further validate this assay as a low-cost discriminatory tool for initial subtype characterization of Listeria species isolates. In comparison, sequencing of the 16S rRNA, the 23S rRNA, and the 16S-23S rRNA intergenic regions has been shown to provide very limited subtype discrimination between Listeria species isolates (13, 22, 23, 54). One previous study also showed that L. monocytogenes isolates representing eight EcoRI ribotypes could be differentiated into 10 sigB allelic types (7), indicating that sigB sequencing provides discriminatory power similar to other, broadly used, but more-expensive subtyping methods. While it is likely that highly discriminatory molecular subtyping, such as pulsed-field gel electrophoresis (PFGE) or MLST, would provide further improved subtype differentiation of the isolates collected here, sigB sequencing allows for subtype discrimination within each Listeria species and provides a highly economical approach (<$15/isolate) for subtyping of large isolate sets. Further characterization of a subset of L. monocytogenes isolates reported here by MLST (including sigB, gap, prs, ribC, purM, inlA, and actA sequencing) and automated EcoRI ribotyping indeed showed improved subtype discrimination (55), but at a considerably higher cost per isolate.
While Listeria is ubiquitous and found in both urban and natural environments, individual Listeria spp. and sigB allelic types differ significantly in their prevalences between urban and natural environments and among specific sample sites and seasons.
Seeliger noted in 1961 (60) that outbreaks of animal listeriosis on multiple continents “gave rise to the assumption of the organisms being globally spread… [and]… that neither geographical nor geomedical borders exist.” Further studies by Seeliger and others subsequently confirmed that isolates classified as L. monocytogenes were indeed widespread among vegetation and soil samples collected from agricultural and nonagricultural sites (69–71); it is important to note, though, that the taxonomy of the genus Listeria has been considerably revised since these early studies were reported. For example, L. innocua and L. seeligeri were defined as separate species in 1977 and 1983, respectively (59). Consistent with previous studies that noted considerable prevalences (often >20%) of Listeria spp. in different environments (2, 10, 18, 19, 69, 71), we also found Listeria prevalences of >20% among samples collected from both urban and natural environments.
Even though there were similar overall prevalences of Listeria in natural and urban samples, the prevalences of individual Listeria spp. and sigB allelic types differed significantly between urban and natural environments. For example, while L. seeligeri and L. welshimeri were significantly overrepresented among isolates from natural environments, L. monocytogenes and L. innocua were overrepresented among isolates from urban environments. Some species also showed significant positive associations with specific sampling areas; these associations did not appear to drive the association between specific species and natural or urban environments, though. Prior to our study reported here, only very few studies (e.g., references 1, 5, 10, 20, and 58) on the distribution of different Listeria spp. in different environments were available. One report from the United Kingdom found that L. ivanovii and L. seeligeri were marginally more common in urban soils than other Listeria spp. (40). Listeria grayi and L. rocourtiae were not identified among our isolates; however, the fact that these species were not detected may reflect poor growth for these species in the standard selective and differential enrichment and plating media used or an inability to amplify these species with our sigB primers rather than an absence of these species in the samples collected (as the enrichment media used have not been validated for detection of these species). We also did not identify L. ivanovii among our isolates. Oxford agar, which we used for primary isolation, has been shown in at least one study (42) to be inhibitory to L. ivanovii.
In addition to significant associations between different Listeria spp. and urban and natural environments, we also found associations between some Listeria sigB allelic types and different environments, indicating the potential existence of ecotypes within a given Listeria species. This is consistent with previous studies indicating the presence of host- or niche-specific L. monocytogenes subtypes (34) as well as with a variety of studies that have identified ecotypes of other bacterial species and genera (9). Ivanek et al. (33) recently, in a comprehensive retrospective analysis of ecological and biogeographic characteristics of the natural environment sample areas included in this study, showed that precipitation occurrence and alternating freezing and thawing temperatures prior to sample collection were key predictors for the isolation of Listeria spp. from a given location (33). Interestingly, analysis of seasonal data showed that Listeria prevalence in natural areas in the summer was the highest (P = 0.000042), while urban areas showed the lowest Listeria prevalence in the summer (P = 0.00017).
While these observations provide a starting point for further studies on potential physiological and genetic differences that might explain ecological preferences of different Listeria spp. and subtypes, future studies in other locations will also be needed to determine whether the findings reported here for New York State can be confirmed elsewhere.
The genus Listeria comprises a high level of genetic diversity, which appears to be higher among urban environments.
Assessment of Listeria sigB allelic diversity showed that the overall level of diversity among Listeria spp. as well as the diversity stratified by species (with the exception of L. marthii) was higher among strains isolated from urban environments. Greater diversity among Listeria isolates from urban areas may suggest that urban environments allow for less-efficient dispersal of bacteria (e.g., due to landscape barriers), hence reducing the likelihood of isolation of the same strain from different sites. Natural environments, on the other hand, may allow for more-efficient bacterial dispersal (e.g., due to free movement of animals), thus increasing the chance of isolating the same strain at different sampling sites. Our findings furthermore could be interpreted as an indication that Listeria isolates from urban and natural environments represent two separate populations influenced by different ecological constraints (27). While we were not able to identify any other studies that evaluated Listeria diversity among isolates from different environments, our data are consistent with an emerging body of literature indicating that L. monocytogenes isolates found in different environments and hosts represent distinct populations (26). The true levels of Listeria diversity in different environments may even be higher than reported here, since traditional culture of Listeria from environmental samples, which requires the use of selective enrichment and plating media (12, 66), may favor recovery of certain Listeria spp. or lineages (6). In particular, even though most selective media designed for the recovery of L. monocytogenes are thought to be adequate for the recovery of other Listeria spp., this has not been thoroughly evaluated (12), and it cannot be excluded that the selective enrichment used here may have prevented or reduced the recovery of certain Listeria sigB allelic types. However, we feel that the use of a single enrichment procedure allowed an adequate initial assessment of Listeria diversity in different environments; selective enrichment procedures were deemed necessary to allow recovery of Listeria spp., which are likely to be found at low levels in environmental samples, which may contain high levels of competing background flora (2).
Certain Listeria spp. and sigB allelic types tend to cluster spatially within both natural and urban environments, and there is evidence that these species and allelic types can persist over time.
Nearest-neighbor spatial analysis (68) showed that a number of Listeria species and allelic types showed clustering within a given area, indicating establishment of area-specific local populations in addition to the presence of widely distributed allelic types. All statistically significant spatial clusters of Listeria species and sigB allelic represented isolates from samples obtained at multiple and in some cases all sampling dates, further supporting that these clusters represent area-specific persistent populations. These findings are consistent with a number of studies that have indicated that specific Listeria subtypes can establish persistent populations in food-processing and retail environments (41, 52, 56), including some instances where a given subtypes seems to have persisted in a specific plant for at least 12 years (34). We propose that the ability of members of the genus Listeria to establish persistent populations in different environments is critical for its widespread and ubiquitous presence. Persistence of Listeria in urban and natural environments also has important implications for studies that use reisolation of a specific Listeria subtype in a processing plant or retail environment as evidence for Listeria persistence in that location (34, 37, 56, 57). Our data specifically indicate that reisolation of a specific Listeria subtype in a food-related environment that it is not well isolated from its surroundings (e.g., retail environments which cannot control outside traffic and install footbaths) may also, in some cases, present reintroduction of a subtype that persists in the surrounding environment.
Conclusions.
Our data not only support that the genus Listeria is widely distributed and highly prevalent in many environments but also show that species and sigB allelic types within this genus appear to show distinct ecological preferences. The observation that L. seeligeri, which is nonpathogenic in mammals (31) but often carries a homologue of the L. monocytogenes prfA virulence gene cluster, is more common among samples from natural environments and is the most common Listeria species isolated there may indicate that possession of these virulence genes provides a selective advantage in these environments over other Listeria spp. (e.g., L. innocua), possibly by facilitating survival of protozoan predation as previously postulated (36). On a more practical level, our data showing species-specific prevalence patterns in different environments suggest that the use of Listeria spp. as an indicator for L. monocytogenes contamination may require refinement. For example, L. innocua may be a better indicator for L. monocytogenes contamination than L. seeligeri or L. welshimeri, which appear to occupy environmental niches distinct from that of L. monocytogenes (e.g., natural environments), while L. monocytogenes is typically associated with other environments (e.g., urban environments). The further characterization of different Listeria ecotypes may also ultimately facilitate development of molecular-subtyping-based source tracking approaches analogous to those developed for Escherichia coli source tracking (49).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Award R01GM63259 (to M.W.). The Pathogen Tracker 2.0 database online public subtyping database is supported by USDA Special Research grants 2001-34459-10296 and 2002-34459-11758 (to M.W.).
We gratefully acknowledge the contributions of those who helped with sample collection, including Courtney Bolger, Mary Pat Craver, Laura Kornstein, Sarita Raengpradub, Hazel Fromm, Zhisheng Her, Fok Moon Lum, Kelly Sauders, and Lewis Sauders, and those who helped with gene sequencing, including Leroy Chan, Kin Hup Tan, and Mariel Fisher.
Footnotes
Published ahead of print 13 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bauwens L, Vercammen F, De Meurichy W. 2001. Occurrence of Listeria spp. in captive antelope herds and their environment. J. Zoo Wildl. Med. 32:514–518 [DOI] [PubMed] [Google Scholar]
- 2. Bernagozzi M, Bianucci F, Sacchetti R, Bisbini P. 1994. Study of the prevalence of Listeria spp. in surface water. Zentralbl. Hyg. Umweltmed. 196:237–244 [PubMed] [Google Scholar]
- 3. Bille J, et al. 1992. API Listeria, a new and promising one-day system to identify Listeria isolates. Appl. Environ. Microbiol. 58:1857–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boerlin P, Rocourt J, Piffaretti JC. 1991. Taxonomy of the genus Listeria by using multilocus enzyme electrophoresis. Int. J. Syst. Bacteriol. 41:59–64 [DOI] [PubMed] [Google Scholar]
- 5. Bouttefroy A, Lemaitre JP, Rousset A. 1997. Prevalence of Listeria sp. in droppings from urban rooks (Corvus frugilegus). J. Appl. Microbiol. 82:641–647 [DOI] [PubMed] [Google Scholar]
- 6. Bruhn JB, Vogel BF, Gram L. 2005. Bias in the Listeria monocytogenes enrichment procedure: lineage 2 strains outcompete lineage 1 strains in University of Vermont selective enrichments. Appl. Environ. Microbiol. 71:961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai S. 2002. DNA sequence based subtyping and molecular evolution of Listeria and Listeria monocytogenes. M.S. thesis Cornell University, Ithaca, NY [Google Scholar]
- 8. Chemaly M, Toquin MT, Le Notre Y, Fravalo P. 2008. Prevalence of Listeria monocytogenes in poultry production in France. J. Food Prot. 71:1996–2000 [DOI] [PubMed] [Google Scholar]
- 9. Cohan FM. 2001. Bacterial species and speciation. Syst. Biol. 50:513–524 [DOI] [PubMed] [Google Scholar]
- 10. Colburn KG, Kaysner CA, Abeyta C, Jr, Wekell MM. 1990. Listeria species in a California coast estuarine environment. Appl. Environ. Microbiol. 56:2007–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collins MD, et al. 1991. Phylogenetic analysis of the genus Listeria based on reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:240–246 [DOI] [PubMed] [Google Scholar]
- 12. Curtis GD, Lee WH. 1995. Culture media and methods for the isolation of Listeria monocytogenes. Int. J. Food Microbiol. 26:1–13 [DOI] [PubMed] [Google Scholar]
- 13. Czajka J, et al. 1993. Differentiation of Listeria monocytogenes and Listeria innocua by 16S rRNA genes and intraspecies discrimination of Listeria monocytogenes strains by random amplified polymorphic DNA polymorphisms. Appl. Environ. Microbiol. 59:304–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. den Bakker HC, Bundrant BN, Fortes ED, Orsi RH, Wiedmann M. 2010. A population genetics-based and phylogenetic approach to understanding the evolution of virulence in the genus Listeria. Appl. Environ. Microbiol. 76:6085–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. den Bakker HC, et al. 2010. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 11:688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dowe MJ, Jackson ED, Mori JG, Bell CR. 1997. Listeria monocytogenes survival in soil and incidence in agricultural soils. J. Food Prot. 60:1201–1207 [DOI] [PubMed] [Google Scholar]
- 17. Felsenstein J. 2004. Inferring phylogenies. Sinauer Associates, Sunderland, MA [Google Scholar]
- 18. Fenlon DR, Wilson J, Donachie W. 1996. The incidence and level of Listeria monocytogenes contamination of food sources at primary production and initial processing. J. Appl. Bacteriol. 81:641–650 [DOI] [PubMed] [Google Scholar]
- 19. Frances N, Hornby H, Hunter PR. 1991. The isolation of Listeria species from fresh-water sites in Cheshire and North Wales. Epidemiol. Infect. 107:235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garrec N, Picard-Bonnaud F, Pourcher AM. 2003. Occurrence of Listeria sp. and L. monocytogenes in sewage sludge used for land application: effect of dewatering, liming and storage in tank on survival of Listeria species. FEMS Immunol. Med. Microbiol. 35:275–283 [DOI] [PubMed] [Google Scholar]
- 21. Gouin E, Mengaud J, Cossart P. 1994. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 62:3550–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Graham T, Golsteyn-Thomas EJ, Gannon VP, Thomas JE. 1996. Genus- and species-specific detection of Listeria monocytogenes using polymerase chain reaction assays targeting the 16S/23S intergenic spacer region of the rRNA operon. Can. J. Microbiol. 42:1155–1162 [DOI] [PubMed] [Google Scholar]
- 23. Graham TA, Golsteyn-Thomas EJ, Thomas JE, Gannon VP. 1997. Inter- and intraspecies comparison of the 16S-23S rRNA operon intergenic spacer regions of six Listeria spp. Int. J. Syst. Bacteriol. 47:863–869 [DOI] [PubMed] [Google Scholar]
- 24. Graves LM, et al. 2010. Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 60:1280–1288 [DOI] [PubMed] [Google Scholar]
- 25. Graves LM, Swaminathan B, Hunter SB. 1999. Subtyping Listeria monocytogenes, p 279–297 In Ryser ET, Marth EH. (ed), Listeria, listeriosis, and food safety, 2nd ed Marcel Dekker, New York, NY [Google Scholar]
- 26. Gray MJ, et al. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grundmann H, Hori S, Tanner G. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hansen CH, Vogel BF, Gram L. 2006. Prevalence and survival of Listeria monocytogenes in Danish aquatic and fish-processing environments. J. Food Prot. 69:2113–2122 [DOI] [PubMed] [Google Scholar]
- 29. Hellstrom S, Kiviniemi K, Autio T, Korkeala H. 2008. Listeria monocytogenes is common in wild birds in Helsinki region and genotypes are frequently similar with those found along the food chain. J. Appl. Microbiol. 104:883–888 [DOI] [PubMed] [Google Scholar]
- 30. Ho AJ, Lappi VR, Wiedmann M. 2007. Longitudinal monitoring of Listeria monocytogenes contamination patterns in a farmstead dairy processing facility. J. Dairy Sci. 90:2517–2524 [DOI] [PubMed] [Google Scholar]
- 31. Hof H, Hefner P. 1988. Pathogenicity of Listeria monocytogenes in comparison to other Listeria species. Infection 16(Suppl. 2):S141–S144 [DOI] [PubMed] [Google Scholar]
- 32. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ivanek R, et al. 2009. Modeling of spatially referenced environmental and meteorological factors influencing the probability of Listeria species isolation from natural environments. Appl. Environ. Microbiol. 75:5893–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kathariou S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811–1829 [DOI] [PubMed] [Google Scholar]
- 35. Korthals M, Ege M, Lick S, von Mutius E, Bauer J. 2008. Occurrence of Listeria spp. in mattress dust of farm children in Bavaria. Environ. Res. 107:299–304 [DOI] [PubMed] [Google Scholar]
- 36. Kreft J, Vazquez Boland JA, Ng E, Goebel W. 1999. Virulence gene clusters and putative pathogenicity islands in listeriae, p 219–232 In Kaper JB, Hacker J. (ed), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, DC [Google Scholar]
- 37. Latorre AA, et al. 2011. Increased in vitro adherence and on-farm persistence of predominant and persistent Listeria monocytogenes strains in the milking system. Appl. Environ. Microbiol. 77:3676–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leclercq A, et al. 2010. Listeria rocourtiae sp. nov. Int. J. Syst. Evol. Microbiol. 60:2210–2214 [DOI] [PubMed] [Google Scholar]
- 39. Levine N. 2002. CrimeStat: a spatial statistics program for the analysis of crime incident locations (v 2.0). Ned Levine & Associates, Houston, TX [Google Scholar]
- 40. MacGowan AP, Bowker K, McLauchlin J, Bennett PM, Reeves DS. 1994. The occurrence and seasonal changes in the isolation of Listeria spp. in shop bought food stuffs, human faeces, sewage and soil from urban sources. Int. J. Food Microbiol. 21:325–334 [DOI] [PubMed] [Google Scholar]
- 41. Miettinen MK, Bjorkroth KJ, Korkeala HJ. 1999. Characterization of Listeria monocytogenes from an ice cream plant by serotyping and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 46:187–192 [DOI] [PubMed] [Google Scholar]
- 42. Monaca NL. 2002. Limitations of Oxford and PALCAM agars for the detection of Listeria ivanovii, abstr PP9.1. Abstr. 28th Annu. Sci. Meet. Aust. Soc. Microbiol Australian Society for Microbiology, Melbourne, Australia [Google Scholar]
- 43. Moshtaghi H, Garg SR, Mandokhot UV. 2003. Prevalence of Listeria in soil. Indian J. Exp. Biol. 41:1466–1468 [PubMed] [Google Scholar]
- 44. Nightingale K, Bovell L, Grajczyk A, Wiedmann M. 2007. Combined sigB allelic typing and multiplex PCR provide improved discriminatory power and reliability for Listeria monocytogenes molecular serotyping. J. Microbiol. Methods 68:52–59 [DOI] [PubMed] [Google Scholar]
- 45. Nightingale KK, et al. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70:4458–4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nightingale KK, Windham K, Wiedmann M. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187:5537–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Odjadjare EE, Obi LC, Okoh AI. 2010. Municipal wastewater effluents as a source of listerial pathogens in the aquatic milieu of the Eastern Cape Province of South Africa: a concern of public health importance. Int. J. Environ. Res. Public Health 7:2376–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paillard D, et al. 2005. Occurrence of Listeria spp. in effluents of French urban wastewater treatment plants. Appl. Environ. Microbiol. 71:7562–7566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ram JL, Ritchie RP, Fang J, Gonzales FS, Selegean JP. 2004. Sequence-based source tracking of Escherichia coli based on genetic diversity of beta-glucuronidase. J. Environ. Qual. 33:1024–1032 [DOI] [PubMed] [Google Scholar]
- 50. Restaino L, Frampton EW, Irbe RM, Schabert G, Spitz H. 1999. Isolation and detection of Listeria monocytogenes using fluorogenic and chromogenic substrates for phosphatidylinositol-specific phospholipase C. J. Food Prot. 62:244–251 [DOI] [PubMed] [Google Scholar]
- 51. Rocourt J. 1999. The genus Listeria and Listeria monocytogenes: phylogenetic position, taxonomy, and identification, p 1–20 In Ryser ET, Marth EH. (ed), Listeria, listeriosis, and food safety, 2nd ed Marcel Dekker, New York, NY [Google Scholar]
- 52. Rørvik LM, Aase B, Alvestad T, Caugant DA. 2003. Molecular epidemiological survey of Listeria monocytogenes in broilers and poultry products. J. Appl. Microbiol. 94:633–640 [DOI] [PubMed] [Google Scholar]
- 53. Rothman KJ, Greenland S. 1998. Approaches to statistical analysis, p 183–199 In Rothman KJ, Greenland S. (ed), Modern epidemiology. Lippincot-Raven, Philadelphia, PA [Google Scholar]
- 54. Sallen B, Rajoharison A, Desvarenne S, Quinn F, Mabilat C. 1996. Comparative analysis of 16S and 23S rRNA sequences of Listeria species. Int. J. Syst. Bacteriol. 46:669–674 [DOI] [PubMed] [Google Scholar]
- 55. Sauders BD, et al. 2006. Molecular characterization of Listeria monocytogenes from natural and urban environments. J. Food Prot. 69:93–105 [DOI] [PubMed] [Google Scholar]
- 56. Sauders BD, et al. 2004. Distribution of Listeria monocytogenes molecular subtypes among human and food isolates from New York State shows persistence of human disease-associated Listeria monocytogenes strains in retail environments. J. Food Prot. 67:1417–1428 [DOI] [PubMed] [Google Scholar]
- 57. Sauders BD, et al. 2009. Prevalence and molecular diversity of Listeria monocytogenes in retail establishments. J. Food Prot. 72:2337–2349 [DOI] [PubMed] [Google Scholar]
- 58. Sauders BD, Wiedmann M. 2007. Ecology of Listeria species and L. monocytogenes in the natural environment, p 21–53 In Ryser ET, Marth EH. (ed), Listeria, listeriosis, and food safety, 3rd ed CRC Press, Boca Raton, FL [Google Scholar]
- 59. Seeliger HP, Jones D. 1986. Listeria, p 1235–1245 In Sneath PHA, Mair NS, Sharpe ME, Holt JG. (ed), Bergey's manual of systematic bacteriology, vol 2 Williams & Wilkins, Baltimore, MD [Google Scholar]
- 60. Seeliger HPR. 1961. Listeriosis. Hafner Publishing, New York, NY [Google Scholar]
- 61. Stuart SE, Welshimer HJ. 1973. Intrageneric relatedness of Listeria Pirie. Int. J. Syst. Bacteriol. 23:8–14 [Google Scholar]
- 62. Stuart SE, Welshimer HJ. 1974. Taxonomic reexamination of Listeria Pirie and transfer of Listeria grayi and Listeria murrayi to a new genus, Murraya. Int. J. Syst. Bacteriol. 24:177–185 [Google Scholar]
- 63. Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland, MA [Google Scholar]
- 64. Thimothe J, et al. 2002. Detection of Listeria in crawfish processing plants and in raw, whole crawfish and processed crawfish (Procambarus spp.). J. Food Prot. 65:1735–1739 [DOI] [PubMed] [Google Scholar]
- 65. Vaneechoutte M, et al. 1998. Comparison of PCR-based DNA fingerprinting techniques for the identification of Listeria species and their use for atypical Listeria isolates. Int. J. Syst. Bacteriol. 48(Pt. 1):127–139 [DOI] [PubMed] [Google Scholar]
- 66. Van Renterghem B, Huysman F, Rygole R, Verstraete W. 1991. Detection and prevalence of Listeria monocytogenes in the agricultural ecosystem. J. Appl. Bacteriol. 71:211–217 [DOI] [PubMed] [Google Scholar]
- 67. Volokhov D, George J, Anderson C, Duvall RE, Hitchins AD. 2006. Discovery of natural atypical nonhemolytic Listeria seeligeri isolates. Appl. Environ. Microbiol. 72:2439–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Waller LA, Gotway CA. 2004. Applied spatial statistics for public health data. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 69. Weis J, Seeliger HP. 1975. Incidence of Listeria monocytogenes in nature. Appl. Microbiol. 30:29–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Welshimer HJ. 1968. Isolation of Listeria monocytogenes from vegetation. J. Bacteriol. 95:300–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Welshimer HJ, Donker-Voet J. 1971. Listeria monocytogenes in nature. Appl. Microbiol. 21:516–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wiedmann M, et al. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.