Abstract
Lactococcin 972 (Lcn972) is a nonlantibiotic bacteriocin that inhibits cell wall biosynthesis by binding to lipid II. In this work, two mutants resistant to Lcn972, Lactococcus lactis D1 and D1-20, with high (>320 arbitrary units [AU]/ml) and low (80 AU/ml) susceptibilities, respectively, have been isolated. Resistance to Lcn972 did not impose a burden to growth under laboratory conditions, nor did it substantially alter the physicochemical properties of the cell surface. However, the peptidoglycan of the mutants featured a higher content of muropeptides with tripeptide side chains than the wild-type strain, linking for the first time peptidoglycan remodelling to bacteriocin resistance. Moreover, L. lactis lacking a functional d,d-carboxypeptidase DacA (i.e., with a high content of pentapeptide side chain muropeptides) was shown to be more susceptible to Lcn972. Cross-resistance to lysozyme and nisin and enhanced susceptibility to penicillin G and bacitracin was also observed. Intriguingly, the Lcn972-resistant mutants were not infected by the lytic phage c2 and less efficiently infected by phage sk1. Lack of c2 infectivity was linked to a 22.6-kbp chromosomal deletion encompassing the phage receptor protein gene pip. The deletion also included maltose metabolic genes and the two-component system (TCS) F. However, a clear correlation between these genes and resistance to Lcn972 could not be clearly established, pointing to the presence of as-yet-unidentified mutations that account for Lcn972 resistance.
INTRODUCTION
Lactococcus lactis is one of the main components of the mesophilic starter cultures used in cheese manufacturing. Thereby, there is a genuine interest in improving robustness to ensure the success of dairy fermentations. L. lactis performance may be compromised by the presence of bacteriophages or other inhibitors such as antibiotics, lysozyme, or bacteriocins in raw milk (12, 27). Many of these antibacterial compounds target cell wall components. Bacteriophages recognize bacterial receptors, mostly of polysaccharide nature, prior to infection (37), while lysozyme acts directly on the cell wall peptidoglycan, hydrolyzing the glycosidic bonds between N-acetylmuramic acid and N-acetylglucosamine. Moreover, an increasing number of bacteriocins, antimicrobial peptides synthesized by bacteria, including the lactococcal lantibiotics nisin and lacticin 3147 and the nonlantibiotic lactococcin 972 (Lcn972), have been reported to inhibit cell wall biosynthesis by binding to the cell wall precursor lipid II (4, 23, 42).
As a Gram-positive bacterium, the cell envelope of L. lactis consists of a cytoplasmic membrane and a thick cell wall. The cell wall is composed mostly of peptidoglycan (PG) made of glycan strands cross-linked by peptides and secondary polymers such as teichoic acids, proteins, and carbohydrates. L. lactis has an A4α-type peptidoglycan with an l-Ala-α-d-Glu-l-Lys-d-Ala as the tetrapeptide and d-Asp in the interpeptide bridge (9). Recent microscopy advances have provided a very detailed knowledge on the PG structure in the L. lactis cell wall (1, 41).
The cell wall in Gram-positive bacteria protects cells from osmotic pressure and acts as an exoskeleton, maintaining cell shape, and as scaffold for anchoring other cell envelope components (see reference 40 and references therein). Thus, monitoring its integrity is crucial for survival. In L. lactis, the response to cell envelope stress is governed by the two-component system CesSR, which has been shown to be triggered by lysozyme, nisin, and Lcn972 (26, 39) and by heterologous protein secretion and phage infection (11, 33). Although the CesR regulon is not fully understood, certain CesR-regulated components are known to contribute positively to L. lactis survival under technological relevant stresses (34).
In this work, we have isolated L. lactis mutants resistant to Lcn972 (Lcn972r) which were characterized with a particular emphasis on cell surface properties, PG composition, and resistance to cell wall-active antimicrobials, such as lysozyme and bacteriophages. Lcn972 is an atypical 66-amino-acid bacteriocin that does not meet the widely accepted criteria of small, heat-resistant hydrophobic peptides. Lcn972 is a highly hydrophilic cationic peptide easily inactivated by heat (25). In contrast to other lipid II-binding bacteriocins, Lcn972 does not form pores in the cytoplasmic membrane and is active exclusively against lactococci (23). These features make Lcn972 a unique candidate to shed light on mechanisms that help L. lactis to cope better with cell wall stress.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and growth conditions.
L. lactis strains (Table 1) were routinely grown in M17 with 0.5% glucose (GM17), statically and at 30°C. When specified, glucose was replaced by 0.5% maltose (MM17) or chemically defined medium (CDM) (29) was used. Escherichia coli strains were grown in 2× YT (35) at 37°C with shaking. When needed, antibiotics erythromycin and ampicillin were used at final concentrations of 5 μg ml−1 and 100 μg ml−1, respectively. Growth rates (μ) were calculated through linear regressions of the plots of ln(optical density at 600 nm [OD600]) versus time during the exponential growth phase. Bacteriophages c2 and sk1 were propagated on L. lactis MG1614. Phage titer was calculated by the standard plate assay. Decimal dilutions in 0.9% NaCl of phage lysates were mixed with 3.5 ml of molten GM17 0.7% agar supplemented with 10 mM CaCl2 and 100 μl of stationary-phase L. lactis MG1614 culture. The mixture was spread on GM17 plates and incubated at 30°C for 16 h until clear lytic plaques were visible. Bacterial cultures were stored at −80°C in the appropriate medium and 10% glycerol (vol/vol). Phage lysates were stored at 4°C.
Table 1.
Bacterial strains, bacteriophages, and plasmids used in this work
| Strain, phage, or plasmid | Descriptiona | Reference/source |
|---|---|---|
| L. lactis | ||
| MG1363 | Plasmid-free and prophage-cured derivative of L. lactis NCDO712 | 13 |
| MG1614 | Strr Rifr derivative of MG1363, Lcn972s | 13 |
| NZ9000 | MG1363, carrying pepN::nisRK | 21 |
| D1 | MG1614, Lcn972 high-resistant mutant, unstable | This work |
| D1-20 | MG1614, Lcn972 low-resistant mutant derived from D1, stable | This work |
| ΔdacA strain | NZ9000 lacking dacA gene | This work |
| ΔdacB strain | MG1363 lacking dacB gene | 9 |
| ΔdltD strain | MG1363 lacking dltD gene | 10 |
| MGRrF | MG1363 pRV300::llrF | 30 |
| E. coli | ||
| DH10B | Plasmid free, cloning host | Invitrogen |
| Phages | ||
| c2 | L. lactis lytic phage belonging to c2 family | E. Bidnenko |
| sk1 | L. lactis lytic phage belonging to 936 family | E. Bidnenko |
| Plasmids | ||
| pORI280 | EmrlacZ+, integrative plasmid | 22 |
| pBL16 | 600-bp dacA flanking regions cloned in pORI280 | This work |
Str, streptomycin; Rif, rifampin; Em, erythromycin.
Standard DNA techniques.
Standard molecular techniques were followed as described elsewhere (35). Chromosomal DNA was isolated with the GenElute bacterial genomic DNA kit (Sigma-Aldrich, Spain). Restriction enzymes were purchased from TaKaRa (Japan) and T4 ligase from Fisher Scientific (Spain). Oligonucleotides were supplied by Sigma-Aldrich (Spain). Standard PCRs were carried out using Pure Taq Ready-to-Go PCR beads (GE Healthcare, United Kingdom). For cloning purposes and PCRs expected to yield long products, the proofreading Phusion high-fidelity DNA polymerase (Fisher Scientific, Spain) was used.
Selection of L. lactis resistant to Lcn972.
The bacteriocin Lcn972 was purified and quantified as described previously (26). Lcn972 dilutions were done in 50 mM sodium phosphate buffer, pH 6.8. To isolate Lcn972-resistant mutants, L. lactis MG1614 was cultivated stepwise in GM17 in the presence of increasing Lcn972 concentrations ranging from 5 AU/ml to up to 400 AU/ml. Overnight cultures grown at the highest Lcn972 concentration were diluted and plated on GM17 to get isolated colonies. A single colony, designated L. lactis D1, was randomly selected. L. lactis D1-20 was colony-isolated after serial passages of L. lactis D1 in GM17 during 200 generations.
Construction of the L. lactis ΔdacA strain.
The flanking 600-bp regions up- and downstream of dacA (llmg2560 in L. lactis MG1363; GenBank AM406671) were amplified and fused by splicing by overlap extension PCR (SOE-PCR) using the primers D1 (5′ AACTGCAGTATTGACAAATGCCG 3′), D2 (5′ AAAAACTTTTGGAGCACTGACAGCAAAGCTCG 3′), D3 (5′ AGTGCTCCAAAAGTTTTTTGG 3′), and D4 (5′ GAAGATCTGCTAAACGTGACCC 3′). The SOE-PCR fragment was cloned into the nonreplicating plasmid pORI280 using the engineered restriction sites PstI and BglII in the far ends of primers D1 and D4, respectively, to generate the plasmid pBL16. Transformation of L. lactis NZ9000 and selection of deletion mutants proceeded according to Leenhouts et al. (22).
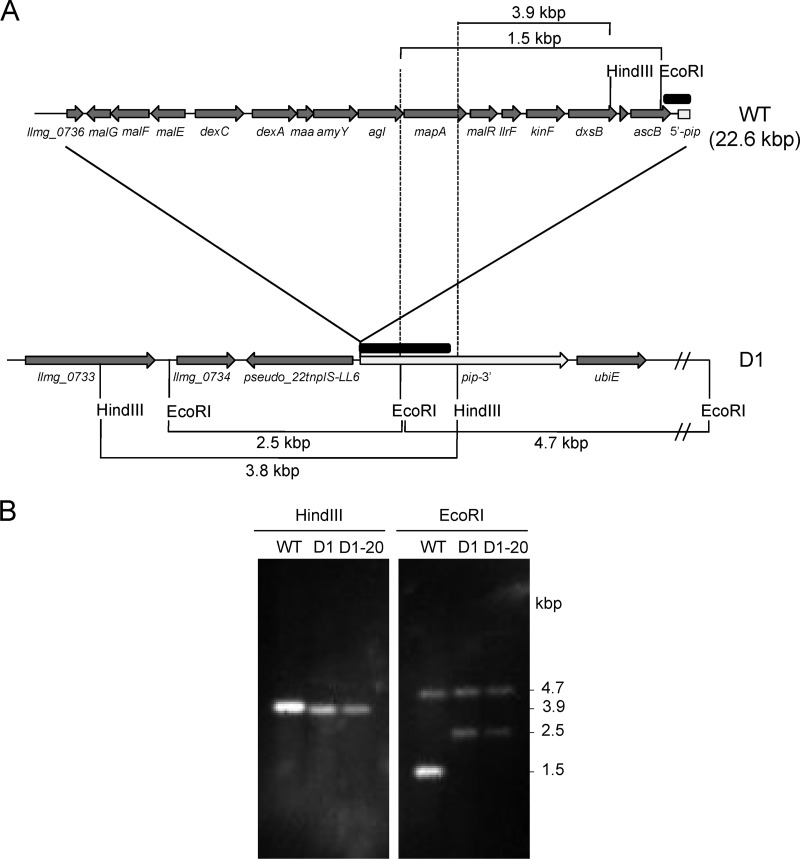
Mapping chromosomal deletion in L. lactis D1 and D1-20.
A forward primer, 0734F (5′ GAAATGGCCCTGCGACGTGTAG 3′), located within the llmg0734 locus, was used in combination with the reverse primer, 0752R (5′ CGCTGCATCCAATGTCACAGTC 3′), in pip for PCR amplification using L. lactis DNA. PCR conditions were 98°C for 30 s; 35 cycles at 98°C for 10 s, 63°C for 30 s, and 72°C for 13 min; and 72°C for 10 min, and Phusion high-fidelity DNA polymerase was used. PCR products were purified with Illustra GFX PCR DNA and the gel band purification kit (GE Healthcare, United Kingdom) and sequenced. Southern hybridization was carried out on 200 ng of total DNA digested with EcoRI and HindIII blotted to a nylon Hybond-N membrane (GE Healthcare, United Kingdom) (35). The DNA probe was labeled by PCR on L. lactis MG1614 DNA using primers pipF (5′ CGGATTCATCTATGTTGACC 3′) and pipR2 (5′ AATTGCTTCTCTTTGTCGG 3′), expanding the 5′ end of pip (see Fig. 3), and labeled dNTPs from PCR DIG labeling mix (Roche, Spain). PCR conditions were 98°C for 30 s; 35 cycles at 98°C for 10 s, 45°C for 30 s, and 72°C for 2 min; and 72°C for 10 min, using Phusion high-fidelity DNA polymerase. The blots were revealed by immunodetection at 20°C using CDP Star (Roche, Spain) by following the manufacturer's recommendations.
Fig 3.
Overview of the 22.6-kbp deletion found in L. lactis D1 and D1-20 resistant to Lcn972. (A) Schematic drawing of L. lactis D1 deleted genes compared to L. lactis MG1614 (WT). Gene annotation according to L. lactis MG1363 (GenBank accession no. AM406671). Only relevant EcoRI and HindIII sites are shown. Note that genes are not at scale. (B) Confirmation of pip truncation (white arrow in panel A) by Southern blot analysis using a PCR probe (black rectangle in panel A) and L. lactis DNA digested with HindIII and EcoRI.
Surface properties.
The electrophoretic mobility was measured using stationary-phase cells concentrated to 107 CFU/ml in 5 mM NaCl as previously described (15). Hydrophobicity was determined following the microbial-adhesion-to-solvents method (MATS) using hexadecane and stationary-phase cells in 0.15 M NaCl adjusted to a final OD600 of 0.8 (2). Each measurement was performed in triplicate, and the assay was carried out twice with independent cultures. Adsorption of Lcn972 to lactococcal cells was performed by mixing 200 μl of exponentially growing cells adjusted to an OD600 of 2.0 with 200 μl of Lcn972 (20 μg/ml) in 50 mM sodium phosphate buffer, pH 6.8. Residual inhibitory activity was measured by the agar diffusion test using L. lactis MG1614 as an indicator (26).
Cell wall composition.
Peptidoglycan (PG) preparations were obtained from 500 ml of exponentially growing (OD600 of 0.3) cultures as described previously (9). Briefly, after SDS-lysis, the crude cell wall preparation containing PG was treated with 48% hydrofluoric acid for 16 h at 4°C to eliminate anionic polymers linked to PG. Two milligrams of purified PG was digested with mutanolysin (Sigma; 2,500 U/ml) for 10 h at 37°C under shaking. Solubilized muropeptides were reduced by sodium borohydride and separated by reverse-phase high-performance liquid chromatography (RP-HPLC) as described previously (9). The percentage of muropeptides with a certain peptide side chain (X = tri, tetra, and penta) with free COOH (donor chain) was calculated according to Glauner et al. (16) as follows: percentage (X) = [Σmonomers(X) + (1/2)Σdimers(X) + (1/3)Σtrimers(X) + (1/4)Σtetramers(X)]/Σall muropeptides. During the isolation of PG, part of the cell wall fraction was kept before the treatment with hydrofluoric acid to determine the sugar composition (6). The cell wall fraction was hydrolyzed with TFA 4 M at 110°C for 3 h and derivatizated with N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) for 30 min at room temperature and analyzed by gas chromatography coupled to mass spectrometry (6). The content of d-alanine esterified to teichoic acids was determined after release by alkaline hydrolysis from 10 mg of dried cells from 300 ml of stationary-phase lactococcal cultures in GM17 as previously described (15).
Antimicrobial susceptibility assays.
The susceptibility of L. lactis cell envelope mutants to Lcn972 was tested by the spot-on-the-lawn method. Overnight cultures were diluted 10-fold in Ringer saline solution (Merck, Germany) and inoculated 1:1,000 (vol/vol) in melted GM17 containing 1.2% agar. Drops (5 μl) of 2-fold dilutions of Lcn972 were spotted on the plates and incubated at 30°C. To determine cross-resistance to cell wall antimicrobials, serial dilutions in GM17 broth of L. lactis D1 and D1-20 and the wild-type MG1614 overnight cultures were spotted on GM17 plates containing 0.5 mg/ml lysozyme, 10 ng/ml nisin, 0.5 μg/ml bacitracin, or 0.05 μg/ml penicillin G. Determinations of Lcn972 MICs were carried out in microtiter plates using either GM17 or MM17 essentially as described previously (5).
Phage assays.
The efficiency of plaquing (EOP) was defined as the phage titer on the tested strain divided by that on the reference L. lactis MG1614. Each c2 and sk1 phage suspension was plated in triplicate. Phage adsorption was determined by mixing exponentially growing lactococcal cultures L. lactis MG1614, L. lactis D1, and L. lactis D1-20 at an OD600 of 0.5 with c2 or sk1 phage suspensions to match a phage/bacteria ratio (multiplicity of infection [MOI]) of 2 × 10−6 for c2 and 1 × 10−3 for ski1 (i.e., 1 ski1 phage per 1,000 bacterial cells). After 10 min of incubation at 30°C, samples were centrifuged, and the phage titer of the supernatant was determined by the standard plaque assay using L. lactis MG1614 as the host. The percentage of adsorption was determined as follows: [1 − (phage titer of the supernatant/phage titer of a control tube without cells)] × 100. The assay was carried out twice with independent cultures, and plating was made in triplicate.
RESULTS
Isolation of L. lactis MG1614 resistant to Lcn972.
Stepwise exposure of L. lactis MG1614 to increasing Lcn972 concentrations resulted in cultures able to multiply in the presence of 400 AU/ml of Lcn972. A single colony, designated L. lactis D1, was randomly selected. This mutant was highly resistant to Lcn972 with a MIC of over 320 AU/ml (Table 2). This high-resistant phenotype was lost in the absence of selective pressure for 130 generations, after which a stable phenotype, L. lactis D1-20, with a MIC of 80 AU/ml (Table 2), could be maintained at least for 70 generations more.
Table 2.
Properties of L. lactis MG1614 and its derivatives resistant to Lcn972
| L. lactis strain | MIC of Lcn972 (AU/ml) | μ (h−1)a |
Lcn972 adsorption (%) | Adhesion to hexadecaneb (%) | d-Ala (ng/mg dried cells) | |||
|---|---|---|---|---|---|---|---|---|
| GM17 | CDM |
|||||||
| Glu | Gal | Mal | ||||||
| MG1614 | 10 | 1.07 | 0.98 | 0.38 | 0.39 | 25.6 ± 3.1 | 75.75 ± 4.55 | 17.7 ± 2.7 |
| D1 | >320 | 0.96 | 0.67 | 0.2 | NG | 13.9 ± 0.8 | 62.90 ± 1.87 | 14.1 ± 0.6 |
| D1-20 | 80 | 0.97 | 0.68 | 0.37 | NG | 14.4 ± 1.6 | 63.60 ± 1.74 | 14.3 ± 1.9 |
Growth was carried out in microtiter plates in GM17 or in tubes in chemically defined medium (CDM) supplemented with glucose (Glu), galactose (Gal), or maltose (Mal) at 0.5%. NG, no growth.
Determined by the MATS method.
Resistance to Lcn972 did not clearly impose a burden to L. lactis, as similar growth rates were observed in GM17 broth at 30°C (Table 2). However, under more limiting nutritional conditions in CDM-glucose, a decrease of 30% in the growth rate was observed for both Lcn972R mutants. Growth on galactose was seriously compromised in L. lactis D1, and maltose did not support growth (Table 2).
Surface properties and cell wall composition of L. lactis MG1614 resistant to Lcn972.
Physicochemical properties of the bacterial cell surface may determine the initial interaction of bacteriocins to the target cell and contribute to resistance. Indeed, adsorption of Lcn972 to Lcn972r cells was reduced by 50% compared to that of the susceptible L. lactis MG1614 strain (Table 2). According to their electrophoretic mobility, L. lactis MG1614 and the mutants were equally negatively charged (data not shown), whereas the Lcn972r strains revealed a less hydrophobic character (P < 0.001) than L. lactis MG1614 (Table 2), anticipating subtle changes in the bacterial surface.
As Lcn972 is active at the cell wall level (23), we looked for alterations in the structure and composition of the cell wall in the Lcn972r mutants. Macroscopically, cells grew in pairs and short chains irrespectively of their phenotype. Large ultrastructural changes such as a thickened cell wall or absence of the surface polysaccharide layer (6) were not observed by electron microscopy (data not shown). Likewise, the overall content of monosaccharides (glucose, galactose, rhamnose), amino sugars (glucosamine), glycerol, and phosphate of the cell wall fraction of the strains did not reveal substantial differences among the strains (data not shown). Teichoic acids, which can be d-alanylated to modulate the negative charges inside the cell wall, were also analyzed. Although the resistant mutants contained less d-Ala than the wild type, the differences were not statistically significant (P > 0.05) (Table 2), suggesting that d-alanylation does not contribute largely to resistance to Lcn972.
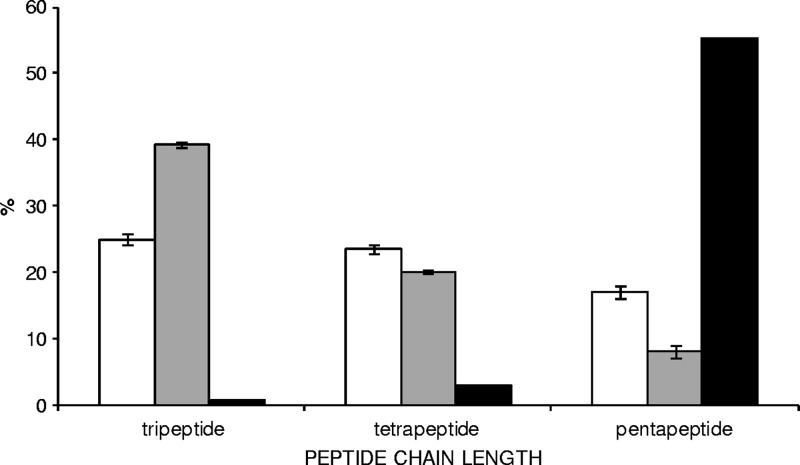
On the contrary, remarkable differences among the strains were observed when the soluble peptidoglycan muropeptides were analyzed. While similar percentages of mono-, di-, tri-, and tetramers were found among the strains, both Lcn972r mutants D1 and D1-20 showed a higher content of muropeptides with tripeptide side chains and a reduced content of pentapeptide side chains than the wild type (Fig. 1). This observation suggested an alteration in the Lcn972r mutants of activities involved in PG maturation.
Fig 1.
Distribution of muropeptides containing tri-, tetra-, or pentapeptide side chains in the Lcn972-susceptible L. lactis MG1614 (white bars), Lcn972-resistant L. lactis D1 (gray bars), and L. lactis dacA (black bars) strains. Values are the means from two independent peptidoglycan extractions, except for the dacA mutant. Error bars indicate standard deviations.
Susceptibility to Lcn972 of cell envelope L. lactis mutants.
To test the putative role of the PG structure on the Lcn972-resistant phenotype, an L. lactis dacA cell envelope mutant was generated. dacA encodes a d-Ala-d-Ala carboxypeptidase putatively involved in trimming away the last d-Ala residue from the PG pentapeptide. The chromosomal deletion of dacA was confirmed by PCR, and absence of the protein was evidenced by the lack of the respective bocillin FL-labeled 45-kDa protein band in membrane protein extracts analyzed by SDS-PAGE (data not shown). L. lactis dacA PG analysis revealed a dramatic decrease of muropeptides with tri- and tetrapeptide chains accompanied with a strong increase of muropeptides with pentapeptide side chains (Fig. 1), confirming a major role of DacA in PG maturation in L. lactis.
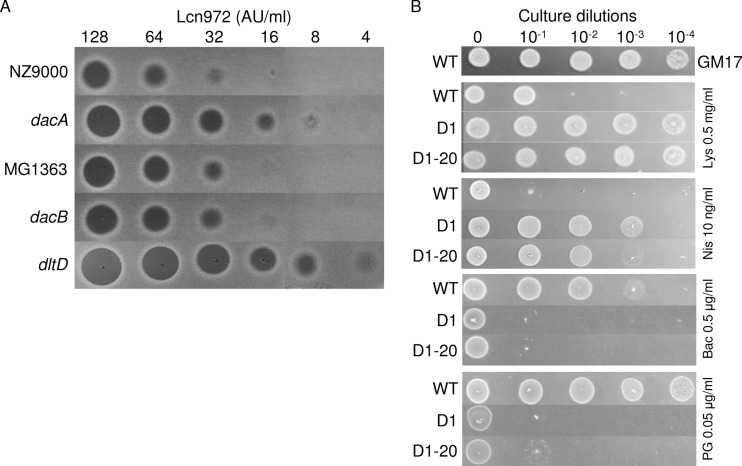
The L. lactis dacA mutant and other available L. lactis strains defective in cell wall modification enzymes (Table 1) were screened for their susceptibilities to Lcn972. The L. lactis dltD strain lacking DltD, a membrane protein involved in LTA d-alanylation, and the L. lactis dacB strain lacking the d,l-carboxypeptidase DacB, which cleaves the bond between l-Lys-d-Ala of the pentapeptide side chain, were assayed (Fig. 2). Despite the fact that no significant differences in the d-Ala content were observed among L. lactis MG1614 and strains D1 and D1-20, the L. lactis dltD strain devoid of d-Ala showed a 4-fold-increased susceptibility to Lcn972. Higher susceptibility was also observed in the case of the L. lactis dacA strain, whereas the L. lactis dacB strain showed no differences compared to the wild-type strain (Fig. 2). These results indicate that the high content of pentapeptide side chains present in dacA PG enhances the antimicrobial activity of Lcn972 and correlated well with their decrease in the Lcn972-resistant mutants (Fig. 1).
Fig 2.
Susceptibility of L. lactis strains to cell wall-active antimicrobials. (A) Susceptibility of different L. lactis cell envelope mutants to the bacteriocin Lcn972. Twofold dilutions of Lcn972 in 50 mM sodium phosphate buffer, pH 6.8, were spotted (5 μl) on L. lactis lawns. (B) Susceptibility of L. lactis MG1614 (WT) and the Lcn972-resistant strains L. lactis D1 and D1-20 to lysozyme (Lys), nisin (Nis), bacitracin (Bac), and penicillin G (PG). Tenfold dilutions of overnight cultures (5 μl) were spotted on GM17 plates containing the indicated concentrations of the antimicrobials.
Cross-resistance to cell wall-active antimicrobials.
In order to ascertain if resistance to Lcn972 in L. lactis could interfere with the activity of other cell wall inhibitors, we checked the susceptibility profile of L. lactis D1 and D1-20 and the parent MG1614 to lysozyme, which hydrolyzes the PG sugar chains; bacitracin, which inhibits recycling of the PG lipid carrier; penicillin G, which inhibits transpeptidation during the last stage of PG synthesis; and nisin, a pore-forming bacteriocin that also prevents cell wall biosynthesis by binding to lipid II. Both Lcn972r strains were more resistant to lysozyme and nisin and more susceptible to bacitracin and penicillin G than L. lactis MG1614 (Fig. 2). It is worth noting that nisin resistance in L. lactis D1 was higher than in D1-20, somewhat correlating with their resistance to Lcn972 (Table 2).
Resistance to bacteriophages.
Although rarely addressed, resistance to phage infection has been observed in nisin-resistant Staphylococcus aureus (24) and in a sakacin P-resistant Listeria monocytogenes (36). Considering the negative impact of bacteriophages in industrial dairy fermentations, the Lcn972r strains were challenged with phages c2 and sk1, two lytic phages belonging to the c2 and 936 families, respectively, and commonly found in the dairy environment. As judged by the efficiency of plaquing (EOP) referred to L. lactis MG1614 as the control, L. lactis D1 and D1-20 were fully resistant to phage c2 (Table 3). Phage sk1 was still able to infect both Lcn972r mutants, although resistance to sk1infection was more pronounced in L. lactis D1, somewhat correlating with the Lcn972-resistant phenotype. The distinct phage-resistant phenotype was further confirmed by the low adsorption of phage c2 to L. lactis D1, whereas in the case of sk1, adsorption to both Lcn972r mutants was not affected (Table 3).
Table 3.
Phage activity on L. lactis MG1614 and the Lcn972-resistant derivatives L. lactis D1 and L. lactis D1-20
| L. lactis strain | Phage susceptibility |
|||
|---|---|---|---|---|
| EOPa |
% adsorption |
|||
| c2 | sk1 | c2 | sk1 | |
| MG1614 | 1.00 ± 0.00 | 1.00 ± 0.00 | 92.6 ± 4.8 | 88.9 ± 4.5 |
| D1 | 0 | 0.23 ± 0.06 | 43.6 ± 8.0 | 93.7 ± 4.5 |
| D1-20 | 0 | 0.60 ± 0.06 | NDb | 94.9 ± 0.7 |
EOP, efficiency of plaquing.
ND, not determined.
Impaired growth on maltose and lack of c2 infection is based on a chromosomal deletion detected in L. lactis D1 and D1-20 resistant to Lcn972.
Prompted by the extreme resistance to phage c2 displayed by the L. lactis Lcn972r strains, we investigated further the molecular basis underlying this phenomenon. In the case of phage c2, the membrane protein Pip (phage infection protein) is required for phage adsorption and DNA injection (14). Thereby, we aimed at identifying mutations in pip which might have occurred upon adaptation to Lcn972. However, attempts to amplify pip in L. lactis D1 and D1-20 by PCR failed, until a forward primer, 0734F, located 20.6 kbp upstream of pip was used in combination with the reverse primer 0752R internal to pip. With these two primers, a 2.0-kbp PCR product was obtained on both L. lactis D1 and D1-20 DNAs. Sequencing of this PCR product revealed a large 22.6-kbp deletion expanding from llmg0736 to pip, which was further confirmed by Southern hybridization (Fig. 3). This deletion encompassed genes involved in maltose metabolism, the two-component system (TCS) F, and the 5′ end of pip and explained both the impaired growth on maltose and the phage-resistant phenotype based on the absence of a functional Pip protein.
Contribution of deleted genes to Lcn972 antimicrobial activity.
Experiments were carried out to confirm whether or not the genes included in the deleted region were directly involved in Lcn972 resistance. When L. lactis MG1614 was growing on maltose, i.e., inducing maltose metabolic genes, the Lcn972 MIC was 20 AU/ml, differing in only one dilution step from the MIC in glucose (10 AU/ml). Thus, activating maltose metabolism seems not to increase susceptibility to Lcn972, and a correlation to Lcn972 activity could not be established. On the other hand, the MIC of Lcn972 for L. lactis MGRrF, which lacks the response regulator LlrF of TCS F, was also two times more resistant than the wild-type L. lactis strain (20 AU/ml), pointing to a marginal role of this TCS in Lcn972 resistance.
DISCUSSION
The potent antimicrobial activity of bacteriocins has supported research for their application as food biopreservatives and as lead molecules for the design of new antibiotics. As development of resistance may impair their efficacy, subsequent studies have been addressed to understand the molecular basis of bacteriocin resistance and its impact in the physiology of otherwise susceptible bacteria. Mutants with different degrees of resistance toward bacteriocins may be easily selected under laboratory conditions after exposure to bacteriocins for several generations (3, 17, 19, 38). Mechanisms behind bacteriocin resistance mostly involve changes at the cell envelope that preclude the bacteriocins from reaching their target, mainly the plasma membrane. In this work, we have been able to isolate Lcn972r L. lactis mutants with reduced susceptibility to this cell wall-active bacteriocin. As described for nisin (19), the acquired Lcn972 high-resistant phenotype was lost quickly in the absence of selective pressure. Lcn972 is known to activate the two-component system CesSR (26), whose regulon comprises other regulatory protein genes. Thereby, an adaptive response which may include CesR-mediated gene activation seemed to be involved in the Lcn972 high-resistance phenotype. However, stable mutations have also occurred during selection, because a stable resistant phenotype (8× MIC) could be maintained in the absence of Lcn972.
Resistance to Lcn972 did not strongly alter the overall surface properties of the cells. A common mechanism described for many cationic antimicrobial peptides, including bacteriocins, consists of d-alanylation of LTA (15, 20, 32) to decrease the negative charge and reduce the electrostastic interactions. However, in the case of Lcn972, it does not play a major role, as no differences were observed both in the d-Ala content and the cell net charge. On the contrary, a remarkable change in the PG structure was observed. We could establish a direct correlation between the length of the peptide side chain and the susceptibility to Lcn972. Lcn972-resistant mutants had a reduced content of pentapeptide muropeptides. Moreover, L. lactis strains lacking the carboxypeptidase DacA and, consequently, with a high content of peptapeptide muropeptides were more susceptible to Lcn972. Changes at the PG structure level have not been approached when studying bacteriocin resistance mutants. Thus, it is not possible to anticipate if this is a common mechanism of resistance among bacteriocins. Nevertheless, it is worth noting that enhanced susceptibility toward beta-lactam antibiotics and bacitracin, both targeting PG biosynthesis, has been previously linked to resistance to lipid II-binding bacteriocins (8, 19). Altered antimicrobial susceptibility has been explained mostly by the role of cell envelope enzymes (e.g., dlt operon and penicillin binding protein genes) in the response to cell envelope stress, specialized ABC exporters, and specific and global regulators (7, 8, 19, 39). In the case of Lcn972, cross-protection to nisin and lysozyme might occur through the activation of CesSR or up-mutations in any of the CesR-regulated genes, but the alteration of the PG could also be important.
Another interesting result was the altered phage susceptibility profile of the Lcn972-resistant mutants. Lack of infectivity of phage c2 was undoubtedly linked to the truncation of pip by the chromosomal deletion detected in the Lcn972-resistant strains. In fact, mutated versions of this protein have been previously correlated with resistance to c2 (28). On the other hand, the lower infectivity of sk1 remained unexplained. Resistance to sk1 has been correlated with the absence of a polysaccharide pellicle in L. lactis MG1363 (6). However, the Lcn972-resistant strains have a gross carbohydrate composition similar to that of the wild type, and the pellicle was visible in the micrographs. Therefore, there must be some other as-yet-unidentified factors that compromise phage infectivity. Tentatively, the altered tripeptide/pentapeptide ratio in the PG of the Lcn972r strains could make it less susceptible to the phage endolysin, which is needed to release the phage progeny. Interestingly, sk1 plaques on the Lcn972r strains were consistently smaller (data not shown).
The deletion found in the Lcn972-resistant strains was rather large and encompassed, besides pip, maltose metabolic genes and TCS F. Compelling data have been gathered connecting carbohydrate metabolism with resistance, namely, to class IIa bacteriocins that target the mannose phosphotransferase system-PTSman (18, 31, 36). Impaired growth on cellobiose has also been correlated with tolerance to Lcn972 in producing strains (5). However, our results indicated that maltose metabolism does not play a major role, as cells are similarly susceptible, even slightly more resistant, when growing on maltose. On the other hand, TCS F has been reported to be relevant under oxidative stress (30). According to our results, only a minor role in Lcn972 resistance could be recognized. Therefore, there must be some other mutations that account for the Lcn972-resistant phenotype. In fact, the histidine kinase KinF has been reported to be essential (30), and it is likely that countermutations have been selected. Transcriptomic analyses are in progress in order to clarify the molecular basis underlying Lcn972 resistance.
ACKNOWLEDGMENTS
The work has been funded by grants BIO2007-65061 and BIO2010-17414 of the Ministerio de Ciencia e Innovación (Spain). C.R. is a recipient of a predoctoral JAE-CSIC fellowship. Work of M.-P.C.-C. and S.K. was supported by an INRA Jeune Equipe grant.
We thank Christine Longin and Sophie Chat for electron microscopy observations with the equipment available on the MIMA2 platform (INRA, Jouy-en-Josas, France) and Romain Briandet (INRA, Micalis, Jouy-en-Josas, France) for his help with electophoretic mobility experiments. We are also grateful to Verónica Pérez (IPLA-CSIC, Spain) for her technical assistance in performing growth curves and to Elena Bidnenko (INRA, France) and Mary O'Connell-Motherway (UCC, Ireland) for supplying L. lactis phages and L. lactis MGRrF, respectively.
Footnotes
Published ahead of print 13 April 2012
REFERENCES
- 1. Andre G, et al. 2010. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat. Commun. 1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bellon-Fontaine MN, Rault J, van Oss CJ. 1996. Microbial adhesion to solvents: a novel method to determine the electron donor/electron acceptor or Lewis acid-base properties of microbial cells. Colloids Surf. B 7:47–53 [Google Scholar]
- 3. Bennik MH, et al. 1997. Interactions of nisin and pediocin PA-1 with closely related lactic acid bacteria that manifest over 100-fold differences in bacteriocin sensitivity. Appl. Environ. Microbiol. 63:3628–3636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breukink E, et al. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364 [DOI] [PubMed] [Google Scholar]
- 5. Campelo AB, et al. 2011. The Lcn972 bacteriocin-encoding plasmid pBL1 impairs cellobiose metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 77:7576–7585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chapot-Chartier M-P, et al. 2010. Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J. Biol. Chem. 285:10464–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collins B, Curtis N, Cotter PD, Hill C, Ross RP. 2010. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various β-lactam antibiotics Antimicrob. Agents Chemother. 54:4416–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cotter PD, Guinane CM, Hill C. 2002. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob. Agents Chemother. 46:2784–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Courtin P, et al. 2006. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an l,d-carboxypeptidase involved in peptidoglycan maturation. J. Bacteriol. 188:5293–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duwat P, Cochu A, Ehrlich SD, Gruss A. 1997. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 179:4473–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fallico V, Ross RP, Fitzgerald GF, McAuliffe O. 2011. Genetic response to bacteriophage infection in Lactococcus lactis reveals a four-strand approach involving induction of membrane stress proteins, d-alanylation of the cell wall, maintenance of proton motive force, and energy conservation. J. Virol. 85:12032–12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garneau JE, Moineau S. 2011. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Fact. 10(Suppl 1):S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gasson MJ. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geller BL, Ivey RG, Trempy JE, Hettinger-Smith B. 1993. Cloning of a chromosomal gene required for phage infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 175:5510–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giaouris E, Briandet R, Meyrand M, Courtin P, Chapot-Chartier M-P. 2008. Variations in the degree of d-alanylation of teichoic acids in Lactococcus lactis alter resistance to cationic antimicrobials but have no effect on bacterial surface hydrophobicity and charge. Appl. Environ. Microbiol. 74:4764–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glauner B, Holtje JV, Schwarz U. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263:10088–10095 [PubMed] [Google Scholar]
- 17. Guinane CM, Cotter PD, Hill C, Ross RP. 2006. Spontaneous resistance in Lactococcus lactis IL1403 to the lantibiotic lacticin 3147. FEMS Microbiol. Lett. 260:77–83 [DOI] [PubMed] [Google Scholar]
- 18. Kjos M, Nes IF, Diep DB. 2011. Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl. Environ. Microbiol. 77:3335–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kramer NE, van Hijum SA, Knol J, Kok J, Kuipers OP. 2006. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob. Agents Chemother. 50:1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kramer NE, et al. 2008. Increased d-alanylation of lipoteichoic acid and a thickened septum are main determinants in the nisin resistance mechanism of Lactococcus lactis. Microbiology 154:1755–1762 [DOI] [PubMed] [Google Scholar]
- 21. Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM. 1998. Quorum sensing controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15–21 [Google Scholar]
- 22. Leenhouts K, et al. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217–224 [DOI] [PubMed] [Google Scholar]
- 23. Martínez B, et al. 2008. Specific interaction of the unmodified bacteriocin lactococcin 972 with the cell wall precursor lipid II. Appl. Environ. Microbiol. 74:4666–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martínez B, Obeso JM, Rodríguez A, García P. 2008. Nisin-bacteriophage cross-resistance in Staphylococcus aureus. Int. J. Food Microbiol. 122:253–258 [DOI] [PubMed] [Google Scholar]
- 25. Martínez B, Suárez JE, Rodríguez A. 1996. Lactococcin 972: a homodimeric lactococcal bacteriocin whose primary target is not the plasma membrane. Microbiology 142:2393–2398 [DOI] [PubMed] [Google Scholar]
- 26. Martínez B, Zomer AL, Rodríguez A, Kok J, Kuipers OP. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64:473–486 [DOI] [PubMed] [Google Scholar]
- 27. Mäyra-Mäkinen A, Bigret M. 2005. Industrial use and production of lactic acid bacteria, p 175–198 In Salminen S, Ouwehand AC, vonWright A. (ed), Lactic acid bacteria, microbiological and functional aspects, 3rd ed Marcel Dekker, New York, NY [Google Scholar]
- 28. Mooney DT, Jann M, Geller BL. 2006. Subcellular location of phage infection protein (Pip) in Lactococcus lactis. Can. J. Microbiol. 52:664–672 [DOI] [PubMed] [Google Scholar]
- 29. Neves AR, et al. 1999. In vivo nuclear magnetic resonance studies of glycolytic kinetics in Lactococcus lactis. Biotechnol. Bioeng. 64:200–212 [DOI] [PubMed] [Google Scholar]
- 30. O'Connell-Motherway M, et al. 2000. Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146:935–947 [DOI] [PubMed] [Google Scholar]
- 31. Opsata M, Nes IF, Holo H. 2010. Class IIa bacteriocin resistance in Enterococcus faecalis V583: the mannose PTS operon mediates global transcriptional responses. BMC Microbiol. 10:224 doi:10.1186/1471-2180-10-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peschel A, et al. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405–8410 [DOI] [PubMed] [Google Scholar]
- 33. Pinto JP, Kuipers OP, Marreddy RK, Poolman B, Kok J. 2011. Efficient overproduction of membrane proteins in Lactococcus lactis requires the cell envelope stress sensor/regulator couple CesSR. PLoS One 6:e21873 doi:10.1371/journal.pone.0021873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roces C, et al. 2009. Contribution of the CesR-regulated genes llmg0169 and llmg2164-2163 to Lactococcus lactis fitness. Int. J. Food Microbiol. 133:279–285 [DOI] [PubMed] [Google Scholar]
- 35. Sambrook J, Maniatis T, Fritsch EF. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Tessema GT, Møretrø T, Snipen L, Axelsson L, Naterstad K. 2011. Global transcriptional analysis of spontaneous sakacin P-resistant mutant strains of Listeria monocytogenes during growth on different sugars. PLoS One 6:e16192 doi:10.1371/journal.pone.0016192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tremblay DM, et al. 2006. Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J. Bacteriol. 188:2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vadyvaloo V, et al. 2004. Cell-surface alterations in class IIa bacteriocin-resistant Listeria monocytogenes strains. Microbiology 150:3025–3033 [DOI] [PubMed] [Google Scholar]
- 39. Veiga P, et al. 2007. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 282:19342–19354 [DOI] [PubMed] [Google Scholar]
- 40. Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149–167 [DOI] [PubMed] [Google Scholar]
- 41. Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ. 2011. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol. Microbiol. 82:1096–1109 [DOI] [PubMed] [Google Scholar]
- 42. Wiedemann I, et al. 2006. The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 61:285–296 [DOI] [PubMed] [Google Scholar]