Abstract
Four types of β-1,3-1,4 glucanase (β-glucanase, EC 3.2.1.73) genes, designated bglA13, bglA16, bglA51, and bglM2, were found in the cDNA library of Neocallimastix patriciarum J11. All were highly homologous with each other and demonstrated a close phylogenetic relationship with and a similar codon bias to Streptococcus equinus. The presence of expansion and several predicted secondary structures in the 3′ untranslated regions (3′UTRs) of bglA16 and bglM2 suggest that these two genes were duplicated recently, whereas bglA13 and bglA16, which contain very short 3′UTRs, were replicated earlier. These findings indicate that the β-glucanase genes from N. patriciarum J11 may have arisen by horizontal transfer from the bacterium and subsequent duplication in the rumen fungus. β-Glucanase genes of Streptococcus equinus, Ruminococcus albus 7, and N. patriciarum J11 were cloned and expressed by Escherichia coli. The recombinant β-glucanases cloned from S. equinus, R. albus 7, and N. patriciarum J11 were endo-acting and had similar substrate specificity, but they demonstrated different properties in other tests. The specific activities and catalytic efficiency of the bacterial β-glucanases were also significantly lower than those of the fungal β-glucanases. Our results also revealed that the activities and some characteristics of enzymes were changed during the horizontal gene transfer event. The specific activities of the fungal β-glucanases ranged from 26,529 to 41,209 U/mg of protein when barley-derived β-glucan was used as the substrate. They also demonstrated similar pH and temperature optima, substrate specificity, substrate affinity, and hydrolysis patterns. Nevertheless, BglA16 and BglM2, two recently duplicated β-glucanases, showed much higher kcat values than others. These results support the notion that duplicated β-glucanase genes, namely, bglA16 and bglM2, increase the reaction efficiency of β-glucanases and suggest that the catalytic efficiency of β-glucanase is likely to be a criterion determining the evolutionary fate of duplicate forms in N. patriciarum J11.
INTRODUCTION
The mixed-linked β-1,3–1,4-glucan (β-glucan) is the predominant cell wall polysaccharide in the endosperm of cereals, such as oat, wheat, and barley, and accounts for up to 5.5% of the dry weight of grains (18). β-1,3-1,4-Glucanase (EC 3.2.1.73; β-glucanase, lichenase) is able to specifically cleave β-1,4-glycosidic linkages adjacent to a 3-O-substituted glucose residue in mixed-linked β-glucans (29). β-Glucanases have received tremendous attention because of their importance in biotechnological applications. The addition of β-glucanases not only reduces brewer mash viscosity and turbidity but also increases yields of extracts, thereby leading to high-quality brewers malt (5). They are also added to poultry feed to eliminate the antinutrimental effects caused by β-glucans in the endosperm of cereals and to improve the conversion efficiency of feed (1). The synergistic action of β-glucanase and β-glucosidase results in improved efficiency of saccharification and fermentation of ethanol from lichenan (23). Enzymes with high catalytic activity could reduce the quantity of enzyme supplements required during industrial processing, potentially leading to an increase in enzyme utilization efficiency through cost reduction. Therefore, the selection of a β-glucanase with high activity is of significant interest to the brewing industry and animal feed industry, as well as other industries.
Anaerobic rumen fungi are members of the microflora in the gastrointestinal tracts of herbivores. They are able to penetrate and weaken plant tissue in vivo and produce a variety of enzymes that degrade plant materials ingested by the host animals (2). However, the flow of digesta through the rumen of ruminants is quick, and studies have demonstrated that large particles in grass leaves and stems are only retained in the rumen of sheep for 11 to 12 h (24). The presence of duplicate genes increases the efficiency of plant debris utilization in the rumen ecosystem because excess proteins are provided (33). Previous studies have shown that multiple mannanase genes in Piromyces sp. and cellulase genes in Orpinomyces sp. strain PC-2 arose by gene duplication (6); however, the evolutionary fate of the duplicated genes in rumen fungi has rarely been examined. In the present study, four duplicate β-glucanase genes with high activities were screened from a cDNA library of Neocallimastix patriciarum J11. Sequence composition and phylogenetic analysis were used to compare their evolutionary relationships. The characteristics of recombinant β-glucanases were further examined, and the evolutionary fate and characteristic diversification of recombinant enzymes of duplicated genes is also discussed.
MATERIALS AND METHODS
Microbial strains and media.
The anaerobic fungus Neocallimastix patriciarum J11 was isolated from the feces of water buffalo as described by Chen et al. (8). N. patriciarum J11 was inoculated in rumen fluid containing medium supplemented with 0.5% (wt/vol) barley-derived β-glucan (Megazyme International Ireland, Ltd.) for cDNA library construction under anaerobic conditions at 39°C for 72 h. Streptococcus equinus BCRC 12578 was purchased from the Bioresource Collection and Research Centre (Shinchu, Taiwan). Ruminococcus albus 7 was kindly provided by Han-Tsung Wang (Department of Animal Science, Chinese Culture University, Taiwan) (30). S. equinus and R. albus 7 were batch cultured in brain heart infusion broth and Scoot & Dehority broth, respectively. Escherichia coli DH5α (Invitrogen, Carlsbad, CA), grown on LB medium (Difco, Detroit, MI), was used as the host for the various plasmid constructions. The resultant vectors were transformed into E. coli BL21(DE3) (Novagen, Inc., Germany) for the production of recombinant proteins.
Cloning and analysis of β-glucanases genes.
Total RNA was extracted by TRIzol reagent (Invitrogen), and mRNAs were purified using a Dynabead mRNA purification kit (Invitrogen). The N. patriciarum J11 cDNA library was constructed using the SMART cDNA library construction kit (BD Bioscience, Palo Alto, CA) packaged with the Gigapack III plus the packing extract kit (Stratagene, San Diego, CA) according to the manufacturer's instructions. The plaques were grown on LB soft agar plates containing 0.1% (wt/vol) barley-derived β-glucan, and the cDNA library was screened for β-glucanase activity using the results of Congo red staining (27). The cDNA plasmids used for directional sequencing and sequential cloning were converted by in vivo excision from the lambda TriplEx2 recombinants in E. coli BM25.8 (BD Bioscience). To construct each expression vector, the fragments of β-glucanases of the rumen fungus were amplified by PCR using the primers Fw (forward, GGATCCAAAARTWTATTATCTATTGC) and Rev (reverse, CTCGAGRTTTCTTGGGGCATCA). Two primer sets—SeFw (CCGGAATTCGAATTTCGTAGCGGAACGATTG)/SeRev (CCGCTCGAGAAATTTATCATAACTAATCCAGTCAT) and Ra7Fw (CGCGGATCCGAACGTTTCAACGGAAGTTATTTC)/Ra7Rev (CCGCTCGAGGTTAACTGCTTCGATATTCCAAAG)—were designed based the reported sequences of S. equinus (accession no. Z92911) and R. albus 7 (accession no. NC_014833.1) and used for the amplification of the β-glucanase gene from genomic DNA samples of S. equinus and R. albus, respectively. The underlined sequences are the cutting sites of the restriction enzymes. Each β-glucanase gene was amplified, purified, digested, and ligated into pET21a (Novagen). The resultant plasmids were transformed into E. coli BL21(DE3) to express recombinant proteins. All recombinant proteins had the T7 tags and six-histidine tags at their N and C termini.
The computer program Bioedit was used to analyze and align the sequences of β-glucanase (15). The ORFinder server (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to estimate the regions of open reading frames (ORFs). β-Glucanase sequences (EC 3.2.1.73) were retrieved from the CAZy database (16). A phylogenetic tree based upon the neighbor-joining (NJ) algorithm was generated using MEGA 4.0 software (26). The resultant unrooted tree topologies were evaluated according to bootstrap analysis of the NJ method based upon 1,000 resamplings using the Seqboot and consensus programs in the MEGA package. Relative synonymous codon usage (RSCU) is defined as the ratio of the observed frequency of a codon to the expected frequency if all of the synonymous codons for that amino acid were used equally (20). The RSCU in the coding domain sequences was analyzed using the GCUA program (22). The RNA secondary structure was predicted using the GeneBee server (3).
Recombinant protein expression and purification.
The β-glucanase gene transformants were grown in 200 ml of LB to an optical density at 600 nm of 0.6 to 0.9 before 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added for induction. After 3 h of induction at 30°C, the cells were harvested by centrifugation (4°C, 4,000 × g, 20 min) and resuspended in 20 ml of CelLytic B reagent for crude β-glucanase preparation. Fusion protein purification was performed by liquid chromatography using CM Sepharose FF and nickel affinity columns (GE Healthcare Bio-Sciences AB). Desalting and buffer exchange of the eluted fraction were performed using a HiTrap Desalting column (GE Healthcare Bio-Sciences AB). The molecular mass and β-glucanase activity of the recombinant enzymes were estimated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and zymography (17). Protein concentrations were determined using a Micro BCA protein assay reagent kit (Pierce Biotechnology, Rockford, IL).
Enzyme activity assays.
The activity of β-glucanase was assayed by measuring the liberation of reducing sugar from 1% (wt/vol) β-glucan dissolved in citrate buffer (50 mM, pH 6). The mixture was incubated in a water bath at 50°C for 10 min. Reactions were terminated by adding dinitrosalicylic acid (DNS) reagent and boiling for 5 min. The absorbance obtained with different amounts of reducing sugar, liberated as a result of enzymatic hydrolysis, was measured using a plate reader at 540 nm. One unit (U) is defined as the amount of enzyme that releases 1 μmol of reducing sugar per min (9). For the determination of Km and kcat values, three different assays were performed at pH 6.0 and 40°C with barley β-glucan as the substrate at concentrations from 0 to 2%. The kinetic parameters of β-glucanase were derived from Lineweaver-Burk plots constructed using the program Enzfitter (version 2.0.18.0; Biosoft, United Kingdom) (9).
Enzyme properties.
The relative activity was determined at several temperatures and several pH levels. The range of buffers used in preparing 1% β-glucan solution for detecting β-glucanase activity comprised citrate buffer (pH 3 to 6), phosphate buffer (pH 6 to 7), and Tris buffer (pH 7 to 9) (10). The buffers mixed with enzyme were incubated at 50°C for 10 min, and the released reducing sugar was measured by the DNS method. The optimal temperature for enzyme activity was determined by performing the standard assay procedure at a range of temperatures (10 to 100°C). All subsequent enzyme assays were performed at the optimum temperature.
Enzymes were also tested for their specificity to hydrolyze a variety of substrates, including 1% (wt/vol) oat-spelt xylan, 1% (wt/vol) lichenan, 1% (wt/vol) pachyman (Megazyme), 1% (wt/vol) β-glucan, 1% (wt/vol) carboxymethyl cellulose (CMC), 1% (wt/vol) starch, and 1% (wt/vol) Avicel (PH101; Asahi, Japan). These substrates were suspended in citrate buffer (50 mM, pH 6). The reaction mixture consisting of equal volumes of substrate solution and enzyme solution was incubated at 50°C for 30 min. Hydrolyzed products were determined by DNS methods (9).
The reaction mixture (250 μl), consisting of equal volumes of a 2% (wt/vol) suspension of β-glucan and the purified enzymes (50 U ml−1) in 50 mM citrate buffer (pH 6.0) was incubated at 40°C for 4 h. Hydrolysis was terminated by boiling for 10 min, and the hydrolyzed products were analyzed by thin-layer chromatography (TLC) on silica gel plates (Merck AG, Darmstadt, Germany). The TLC plates were developed twice at room temperature with a solvent system comprising ethyl acetate, acetic acid, formic acid, and water (9:3:1:4 [vol/vol]). Spots were stained by spraying the plates with orcinol-sulfuric acid reagent and then heating them at 100°C for 5 min (10). Glucose, cellulobiose, and cello-oligosaccharides were selected for the TLC standards. Each experiment was conducted in triplicate. All chemicals in the present study were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the β-glucanase genes were deposited in GenBank, and the accession numbers are shown in Table 1.
Table 1.
Composition of N. patriciarum β-1,3-1,4-glucanase cDNAs
| Gene (accession no.) | No. of transcript copies | Length (bp) | G+C (%) | CDSa |
5′UTR |
3′UTR |
Poly(A) tail length (bp) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | G+C (%) | Similarityb (%) | Length (bp) | G+C (%) | Length (bp) | G+C (%) | |||||
| bglA13 (JN376084) | 3 | 940 | 29.04 | 732 | 35.52 | 100 | 154 | 8.44 | 24 | 1.85 | 30 |
| bglA16 (JN376085) | 6 | 1,405 | 22.07 | 732 | 34.84 | 97.1 | 159 | 7.55 | 483 | 8.37 | 30 |
| bglA51 (JN376083) | 2 | 955 | 29.11 | 732 | 35.56 | 94.8 | 172 | 9.88 | 58 | 3.92 | 26 |
| bglM2 (JN376082) | 7 | 1,091 | 25.67 | 732 | 34.84 | 96.7 | 185 | 7.61 | 144 | 7.61 | 30 |
CDS, coding domain sequence.
Percent similarity values were calculated according to the similarity to bglA13.
RESULTS
Screening of β-glucanase genes.
The cDNA library consisting of 106 titers was constructed and screened for plaques hydrolyzing β-glucan. Eighteen plaques showing specific activity to β-glucan were retained. The plasmids with cDNA sequences encoding β-glucanase were obtained by in vivo excision from the selected plaques. Sequencing of both ends of the cDNAs inserted in plasmids showed that four types of β-1,3-1,4-glucanase sequences, designated bglA13, bglA16, bglA51, and bglM2, were found in the 18 sequenced plasmids. After sequencing, three, six, two, and seven inserts showed identical results with the sequences of bglA13, bglA16, bglA51, and bglM2, respectively.
The lack of introns in the β-glucanase genes was confirmed by size comparison of ORF regions that had been amplified from genomic DNA and cDNAs, respectively (data not shown). 5′-Untranslated regions (5′UTRs), ORFs, 3′UTRs, and poly(A) tails present in a complete transcript of eukaryote DNA were found in the sequences of four β-glucanase genes (Table 1). This finding indicates that four cDNAs were transcribed from different β-glucanase loci and did not arise from alternative splicing of mRNA transcribed from one β-glucanase gene (19).
Nucleotide and deduced amino acid sequences of β-glucanase genes.
The total length of bglM2, bglA13, bglA16, and bglA51 ranged from 940 to 1,405 bp. As shown in Table 1, all contained ORFs encoding 243 amino acids with an estimated molecular mass of 27 kDa. The sequences encoding β-glucanase could be found in the four ORFs, but those encoding the conserved region of the carbohydrate-binding domain or docking domain did not locate in these genes. The G+C ratios of the ORFs of the four genes were close to 35%, as shown in Table 1. Noncoding regions, including 5′UTRs and 3′UTRs, demonstrated extremely low G+C nucleotide contents. The sequences of putative coding regions show a particularly high homology with each other. The highly conserved nucleotide sequence in the coding domain sequences suggests that the four β-glucanase genes apparently arose by gene duplication (32).
Most regions of the four β-glucanase genes are similar, but they vary significantly in the lengths of the 3′UTRs. The 3′UTR lengths in bglA16 and bglM2 were much longer than those in bglA13 and bglA51 (Table 1). The terminal codon sequences of the four β-glucanase genes were aligned, and differences in the 3′UTRs among the four β-glucanase genes were found. Premature stop codons (PSCs), stop codons in the second and third reading frames of a protein-coding region, were found in the 5′ terminals of each 3′UTR of the β-glucanase genes (see Fig. S1 in the supplemental material). The poly(A) tails of the mRNAs were very similar to the PSC regions of bglA13 and bglA51, whereas the PSC regions and poly(A) tails of bglA16 and bglM2 were separated by 474- and 132-bp sequences, respectively. Several complementary sequences such as the regions shown in Fig. S1 in the supplemental material were found in the 3′UTRs of bglA16 and bglM2, and RNA loops possibly involved in translational regulation were predicted in these regions. In contrast, the lengths between the stop codon and the poly(A) tail are short in the 3′UTRs of bglA13 and bglA51, and no secondary structure formation could be predicted in these regions. These results indicate that duplicate β-glucanase genes might be regulated diversely.
Analysis of β-glucanase relatedness.
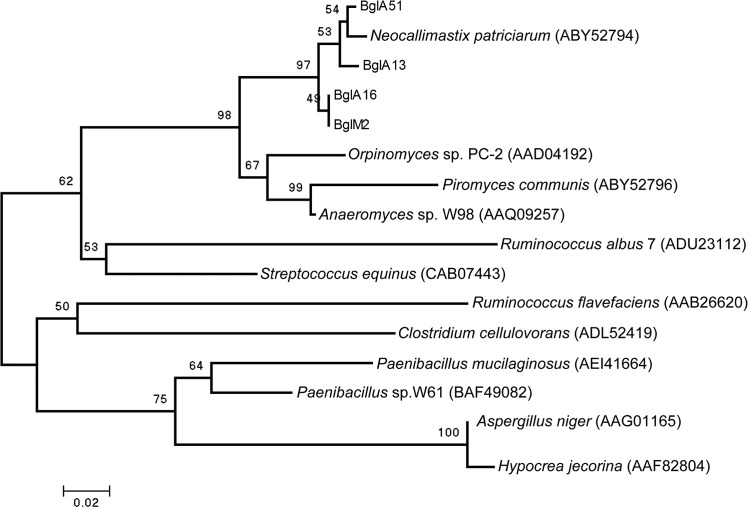
Minimum-evolution-based algorithms were used in the phylogenetic analyses as mentioned above. Figure 1 depicts the relationships between 16 β-glucanase amino acid sequences found in bacteria and fungi. As shown in Fig. 1, β-glucanases from rumen fungi are more phylogenetically related to those of bacteria such as S. equinus and R. albus 7 than to that of Aspergillus niger, an aerobic fungus. The results suggest that four β-glucanases of N. patriciarum J11 have a bacterial origin.
Fig 1.
Relatedness tree based upon conserved amino acid sequences of 16 β-1,3-1,4-glucanases from bacteria and fungi. An unrooted consensus tree was generated using the minimum-evolution algorithm. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown above the branches. The accession numbers of amino acid sequences are listed at the end of the scientific name of the microorganisms.
The values of RSCU in the coding regions of four β-glucanase genes from N. patriciarum J11 and the genes from S. equinus and R. albus 7 are listed in Table S2 in the supplemental material. The codon bias is significant and the codon usage shows a higher correlation with A+T abundance in the genome of Neocallimastix, as described previously (4). Four β-glucanase genes demonstrated highly similar preferences for one of the several codons that encode the same amino acid. The β-glucanase genes from N. patriciarum J11 and S. equinus demonstrated similar codon bias; nevertheless, codon bias differed significantly between the β-glucanase gene from R. albus 7 and the β-glucanase genes from N. patriciarum J11 and S. equinus. These results indicate that the β-glucanase genes of N. patriciarum J11 are more closely related to that of S. equinus than to the R. albus 7 gene.
The β-glucanases belonging to N. patriciarum were clustered together in a specific clade and were divided into two groupings for the analysis presented in Fig. 1. BglM2 and BglA16 were grouped in one subgroup, while BglA13 and BglA51 were grouped into another subgroup. Moreover, β-glucanase obtained from another isolate of N. patriciarum also belonged to the subgroup containing BglA13 and BglA51. The result indicates that evolutionary divergence was generated in these two subgroups of duplicate β-glucanase genes from N. patriciarum J11 and that the level of divergence exceeded the individuality between strains of the same species.
Activities of recombinant β-glucanases.
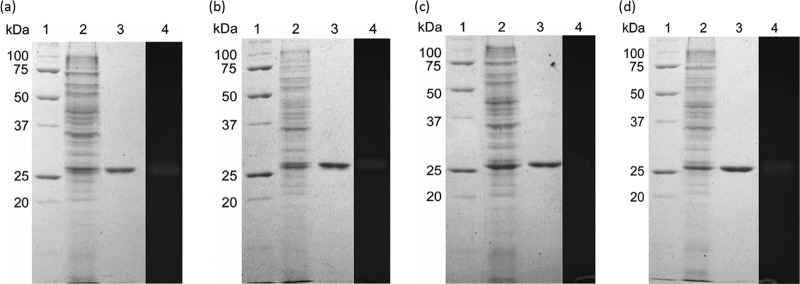
The recombinant β-glucanases were expressed by E. coli BL21(DE3). The crude enzyme was collected from the cell lysate by centrifugation and then purified. After cation exchange and affinity chromatography, fractions containing β-glucanase activity were collected and separated by SDS-PAGE, resulting in distinct bands with an estimated molecular mass of 27 kDa in each gel (Fig. 2). The sizes of BglA13, BglA16, BglA51, and BglM2 were all close to their predicted molecular masses. The crude enzymes also demonstrated β-glucanase activities against β-glucan in the zymogram analyses (Fig. 2) and formed distinct bands with apparent molecular masses of 27 kDa. These results indicated that four β-glucanases were successfully expressed and purified from transformants.
Fig 2.
Coomassie blue-stained gels and zymograms of recombinant β-glucanases expressed by E. coli BL21(DE3) harboring pET21a vectors containing bglA13 (a), bglA16 (b), bglA51 (c), and bglM2 (d) genes. Lane 1, protein standards; lane 2, cell lysate of E. coli BL21(DE3); lane 3, purified β-glucanase; lane 4, location of the β-glucanase activity of crude enzymes expressed by E. coli transformants on the zymogram gel. Proteins were separated on the same 10% acrylamide gels with additional 1% barley β-glucan. The gels for zymogram analysis were stained with 0.1% Congo red solution after citrate buffer (50 mM, pH 6) equilibration.
The specific activities of purified BglA13, BglA16, BglA51, and BglM2 were calculated to be 32,095, 39,524, 26,529, and 41,209 U mg of protein−1, respectively, when reacted with β-glucan (Table 2). β-Glucanase genes cloned from S. equinus (designated bglS) and R. albus 7 (designated bglR) were also expressed. The specific activities of the purified BglS and BglR were 405 and 267 U mg of protein−1, respectively.
Table 2.
Enzymatic activity and kinetic parameters of β-1,3-1,4-glucanase genes expressed in E. coli
| Parameter | Enzyme |
|||||
|---|---|---|---|---|---|---|
| BglA13 | BglA16 | BglA51 | BglM2 | BglS | BglR | |
| Enzyme sp act (U mg of protein−1) | 32,095 | 39,524 | 26,529 | 41,209 | 405 | 267 |
| Kinetics | ||||||
| Km (mM ml−1) | 0.86 | 0.90 | 1.19 | 0.94 | 2.78 | 4.45 |
| Vmax (mM min−1) | 1.26 | 1.30 | 1.85 | 1.57 | 0.41 | 0.50 |
| kcat (min−1) | 3,783.6 | 5,033.4 | 4,091.3 | 7,996.4 | 378.6 | 381.5 |
| kcat/Km (ml mg−1 min−1) | 439.6 | 559.7 | 342.6 | 848.7 | 136.2 | 85.8 |
The results of kinetic analysis are listed in Table 2. A comparison of the Km values suggested that BglA13, BglA16, and BglM2 were similar for barley β-glucan, indicating a similar affinity. Nevertheless, the kcat value for BglM2 was considerably higher than the kcat values for other β-glucanases, indicating more rapid hydrolysis by BglM2. This also leads to greater catalytic efficiency of BglM2 toward β-glucan when judged by the kcat/Km values. BglA16 also demonstrated higher kcat and kcat/Km values than BglA13 and BglA51. Values of 2.78 mM ml−1 and 378.6 min−1 were determined for the Km and kcat of β-glucanases from S. equinus. Values of 4.45 mM ml−1 and 381.5 min−1 were calculated for Km and kcat of β-glucanases from R. albus. These results revealed that the substrate affinity and catalytic rate of the recombinant bacterial β-glucanases were significantly lower than those of the recombinant fungal β-glucanases.
Properties of recombinant β-glucanases.
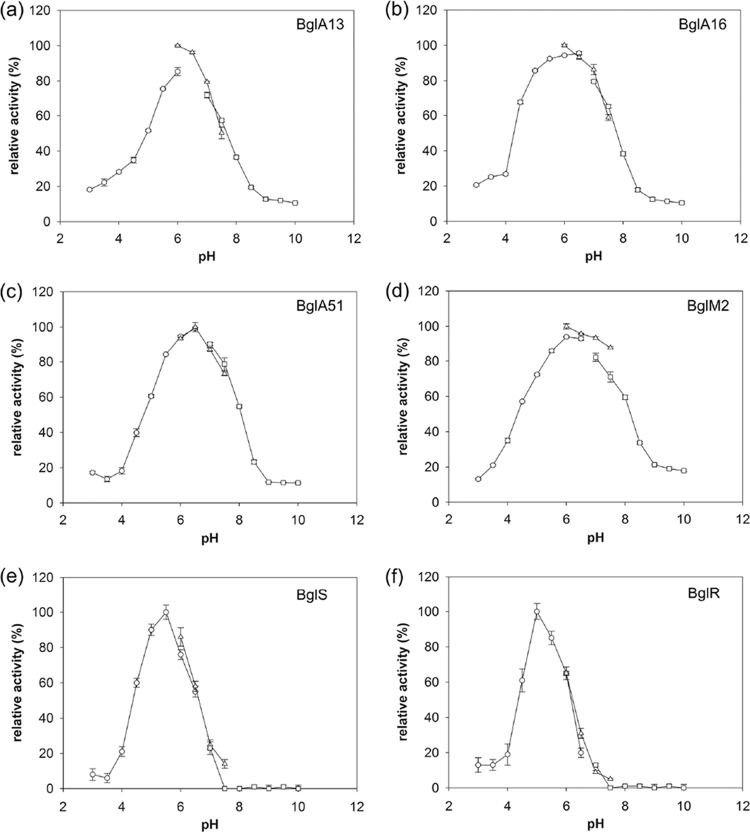
The effects of temperature variation upon the activity of six recombinant β-glucanases are illustrated in Fig. 3. BglA13, BglA16, and BglA51 exhibited maximum activity at 45°C in a 30-min assay with β-glucan as the substrate, and the activity was significantly decreased when the temperature was higher than 50°C. BglM2 showed an optimal activity at 50°C, and the activity was markedly decreased when the temperature was >55°C. The optimal reaction temperatures of BglS and BglR were 40 and 50°C, respectively. The activity was quickly reduced when the temperature reached above 60°C. Examination of the effect of pH upon β-glucanases activities revealed the following features: the optimum pH values were pH 6.0 for BglA13, BglA16, and BglM2, and the pH value for optimal reaction was pH 6.5 for BglA51 (Fig. 4). The optimal reaction pH value for BglS and BglR was pH 5.
Fig 3.
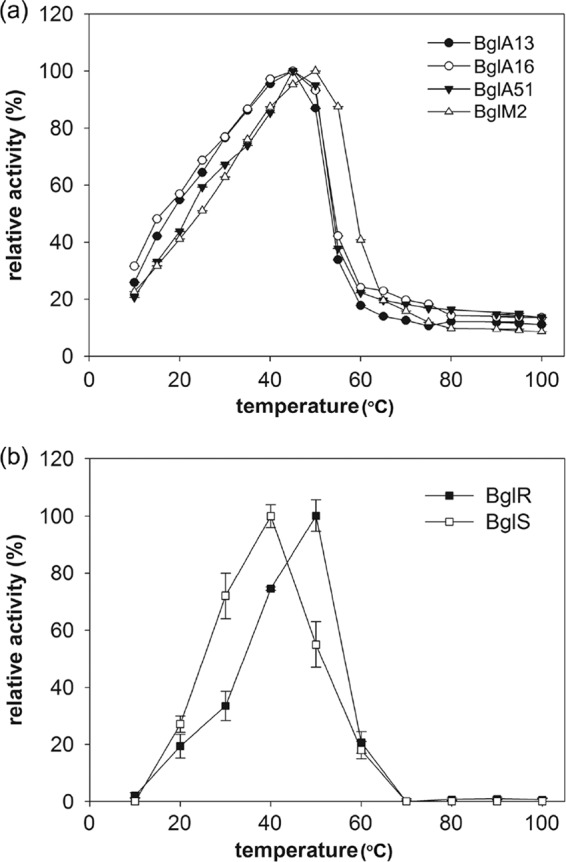
Optimal temperature of recombinant β-glucanases. (a) BglA13, BglA16, BglA51, and BglM2; (b) BglR and BglS. Purified enzymes in substrate solution were reacted at a range of temperatures (10 to 100°C) in order to examine the pH optima. Enzyme activities are compared to the largest activity value attained (i.e., 100%).
Fig 4.
Effects of pH on β-glucanases expressed by Escherichia coli BL21(DE3) harboring pET21a vectors containing the bglA13 (a), bglA16 (b), bglA51 (c), bglM2 (d), bglS (e), and bglR (f) genes. The optimal reaction pH values of the β-glucanases were analyzed in different buffers with specific pH values (pH 3 to 10). Enzyme activities are compared to the largest activity value attained (i.e., 100%).
The hydrolysis products released from barley β-glucan by recombinant β-glucanases were separated by TLC. A series of cellulo-oligosaccharides were found on the TLC plate. The predominant products of β-glucan hydrolysis were cellobiosyltriose and cellotriosyltetraose. Cellotetrasylopentaose, cellopentasylhexaose, cellopentaose, and cellohexaose were also found on the TLC plate. Glucose could be found in the reaction mixture hydrolyzed by BglS (see Fig. S2 in the supplemental material). The hydrolysis patterns revealed that the mode of action of all recombinant β-glucanases belongs to endo-type β-glucanases (29).
The substrate specificity of recombinant β-glucanases against different polysaccharide substrates is listed in Table S2 in the supplemental material. BglA13, BglA16, BglA51, BglM2, BglS, and BglR had the highest activity against barley β-glucan. Lichenan was also hydrolyzed by these β-glucanases. All β-glucanases were inactive against pachyman, laminarin, starch, CMC, Avicel, and xylan. According to previous studies of substrate specificity, the four recombinant β-glucanases specifically act on substrates with β-1,4-glycosidic linkages adjacent to a 3-O-substituted glucose residue in mixed-linked β-glucans. These results reveal that the properties of enzymes appear to be insignificantly altered according to the variations in the sequences of β-glucanases, although their original function persists.
DISCUSSION
The glycosyl hydrolases (GHs) of rumen fungi play important roles in the degradation of plant polysaccharides. Most GH genes from rumen fungi are intronless, and sequences of these genes are also homologous to those of GH bacterial genes (13). Thus, GH genes from rumen fungi have been suggested to be acquired by horizontal gene transfer events (14). The introduction of novel genes could permit the rapid exploitation of new environmental niches (14). The high microbial population density, the bacteriophage population of the rumen, and the existence of communities attached to substrate particles or the gut epithelium or inside the protozoal food vacuoles could provide favorable environments for gene transfer between a wide range of microorganisms within the rumen (13). S. equinus is the main amylolytic bacterium in the rumen. The strain could produce β-glucanase to degrade β-glucans in the endosperm of cereals and to improve its acquirement of starch (25). In the present study, genes encoding β-glucanase were cloned from rumen fungi. The deduced amino acids of four β-glucanases are phylogenetically more closely related to the β-glucanases in S. equinus than to A. niger or H. jecorina, aerobic fungi (Fig. 1). A bacterial β-glucanase motif “DEIDIE” also exists in β-glucanases (7). The RSCU values of β-glucanase genes from rumen fungi and S. equinus are quite similar (see Table S1 in the supplemental material). These results suggest that β-glucanase genes of rumen fungi have a bacterial origin and might be obtained from S. equinus. This gene transfer contributed beneficial phenotypic capabilities that allowed these fungi to colonize among different plant tissues intake by ruminants.
The length of the 3′UTR increased with evolutionary age and with organism complexity. This evolutionary expansion of the 3′UTR suggests that there is substantial potential for 3′UTR-based translational regulation and that such control mechanisms might be significant in determining differences between species (21). In the present study, the presence of expansion in the 3′UTRs of bglA16 and bglM2 was noted, and several predicted secondary structures were also embedded within the 3′UTRs of bglA16 and bglM2 (see Fig. S1 in the supplemental material). According to the results of relativity analysis as depicted in Fig. 1, the subgroup containing BglA16 and BglM2 also demonstrated evolutionary divergence with the subgroup containing BglA13 and BglA51. These results indicate that although four β-glucanase genes may have originated from bacteria, bglA16 and bglM2 might have been duplicated more recently than bglA13 and bglA51. Differences in composition (Table 1) and in the predicted secondary structures (see Fig. S1 in the supplemental material) in the 3′UTR of duplicate β-glucanase genes reveal a divergence in the regulatory network.
Gene duplication has generally been regarded as a major source for the enrichment of genetic novelty in the genomes of organisms. In addition, expression of duplicated genes results in extra amounts of protein products (11, 33). Protein pairs derived from gene duplication events make up 13% of all yeast proteins (31). Thioredoxin isoforms were encoded by a triplicated gene set in the ascomycete fungus Podospora anserine, showing that gene duplications can indeed generate new genes in P. anserine (12). In the present study, four duplicate β-glucanase genes with high similarity were found in the cDNA library of N. patriciarum J11, and the acceleration of β-glucan utilization for a rumen fungus might be achieved via gene duplication. Duplicate genes could be retained or lost during evolution. Furthermore, many fixed duplicated genes become nonfunctional pseudogenes. The fate of duplicated genes can be judged by their functions in evolution (33). In our study, four recombinant enzymes expressed by E. coli harboring β-glucanase genes demonstrated similar pH and temperature optima (Fig. 3), substrate specificity (see Table S2 in the supplemental material), substrate affinity (Table 2), and hydrolysis patterns (see Fig. S2 in the supplemental material). However, BglA16 and BglM2, two recently duplicated β-glucanases, possessed significantly higher kcat values than other β-glucanases. These results indicated that the two β-glucanases, BglA16 and BglM2, evolved toward increased efficiency with substrates and suggested that enzyme efficiency is a criterion for determining the evolutionary fate of duplicate β-glucanase genes in N. patriciarum J11.
Anaerobic fungi are responsible for degrading plant structural polysaccharides in the digestive tracts of ruminants (28). The β-glucanases from N. patriciarum J11 demonstrated high efficiency in the hydrolysis of β-glucan (Table 2). The reaction conditions of β-glucanases are close to the rumen environment (Fig. 3 and 4). These properties could help N. patriciarum J11 to survive and function well in the rumen. The reaction conditions of β-glucanases from N. patriciarum J11 are also similar to the environments of malt saccharification and the digestive tracts of livestock. Therefore, the β-glucanase genes from rumen fungi provide an attractive candidate to be applied in the brewer and feeding enzyme industries (1, 5).
In the present study, four types of β-glucanase genes were screened from the cDNA library of N. patriciarum J11. The highly conserved nucleotide sequences in the coding regions of four β-glucanase genes and their close phylogenetic relationship to S. equinus suggest that the genes originated in rumen bacteria and were subsequently duplicated in the fungus. Gene duplication results in limited biochemical divergence; however, the recently duplicated β-glucanases demonstrated a significantly better efficiency than the β-glucanases duplicated early, which suggests that the evolutionary fate of duplicate β-glucanase genes in N. patriciarum J11 may be determined according to their catalytic efficiency.
Supplementary Material
ACKNOWLEDGMENTS
The financial support of the National Science Council of Taiwan (grants NSC99-2313-B-020-006-MY3, NSC 99-ET-E-020-002-ET, and NSC 100-ET-E-020-002-ET) is greatly appreciated.
Footnotes
Published ahead of print 6 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Beckmann L, Simon O, Vahjen W. 2006. Isolation and identification of mixed linked beta-glucan degrading bacteria in the intestine of broiler chickens and partial characterization of respective 1,3-1,4-beta-glucanase activities. J. Basic Microbiol. 46:175–185 [DOI] [PubMed] [Google Scholar]
- 2. Borneman WS, Akin DE, Ljungdahl LG. 1989. Fermentation products and plant cell wall-degrading enzymes produced by monocentric and polycentric anaerobic ruminal fungi. Appl. Environ. Microbiol. 55:1066–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brodskii LI, et al. 1995. GeneBee-NET: an Internet based server for biopolymer structure analysis. Biokhimiia 60:1221–1230 [PubMed] [Google Scholar]
- 4. Brownlee AG. 1989. Remarkably AT-rich genomic DNA from the anaerobic fungus Neocallimastix. Nucleic Acids Res. 17:1327–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Celestino KR, Cunha RB, Felix CR. 2006. Characterization of a beta-glucanase produced by Rhizopus microsporus var. microsporus, and its potential for application in the brewing industry. BMC Biochem. 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen H, Li XL, Blum DL, Ljungdahl LG. 1998. Two genes of the anaerobic fungus Orpinomyces sp. strain PC-2 encoding cellulases with endoglucanase activities may have arisen by gene duplication. FEMS Microbiol. Lett. 159:63–68 [DOI] [PubMed] [Google Scholar]
- 7. Chen H, Li XL, Ljungdahl LG. 1997. Sequencing of a 1,3-1,4-β-d-glucanase (lichenase) from the anaerobic fungus Orpinomyces strain PC-2: properties of the enzyme expressed in Escherichia coli and evidence that the gene has a bacterial origin. J. Bacteriol. 179:6028–6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen YC, et al. 2007. Caecomyces sympodialis sp. nov., a new rumen fungus isolated from Bos indicus. Mycologia 99:125–130 [DOI] [PubMed] [Google Scholar]
- 9. Cheng HL, Tsai CY, Chen HJ, Yang SS, Chen YC. 2009. The identification, purification, and characterization of STXF10 expressed in Streptomyces thermonitrificans NTU-88. Appl. Microbiol. Biotechnol. 82:681–689 [DOI] [PubMed] [Google Scholar]
- 10. Cheng HL, Wang PM, Chen YC, Yang SS, Chen YC. 2008. Cloning, characterization, and phylogenetic relationships of stxI, a endoxylanase-encoding gene from Streptomyces thermonitrificans NTU-88. Bioresour. Technol. 99:227–231 [DOI] [PubMed] [Google Scholar]
- 11. Choudhary M, Fu YX, Mackenzie C, Kaplan S. 2004. DNA sequence duplication in Rhodobacter sphaeroides 2.4.1: evidence of an ancient partnership between chromosomes I and II. J. Bacteriol. 186:2019–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Espagne E, et al. 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flint HJ. 1994. Molecular genetics of obligate anaerobes from the rumen. FEMS Microbiol. Lett. 121:259–267 [DOI] [PubMed] [Google Scholar]
- 14. Garcia-Vallve S, Romeu A, Palau J. 2000. Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Mol. Biol. Evol. 17:352–361 [DOI] [PubMed] [Google Scholar]
- 15. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis. Nucleic Acids Symp. 41:95–98 [Google Scholar]
- 16. Henrissat B, Bairoch A. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316(Pt 2):695–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu CY, Chi DJ, Chen SS, Chen YC. 2011. The direct conversion of xylan to lactic acid by Lactobacillus brevis transformed with a xylanase gene. Green Chem. 13:1729–1734 [Google Scholar]
- 18. Hua C, Yan Q, Jiang Z, Li Y, Katrolia P. 2010. High-level expression of a specific β-1,3-1,4-glucanase from the thermophilic fungus Paecilomyces thermophila in Pichia pastoris. Appl. Microbiol. Biotechnol. 88:509–518 [DOI] [PubMed] [Google Scholar]
- 19. Joziasse DH, Shaper NL, Kim D, Van den Eijnden DH, Shaper JH. 1992. Murine α-1,3-galactosyltransferase. A single gene locus specifies four isoforms of the enzyme by alternative splicing. J. Biol. Chem. 267:5534–5541 [PubMed] [Google Scholar]
- 20. Martin-Galiano AJ, Wells JM, de la Campa AG. 2004. Relationship between codon biased genes, microarray expression values and physiological characteristics of Streptococcus pneumoniae. Microbiology 150:2313–2325 [DOI] [PubMed] [Google Scholar]
- 21. Mazumder B, Seshadri V, Fox PL. 2003. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 28:91–98 [DOI] [PubMed] [Google Scholar]
- 22. McInerney JO. 1998. GCUA: general codon usage analysis. Bioinformatics 14:372–373 [DOI] [PubMed] [Google Scholar]
- 23. Menon V, Divate R, Rao M. 2011. Bioethanol production from renewable polymer lichenan using lichenase from an alkalothermophilic Thermomonospora sp. and thermotolerant yeast. Fuel Process. Technol. 92:401–406 [Google Scholar]
- 24. Poppi DP, Minson DJ, Ternouth JH. 1981. Studies of cattle and sheep eating leaf and stem fractions of grasses. III. The retention time in the rumen of large feed particles. Aust. J. Agric. Res. 32:123–137 [Google Scholar]
- 25. Stewart CS, Flint HJ. 1997. The rumen bacteria: the rumen microbial ecosystem, 2nd ed, p 10–55 Blackie Academic & Professional, London, United Kingdom [Google Scholar]
- 26. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 27. Teather RM, Wood PJ. 1982. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43:777–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trinci APJ, et al. 1994. Anaerobic fungi in herbivorous animals. Mycol. Res. 98:129–152 [Google Scholar]
- 29. Varghese JN, et al. 1994. Three-dimensional structures of two plant β-glucan endohydrolases with distinct substrate specificities. Proc. Natl. Acad. Sci. U. S. A. 91:2785–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang HT, Chen IH, Hsu JT. 2012. Production and characterization of a bacteriocin from ruminal bacterium Ruminococcus albus 7. Biosci. Biotechnol. Biochem. 76:34–41 [DOI] [PubMed] [Google Scholar]
- 31. Wolfe KH, Shields DC. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708–713 [DOI] [PubMed] [Google Scholar]
- 32. Yan Y, Smant G, Davis E. 2001. Functional screening yields a new β-1,4-endoglucanase gene from Heterodera glycines that may be the product of recent gene duplication. Mol. Plant-Microbe Interact. 14:63–71 [DOI] [PubMed] [Google Scholar]
- 33. Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol. Evol. 18:292–298 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.