Abstract
Sinorhizobium meliloti is a symbiotic nitrogen-fixing bacterium that elicits nodule formation on roots of alfalfa plants. S. meliloti produces two exopolysaccharides (EPSs), termed EPS I and EPS II, that are both able to promote symbiosis. EPS I and EPS II are secreted in two major fractions that reflect differing degrees of subunit polymerization, designated high- and low-molecular-weight fractions. We reported previously that EPSs are crucial for autoaggregation and biofilm formation in S. meliloti reference strains and isogenic mutants. However, the previous observations were obtained by use of “domesticated” laboratory strains, with mutations resulting from successive passages under unnatural conditions, as has been documented for reference strain Rm1021. In the present study, we analyzed the autoaggregation and biofilm formation abilities of native S. meliloti strains isolated from root nodules of alfalfa plants grown in four regions of Argentina. 16S rRNA gene analysis of all the native isolates revealed a high degree of identity with reference S. meliloti strains. PCR analysis of the expR gene of all the isolates showed that, as in the case of reference strain Rm8530, this gene is not interrupted by an insertion sequence (IS) element. A positive correlation was found between autoaggregation and biofilm formation abilities in these rhizobia, indicating that both processes depend on the same physical adhesive forces. Extracellular complementation experiments using mutants of the native strains showed that autoaggregation was dependent on EPS II production. Our results indicate that a functional EPS II synthetic pathway and its proper regulation are essential for cell-cell interactions and surface attachment of S. meliloti.
INTRODUCTION
Sinorhizobium meliloti is a Gram-negative alphaproteobacterium found in soil that, under nitrogen limitation conditions, is able to engage in a symbiotic association with the agriculturally important legume Medicago sativa (alfalfa). In nature, the bacterium plays an important role in the conversion of atmospheric nitrogen into forms that can be utilized by the plant. This process of nitrogen fixation is carried out in specialized structures called nodules that are formed in the legume roots. The interaction of the bacteria (termed rhizobia) and the plants shows a high degree of host specificity (8), and the successful infection of the roots is dependent upon a reciprocal molecular dialogue between the host plant and the rhizobia (11).
Biofilms are defined as bacterial communities surrounded by a self-produced polymeric matrix and reversibly attached to an inert or a biotic surface (7). Bacteria may develop on plant roots as isolated cells, microcolonies, bacterial aggregates, or biofilms (31). Bacterial surface components, particularly exopolysaccharides (EPSs), flagella, and lipopolysaccharides (LPSs), in combination with bacterial functional signals, are crucial for the formation of rhizobial biofilms in all species studied so far (39). Rhizobial surface polysaccharides play important roles in symbiosis and the formation of active root nodules. Mutants defective in the production of EPSs, LPSs, and capsular polysaccharides usually show a reduced induction of effective nodules and are particularly affected in the process of infection through infection threads (18). S. meliloti produces two different EPSs, succinoglycan (also known as EPS I) and galactoglucan (EPS II) (22), which are both able to promote symbiosis. The perceptions of EPSs in the two basic types of nodule ontogeny (determinate versus indeterminate) appear to display differing rhizobial EPS requirements; e.g., EPS mutants of Rhizobium loti (in which LPSs are conserved) are fully effective with a determinate nodulating host but ineffective with an indeterminate nodulating host (20).
EPS I, the best-understood symbiotically important EPS, is required for the invasion of alfalfa roots by S. meliloti strain Rm1021. EPS I is a polymer of repeating octasaccharide subunits (seven glucoses and one galactose), bearing succinyl, acetyl, and pyruvyl substituents (36). Mutations affecting EPS I biosynthesis result in a variety of developmental abnormalities during nodule formation, including delayed root hair curling, defective or aborted infection threads, and empty nodules with no bacteria or bacteroids. These findings suggest that EPS I has a signaling function (12, 26). EPS II is composed of alternating glucose and galactose residues that are acetylated and pyruvylated, respectively (47). EPSs are produced in dual forms having high molecular weight (HMW) versus low molecular weight (LMW). The LMW fraction is an active biological form of EPS that is essential for the successful infection of leguminous plants that form indeterminate-type nodules (45). Under nonstarvation conditions in the laboratory, wild-type S. meliloti Rm1021 produces detectable quantities of succinoglycan but does not produce EPS II. The production of EPS II was observed under low-phosphate conditions (54) and in a mucR mutant (23). Strain Rm1021 carries an insertional mutation within the expR gene (35) that prevents EPS II production under standard culture conditions. The presence of a functional expR open reading frame (ORF) on a plasmid or in the genome is sufficient to promote the production of symbiotically active EPS II, e.g., in strain Rm8530, which has an intact expR and is termed expR+ (17). EPS II-producing strain Rm8530, which has a mucoid phenotype, displays a highly structured architectural biofilm, in contrast to the unstructured one formed by non-EPS II-producing strain Rm1021. In experiments with M. sativa (alfalfa), the Rm8530 expR+ strain formed biofilms that covered the entire surface of the root, including root hairs, whereas strain Rm1021 formed clusters of cells that adhered mostly to the main root (40). Wild-type S. meliloti reference strains carrying nonfunctional expR loci (and therefore unable to synthesize EPS II) fail to autoaggregate and develop a relatively small biomass attached to plastic surfaces.
Bacterial autoaggregation is a process whereby bacteria physically interact with each other and settle to the bottom in a static liquid suspension (33, 46). The adhesion of bacteria to various surfaces, and their self-aggregation, may be modulated by the regulation of EPS synthesis (38). The presence of a functional copy of the expR regulator gene is necessary for autoaggregation. LMW EPS II, either alone or in combination with the HMW fraction, may function as a polymeric extracellular matrix that agglutinates bacterial cells (46).
Laboratory strains of S. meliloti, such as Rm1021, apparently often carry mutations resulting from successive passages under unnatural conditions. Two known examples in this strain are mutations in regulatory genes that control the expression of several genes, such as expR (16), and a mutation in the pstC gene that causes increases in the expression levels of eight genes related to phosphate deficiency stress (24).
For the purpose of characterizing indigenous, undomesticated S. meliloti strains, we isolated bacteria from root nodules of alfalfa plants growing in fields that had not previously undergone inoculation procedures. We then examined the correlation between biofilm formation and autoaggregation in these native strains. The results of our analysis showed that EPS II plays a crucial role in cell-cell interactions in both sessile and planktonic bacterial cells.
MATERIALS AND METHODS
Bacterial strains.
Wild-type reference S. meliloti strains Rm1021 (30) and Rm8530 (17) were grown as described previously (46). Native alfalfa microsymbionts were obtained from plants growing in agricultural fields with no previous known inoculation procedures. Root nodules were taken from 10 randomly chosen plants in each of four geographically distinct sites in Argentina (El Cerrito, San Rafael, Mendoza [SR]; UNRC field, Río Cuarto, Córdoba [CU]; La Escondida field, Río Cuarto, Córdoba [LE]; and Paso de los Indios, Chubut [PI]). The nodules were surface sterilized and crushed, and their contents were plated onto petri dishes with tryptone yeast extract (TY) medium (50). Pure cultures were used in further experiments and were grown in TY medium on a rotary shaker (200 rpm) at 30°C. The final concentrations of antibiotics used were as follows: streptomycin at 500 μg/ml, neomycin at 200 μg/ml, and gentamicin at 40 μg/ml. The strains and phages used are listed in Table 1.
Table 1.
Bacterial strains and phage used in this study
| Strain or phage (GenBank accession no.) | Description or origin | Reference |
|---|---|---|
| Reference S. meliloti strains | ||
| Rm1021 | SU47 str21 expR102::ISRm2011-1 | 30 |
| Rm8530 | SU47 str21 expR101 (expR+) | 17 |
| Rm8530 exoY mutant | exoY210::Tn5 | 46 |
| Rm1021 expA mutant | expA3::Tn5-233 | 17 |
| Rm8530 expA mutant | expA3::Tn5-233 | 46 |
| Rm8530 expA exoY mutant | expA3::Tn5-233 exoY210::Tn5 | 46 |
| Rm1021 mucR mutant | mucR31::Tn5 | 46 |
| Phage ϕM12 | Generalized transducing phage for S. meliloti | 10 |
| Indigenous S. meliloti strains | ||
| PI1 (JQ666174) | Paso del Indio | Present study |
| PI2 (JQ666175) | Paso del Indio | Present study |
| CU4 (JQ666176) | Campus UNRC | Present study |
| CU5 (JQ666177) | Campus UNRC | Present study |
| CU9 (JQ666178) | Campus UNRC | Present study |
| CU10 (JQ666179) | Campus UNRC | Present study |
| LE7 (JQ666180) | La Escondida | Present study |
| LE16 (JQ666181) | La Escondida | Present study |
| LE17 (JQ666182) | La Escondida | Present study |
| SR1 (JQ666183) | San Rafael | Present study |
| SR2 (JQ666184) | San Rafael | Present study |
| SR3 (JQ666185) | San Rafael | Present study |
| SR4 (JQ666186) | San Rafael | Present study |
| SR6 (JQ666187) | San Rafael | Present study |
| SR7, (JQ666188) | San Rafael | Present study |
| SR8 (JQ666189) | San Rafael | Present study |
| SR9 (JQ666190) | San Rafael | Present study |
| SR10 (JQ666191) | San Rafael | Present study |
| SR11 (JQ666192) | San Rafael | Present study |
| SR15 (JQ666193) | San Rafael | Present study |
Plant nodulation tests.
The nodulation phenotype was tested by inoculation with native strains. Seeds of the alfalfa (M. sativa) “Pampeana” cultivar from INTA (Instituto Nacional de Tecnología Agropecuaria, Argentina) were surface sterilized, germinated, and grown in a chamber at 28°C under a 16-h light/8-h dark regimen, supplied with nitrogen-free Hoagland solution as needed (28). Thirty days after planting, inoculated and uninoculated (control) plants were harvested. Nodules were separated from the roots, and the external morphology of the nodules was examined.
DNA extraction.
Colonies were suspended in 500 μl sterile physiological saline solution and centrifuged at 10,000 rpm for 10 min. The supernatant was removed, and the pellet was suspended in 500 μl InstaGene Matrix (Bio-Rad, Hercules, CA) (6). The suspension was incubated for 30 min at 56°C and then heated for 10 min at 100°C. The supernatant was used as a bacterial DNA template for PCR analysis.
Identification of isolated bacterial strains by partial 16S rRNA gene sequencing.
Direct PCR was performed by utilizing 1 μl DNA template in a 20-μl PCR mixture containing the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′) (25); numbering is based on the Escherichia coli 16S rRNA gene (3). Amplification was conducted for 35 cycles, at 94°C for 45 s, 55°C for 60 s, and 72°C for 60 s. Purified PCR products of approximately 1,400 bp were sequenced with an Applied Biosystems model 3730XL automated DNA sequencing system (Applied Biosystems) by Macrogen Inc. Laboratories (South Korea). The 16S rRNA gene sequences were subjected to a BLAST search program (National Center for Biotechnology Information) (1) to find identities between sequences.
Phylogenetic tree construction.
Phylogenetic analyses were conducted by using MEGA, version 4, in order to produce a phylogenetic tree reflecting the evolutionary relationship between alfalfa-nodulating strains and reference strains by the neighbor-joining method (42), using the Kimura two-parameter model.
Diagnostic PCR analysis of the expR gene.
Diagnostic PCR analysis of the expR gene was conducted by using a procedure described previously by Pellock et al. (35), with minor modifications. The two primers used to amplify the expR region were RmndvA5′out (5′-GCGAGGAGATCCTGCCCGAG-3′) and Rmpyc5′out (5′-AGAGTGGCGTGAACATTCGG-3′). We used 1 μl DNA template in a 20-μl PCR mixture containing 2.5 U Taq polymerase (Invitrogen) under the manufacturer-recommended buffer conditions. Primers and deoxynucleoside triphosphates were used at concentrations of 1 μM and 200 μM, respectively. The PCR program used was as follows: (i) 95°C for 5 min, (ii) 94°C for 30 s, (iii) 65°C for 30 s, (iv) 72°C for 5 min, and (v) a hold at 4°C. Steps 2 to 4 were repeated 29 times. The reaction was performed with a final volume of 25 μl. The PCR product was analyzed by electrophoresis in a 0.8% (wt/vol) agarose gel, with ethidium bromide (1 mg/ml), at 90 V for 45 min.
Phage transduction.
The expA::Tn5 mutant alleles were transferred from the Rm1021 expA::Tn5 strain to recipient strains SR4, SR6, and SR9 by using a generalized transduction method described previously by Finan et al. (10), with some modifications. The cotransduction of the resistance markers (neomycin) and the dry-colony phenotype were verified for each transductant strain. Donor and recipient strains were included as controls.
Autoaggregation assay.
The bacteria were grown in 2 ml TY medium supplemented with appropriate antibiotics, incubated for 24 h at 30°C, diluted 1/100 in TY, and incubated for 48 h under the same conditions. The bacterial suspensions (5 ml) were then transferred into a glass tube (10 by 70 mm) and allowed to settle for 24 h at 4°C. A 0.2-ml aliquot of the upper portion of the suspension was transferred onto a microtiter plate, and the final optical density at 600 nm (OD600) (ODfinal) was measured. A control tube was vortexed for 30 s, and the initial OD600 (ODinitial) was determined. The autoaggregation percentage was calculated as follows: 100 × [1 − (ODfinal/ODinitial)]. For both homologous and heterologous autoaggregation assays, cultures were centrifuged at 4,200 × g for 20 min prior to the settling period. For homologous assays, the pellet of a given strain was resuspended in the cell-free supernatant from an independent culture of the same strain. For heterologous assays, the pellet was resuspended in the cell-free supernatant from a culture of a different strain.
Biofilm formation assay.
Biofilm formation was determined macroscopically by a quantitative assay with 96-well microtiter dishes, whereby biofilms were stained with crystal violet (CV) based on a method described previously by O'Toole and Kolter (34), with modifications (14). The bacteria were grown in 2 ml TY medium supplemented with appropriate antibiotics and incubated with agitation for 48 h at 30°C. The cultures were diluted with fresh medium to give an OD600 of 0.1. One hundred microliters of the suspension was added to each well and incubated with agitation for 24 h at 30°C. Bacterial growth was quantified by measuring the OD600. Planktonic cells were gently removed, 180 μl CV aqueous solution (0.1%, wt/vol) was added, and staining proceeded for 15 min. Each CV-stained well was rinsed thoroughly and repeatedly with water and then scored for biofilm formation by the addition of 150 μl 95% ethanol. The OD560 of solubilized CV was measured with a MicroELISA Auto Reader (series 700 microplate reader; Cambridge Technology). Parallel, sterile control cultures were made with TY medium.
Quantification of rhizobial adsorption to roots.
For the quantification of rhizobial adsorption to roots, we followed a protocol described previously by Caetano-Anollés and Favelukes (4), except that our experimental unit consisted of a group of 15 alfalfa plants in which the total number of adsorbed microcolonies was counted. Under each experimental condition, at least 4 independent experiments were performed.
Statistical analysis.
The autoaggregation assays were performed in quintuplicate. For the biofilm assays, each strain was plated onto at least 8 wells of each microtiter dish. The data were subjected to a one-way analysis of variance (ANOVA), followed by a comparison of multiple treatment levels with the control by using post hoc Fisher's least-significant-difference (LSD) test. All statistical analyses were performed by using Infostat, version 1.0.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rRNA gene from alfalfa-nodulating strains PI1, PI2, CU4, CU5, CU9, CU10, LE7, LE16, LE17, SR1, SR2, SR3, SR4, SR6, SR7, SR8, SR9, SR10, SR11, and SR15 determined in this study have been deposited in the GenBank nucleotide sequence database under accession numbers JQ666174, JQ666175, JQ666176, JQ666177, JQ666178, JQ666179, JQ666180, JQ666181, JQ666182, JQ666183, JQ666184, JQ666185, JQ666186, JQ666187, JQ666188, JQ666189, JQ666190, JQ666191, JQ666192, and JQ666193, respectively (Table 1).
RESULTS
Isolation and phylogenetic analysis of alfalfa-nodulating strains.
Native alfalfa microsymbionts were able to develop highly mucoid colonies after a 24-h incubation period in YEMA (yeast extract mannitol) or TY medium. Acidification and a lack of adsorption of Congo red were observed when strains were grown in YEMA medium supplemented with bromothymol blue and Congo red, respectively. In order to confirm the symbiotic nature of the isolates, the nodulation phenotype was tested by inoculating the bacteria onto sterile alfalfa seeds. After 30 days, all isolates elicited characteristic root nodules in the host plant.
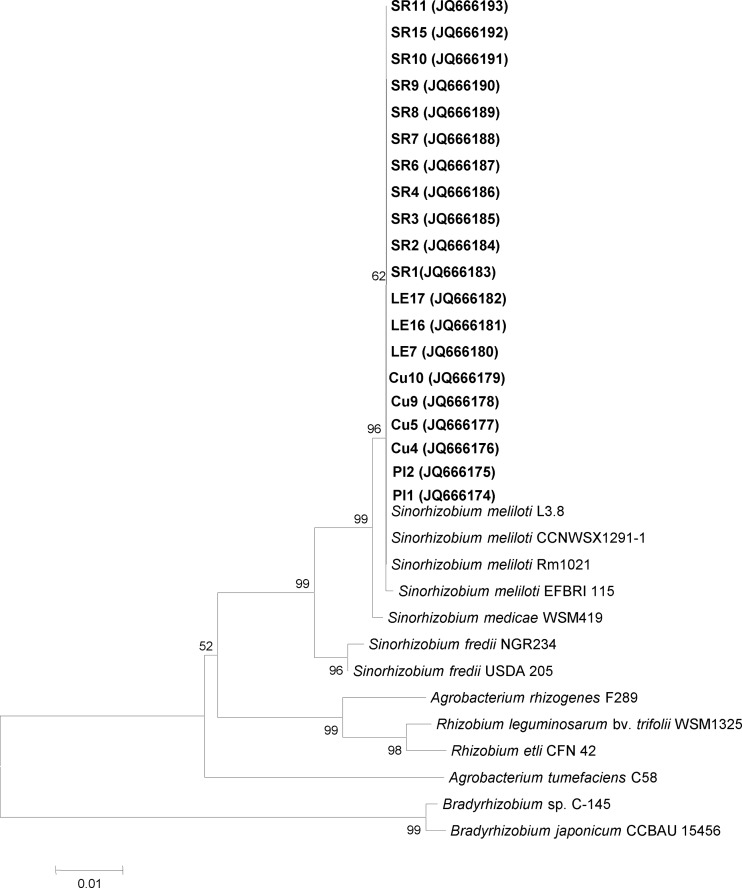
Phylogenetic analysis of 16S rRNA gene sequences grouped all the isolates with S. meliloti reference strains (Fig. 1). High percent identities were obtained when each isolate was compared with the sequenced strain S. meliloti Rm1021. The genetic relationships between different strains can be determined by a comparative analysis of the 16S rRNA gene sequence. This method is useful for taxonomic analyses of bacteria, because there are few variations in the evolutionary level, and the gene product is universally essential and functionally conserved. When closely related strains are compared, the differences in gene sequences are minimal. By use of the criteria described previously by Stackebrandt and Goebel (48), the majority of the strains were identified to the species level, as their sequences showed >97% identity with the 16S rRNA gene sequences of S. meliloti available in the EMBL database.
Fig 1.
Phylogenetic tree based on 16S rRNA gene sequences, showing the evolutionary relationships between alfalfa-nodulating native and reference strains. The tree was constructed by using the neighbor-joining method. Alfalfa-nodulating native strains are indicated by boldface type. GenBank accession numbers are listed in parentheses.
Determination of biofilm formation and autoaggregation.
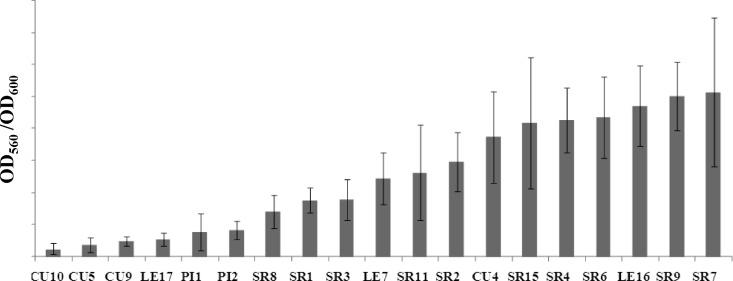
The ability of the native rhizobial strains to attach and develop a sessile biomass on a plastic surface was assessed by growing them on polystyrene microtiter dishes and by using the CV method to indirectly quantify the sessile biomass. The observed biofilm formation abilities ranged from strains with a low attachment ability to strains that showed a high attachment ability and developed a biofilm biomass on the plastic surfaces (Fig. 2).
Fig 2.
Quantitative comparison of biofilm formation in isolated native strains of Sinorhizobium meliloti based on a CV assay. Bars represent standard deviations of the means based on four or more independent experiments with seven replicates each.
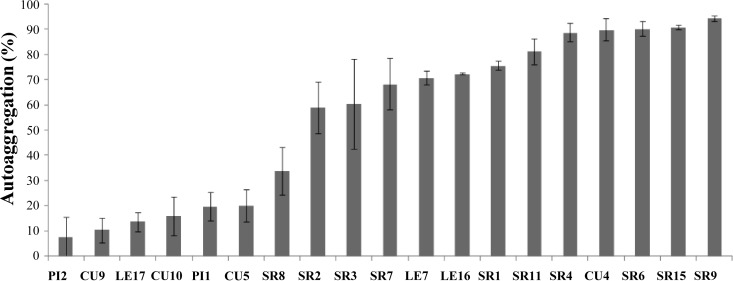
Planktonic autoaggregative behavior was quantified as described in Materials and Methods. Similarly to the range of biofilm formation abilities, we observed a broad heterogeneity in the autoaggregative phenotypes; some strains displayed strong autoaggregation, while others were much weaker (Fig. 3).
Fig 3.
Quantitative autoaggregation assay of isolated native strains of S. meliloti. Bars represent standard deviations from four or more independent experiments with four replicates each.
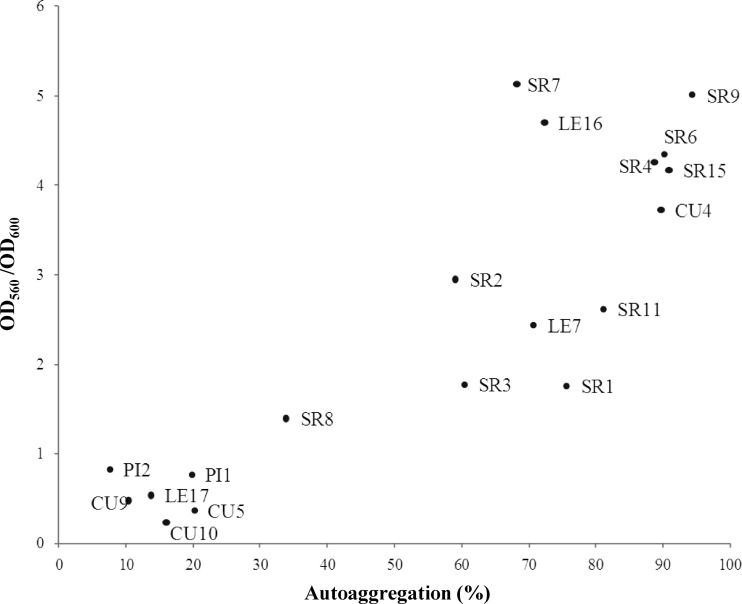
We hypothesized that cell-cell interactions of both biofilm populations and planktonic aggregates depend, to some extent, on the same physical adhesive forces. To test this hypothesis, we conducted a correlation analysis to determine whether the planktonic autoaggregation and biofilm formation abilities of the strains in our collection were quantitatively related phenotypes. A scatter plot was generated (Fig. 4), and the Pearson correlation coefficient (r) was calculated. A statistically significant correlation was observed between the two phenotypes (r = 0.78; P ≤ 0.05).
Fig 4.
Scatter plot of two variables: autoaggregation (percent) and relative biofilm formation ability (OD560/OD600). The circles are ordered pairs that represent different isolates.
Analysis of the expR gene in native and reference strains.
The expression of a functional expR gene regulator is important for the production of the symbiotically important EPS II (35) and therefore for the colony phenotype, biofilm formation, and planktonic autoaggregation (40). We tested for the possible presence of a functional expR locus in our collection of native rhizobia using primers designed to PCR amplify the complete ORF of this gene (35).
The presence of a nonfunctional gene was demonstrated previously by PCR analysis of wild-type reference strains. The size of the PCR product from strain Rm8530 (Rm1021 expR+ [formerly expR101]) is 0.9 kb, while that of the PCR product from wild-type strain Rm1021 is 2.2 kb. Sequence analysis of the 2.2-kb PCR product indicated that the expR ORF was disrupted in Rm1021 by a copy of ISRm2011-1, a previously described 1,319-bp insertion sequence (IS) element (35, 44). Amplification products were detected in all strains, and as expected, size-fragment analysis revealed the presence of 0.9-kb amplicons similar to that obtained from an Rm8530 template (data not shown). This finding indicates that there is no IS element similar to ISRm2011-1 interrupting the expR gene. However, this finding does not provide direct evidence that the sequence of the gene is intact. For example, a previous study showed that although the expR region of S. meliloti strain 102F34 does not contain an IS element, the 102F34 expR ORF has an 11-bp deletion in its coding sequence (35). This deletion is consistent with the dry-colony morphology of strain 102F34, which is distinct from the typical mucoid phenotype of the native strains isolated in the present study. The combination of phenotypic and genotypic results, as described above, supports the presence of a functional expR gene in all the native strains assayed in the present study.
Role of EPSs in cell-cell interactions.
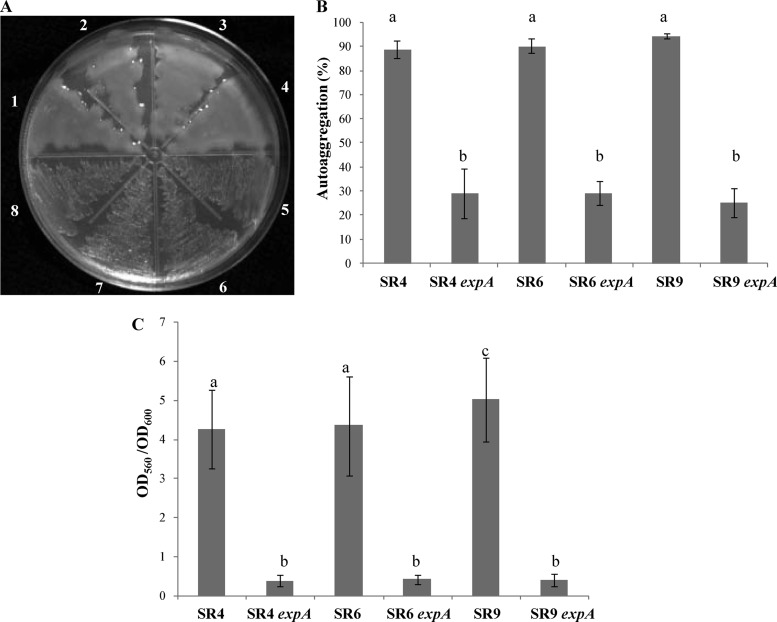
Autoaggregation, a mucoid phenotype, and biofilm formation are three traits that were shown previously to depend on EPS II production (40, 46). In order to determine whether the adhesive and mucoid phenotypes in our collection of indigenous strains also depend on EPS II production, we utilized a genetic approach involving the transduction of the expA::Tn5 mutant allele into indigenous isolates SR4, SR6, and SR9 (which displayed high autoaggregative and biofilm formation abilities), followed by a phenotypic evaluation of the transductant strains. In contrast to the parental strains, the transductant daughter SR4 expA, SR6 expA, and SR9 expA mutants displayed dry-colony phenotypes (Fig. 5A), drastic reductions in autoaggregation percentages (Fig. 5B), and low levels of biofilm formation on a plastic surface (Fig. 5C). These findings indicate that the adhesive phenotypes of these native rhizobia, similarly to those of the reference strains, are closely related to EPS II production.
Fig 5.
Colony phenotype, autoaggregation, and biofilm formation of native strains and their mutants. (A) Appearance of native and mutant strains deficient in EPS II production (expA) following 48 h of incubation in TY medium. (1) Rm8530; (2) SR4; (3) SR6; (4) SR9; (5) Rm8530 expA mutant; (6) SR4 expA mutant; (7) SR6 expA mutant; (8) SR9 expA mutant. (B) Quantitative autoaggregation of native strains SR4, SR6, and SR9 and their respective expA mutants (non-EPS II producers). (C) Relative biofilm formation abilities of native strains and their respective expA mutants determined by using the CV assay. Bars represent standard deviations from three or more independent experiments performed in triplicate. Different letters indicate significant differences (P ≤ 0.05) according to Fisher's LSD test.
Extracellular complementation assays.
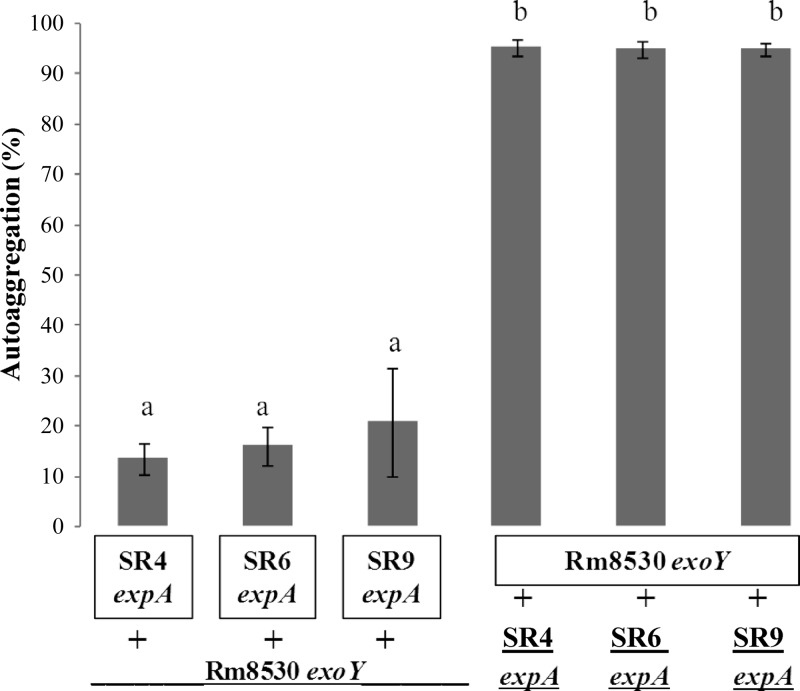
Since the expA mutants showed low autoaggregative behavior, we speculated that the deficiency in biofilm formation and autoaggregation observed for some of the native rhizobial strains could be explained in terms of EPS II production and/or an abnormal EPS II-bacterial surface interaction. Extracellular complementation experiments were performed in order to distinguish between these two possibilities. Rhizobial pellets from the native strains were resuspended in a bacterium-free culture supernatant from the Rm8530 exoY mutant (containing EPS II), and the resulting suspensions were subjected to a quantitative autoaggregation assay (Fig. 6). The supernatant containing EPS II induced significantly more autoaggregative behavior in all the tested strains (Fig. 6), whereas the supernatants from native expA mutant cells did not promote the autoaggregation of the Rm8530 exoY mutant. These findings provide strong evidence for the role of EPS II in bacterial cell-cell interactions.
Fig 6.
Extracellular complementation of autoaggregation assay of expA mutants of native strains. The first 3 bars indicate pellets from cultures of the autoaggregative Rm8530 exoY mutant that were resuspended in the cell-free supernatant from expA mutant cultures. The last 3 bars indicate pellets from expA mutant cultures the were resuspended in the cell-free supernatant from Rm8530 exoY cells. Bars represent standard deviations from two or more independent assays with four replicates each. Different letters indicate significant differences (P ≤ 0.05) according to Fisher's LSD test.
The S. meliloti Rm1021 mucR mutant has a mucoid phenotype and synthesizes EPS II but only in the form with a high degree of polymerization (HMW fraction). This mutant does not develop architecturally complex biofilms and does not display efficient autoaggregation, indicating that the LMW fraction is the form of EPS II essential for both these processes (40, 46). Surprisingly, several native rhizobial strains showing low autoaggregative and biofilm formation abilities (CU5, CU10, PI1, and LE17) were highly mucoid and therefore likely to produce EPS II. In order to elucidate the reason for this apparent discrepancy, we transduced the expA::Tn5 allele into these four native strains. As expected, the expA mutation induced a strong reduction in the mucoid phenotype, and our daughter mutant strains displayed a dry appearance when grown on TY solid medium, similar to the colony phenotypes of other expA mutants. Our expA mutants also did not display efficient autoaggregation (data not shown). These findings strongly support the existence of an EPS II-associated mucoid phenotype in the native isolates.
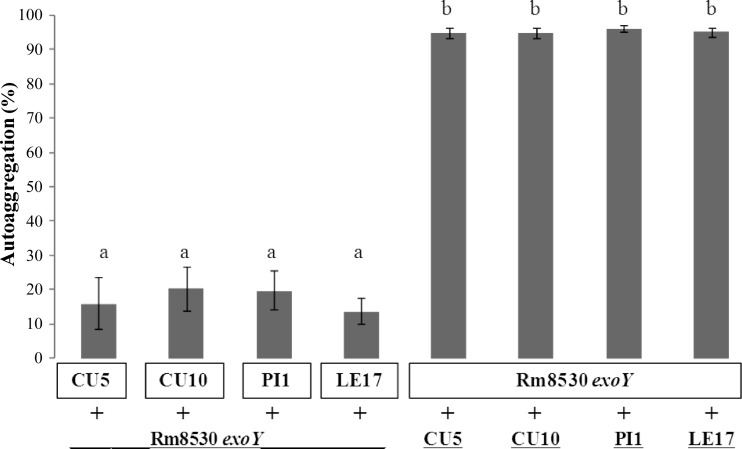
Heterologous autoaggregation assays were also conducted in order to explain the low autoaggregative ability of native strains CU5, CU10, PI1, and LE17. As mentioned above, the expA mutation in these strains induced a strong reduction in the mucoid property, indicating an EPS II-associated phenotype. Treatment with a culture supernatant of the Rm8530 exoY mutant increased the autoaggregation of these four strains, whereas culture supernatants of the four strains did not induce the autoaggregation of the Rm8530 exoY mutant (Fig. 7). The surfaces of CU5, CU10, LE17, and PI1 cells were presumably able to interact normally with the EPS II present in the Rm8530 exoY supernatant. Taken together, these observations suggest that the native rhizobial strains showing low autoaggregative and biofilm formation abilities do not produce the extracellular factors required for strong autoaggregation, presumably because of a difference in the HMW/LMW ratio of EPS II in the extracellular medium.
Fig 7.
Extracellular complementation of autoaggregation assays of native strains with low autoaggregation abilities. The last 3 bars indicate pellets of native strains that were resuspended in the cell-free supernatant from autoaggregative Rm8530 exoY cells. The first 3 bars indicate pellets of the Rm8530 exoY mutant that were resuspended in cell-free supernatants of native strains. Bars represent standard deviations from two or more independent assays with four replicates each. Different letters indicate significant differences (P ≤ 0.05) according to Fisher's LSD test.
To evaluate the role of S. meliloti EPSs in early interactions with alfalfa roots, adsorption assays were performed by using S. meliloti mutants with specific defects in EPS synthesis (Table 2). The results suggest that EPS II partially inhibits rhizobial adhesion to roots, presumably through a “shielding effect.” The mucR mutant (which secretes only the HMW fraction of EPS II) attached to roots in higher numbers than did Rm8530 (which synthesizes both EPS II fractions), suggesting that the LMW fraction of EPS II may partially block rhizobial attachment to roots. Because planktonic rhizobia were incubated with alfalfa plants for 4 h (4), these findings reflect the role that EPSs may play during the initial access to the root; this test should therefore not be interpreted as a biofilm assay. Additional experiments are needed to better clarify the associations between biofilm formation and other adhesion phenotypes.
Table 2.
S. meliloti adsorption to alfalfa roots
| Strain | Mean adsorption (‰) ± SEa |
|---|---|
| Rm8530 | 0.4 ± 0.1A |
| Rm8530 exoY mutant | 0.5 ± 0.1A |
| Rm8530 expA mutant | 2.8 ± 0.1B |
| Rm8530 expA exoY mutant | 2.7 ± 0.1B |
| Rm1021 mucR mutant | 2.6 ± 0.1B |
| Rm1021 | 2.7 ± 0.1B |
Shown are data for the adsorption (per mille) of S. meliloti mutant and wild-type strains to alfalfa roots (groups of 15 plants). Different letters indicate significant differences (P ≤ 0.05) according to Fisher's LSD test.
DISCUSSION
The inoculation of legume crop plants with selected, highly efficient rhizobia is an important method for the improvement of symbiotic nitrogen fixation in agricultural ecosystems and constitutes a major strategy for the sustainable input of nitrogen into agricultural soils (27). However, the native rhizobial populations present in soils often display a superior competitive ability over inoculated strains on the basis of their large population size, positional advantage, and/or superior adaptation to local environmental conditions (2, 49). The selection of efficient rhizobial strains based on their adaptation to local ecological conditions can therefore lead to the increased grain production of crops (27, 32).
We used several approaches to evaluate the rhizobial strains present in root nodules of alfalfa plants growing in fields in Argentina that had no history of inoculation procedures. The isolated strains showed a mucoid phenotype when grown on petri dishes. Such a phenotype was indicative of EPS II synthesis in previously characterized reference strains.
16S rRNA gene analyses of all the isolates revealed a high degree of identity (approximately 98%) with reference S. meliloti strains, corresponding to a value of sequence divergence less than the 3.0% required for differentiation between species (48). PCR analysis of the chromosomal expR gene of these isolates revealed that this gene is not interrupted by an IS element, as is also the case for reference strain Rm8530 (35). ExpR is a LuxR homologue, whose functions include the activation of EPS II production in the presence of N-acyl-homoserine lactone (AHL), which is produced by the sinR/sinI system in S. meliloti. Strain Rm1021 displays a dry (as opposed to mucoid) phenotype because its expR gene is interrupted by an IS element, and it therefore cannot produce EPS II. EPS II-producing strain Rm8530 displays a highly mucoid phenotype. Rm8530 and the native strains isolated in this study harbor an intact (not interrupted) copy of the expR gene, giving a 0.9-kb PCR product. Rm1021 yields a larger amplicon (2.2-kb PCR product) because the expR ORF is disrupted by a copy of ISRm2011-1, a 1,319-bp IS element.
We have shown previously that rhizobial cell surface components such as EPSs, in combination with bacterial functional signals, are essential for the processes of autoaggregation (46) and biofilm formation (39). Both processes play important ecological roles in the survival of rhizobia in their natural soil environment and probably in the nitrogen-fixing symbiosis that occurs within root nodules, in which EPSs are essential for early stages of infection (12). The findings of the present study illustrate a great variability in both autoaggregation and biofilm formation abilities among native soil isolates. This phenotypic diversity may result from differential selective pressures in the soil microenvironment or in the root nodules. Interestingly, a correlation analysis of autoaggregation and biofilm formation abilities gave a Pearson correlation coefficient of 0.78, indicating a positive correlation between these two variables. These findings suggest that the two processes are related and that cell-cell interactions in the context of both biofilm populations and planktonic aggregates depend, at least under the conditions of our assays, on the same physical adhesive forces. A similar positive correlation between autoaggregation and biofilm formation abilities was shown previously for Myroides odoratus, a Gram-negative bacillus (21).
The results of the transduction of the expA::Tn5 mutation into native strains displaying strong autoaggregation and biofilm formation abilities showed that these processes, and the expression of mucoid phenotypic characteristics, depend mainly on EPS II synthesis. This expA::Tn5 mutation also abolished the expression of mucoid colonies in four native strains (Cu5, Cu10, PI1, and LE17) that showed weak autoaggregation and biofilm formation abilities, indicating that the mucoid phenotype depends on EPS II production even in these strains. Autoaggregation in these four strains and their expA mutants could be complemented by the exogenous addition of culture supernatants from Rm8530 exoY cells, indicating that the cell surfaces of these strains can interact normally with EPS II. These findings, taken together, suggest that the low autoaggregation and biofilm formation abilities of some of the isolates that showed a mucoid phenotype were due to a low LMW/HMW ratio of EPS II. Further experiments, including the direct testing of purified EPS II fractions, will be necessary to test this hypothesis.
Bacterial surface components, particularly EPSs, are crucial for biofilm formation in rhizobia (39). S. meliloti has been the subject of studies of the effects of nutritional and environmental conditions (37), EPSs and flagella (13), ExoR with an ExoS-ChvI two-component system (51), nod genes (15), and the regulation of EPS biosynthesis (40). However, for other rhizobial species, a connection between EPS production and biofilm formation ability is not clear. EPS production in Rhizobium sp. YAS34 is not essential for biofilm formation on inert supports or on roots of Arabidopsis thaliana or Brassica napus (43). Rhizobium leguminosarum mutants defective in the synthesis of acid EPSs were also deficient in biofilm formation (41) and showed alterations of the pattern of adherence to pea roots (52). R. leguminosarum mutants defective in the synthesis of glucomannan, another EPS, attached and formed normal biofilms in vitro but did not display normal attachment or biofilm formation on root hairs (52).
It is likely that different polymer types mediate attachment depending on differing substrate chemistries and medium compositions. For example, polymers with nonpolar sites, such as LPSs, may dominate in binding to hydrophobic surfaces, whereas polymers capable of hydrogen bonding or electrostatic interactions, such as polysaccharides, may dominate in binding to hydrophilic surfaces. Different polymer types may act cooperatively in binding to a surface to stabilize the adhesive interaction; e.g., a Pseudomonas fluorescens mutant that lacks the O antigen of the LPS, with the consequent increased exposure of the lipid moiety of the LPS, displays increased adhesion to hydrophobic substrates (53). In S. meliloti, the lpsB mutant lacks glycosyltransferase I, which is responsible for the biosynthesis of the LPS core (5), while the bacA mutant is defective in the distribution of fatty acids on the lipid A component of LPS (9). The lpsB mutation resulted in a slight reduction of biofilm formation compared with that of the wild type, whereas the bacA mutation resulted in a roughly 50% reduction of biofilm formation (19). In view of these observations, it would be very interesting to evaluate the contributions of LPSs (by themselves or in combination with EPSs) to the adhesion properties of the native strains used in the present study. In the case of our subgroup of mucoid isolates that displayed weakly autoaggregative and poor biofilm formation phenotypes, a complete complementation of autoaggregation was observed when the isolates were resuspended in cell-free EPS II-containing supernatants from Rm8530 exoY cells. This finding indicates that under our experimental conditions, all bacterial surfaces are equally effective for EPS II autoaggregative interactions and that possible LPS structural heterogeneity among the strains has no impact on planktonic autoaggregation.
An increased knowledge of the genotypic and phenotypic characteristics of rhizobial populations will help improve agricultural legume production worldwide, through the application of inoculation strategies and other sustainable management practices (29). In view of the economic importance of alfalfa production in Argentina and its status as the most extensively cultivated forage legume worldwide, it is essential to better understand the factors that affect the growth of this crop, including its associated nodulating rhizobial populations. Further detailed studies on genotypic and phenotypic compositions, seasonal shifts in populations, and effects of rhizobia on different varieties of alfalfa, in combination with biogeographic analyses, will clarify the behavior of local rhizobial populations and have direct applications for improved agricultural production.
ACKNOWLEDGMENTS
This work was supported by grants from the Secretaría de Ciencia y Técnica de la UNRC, Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) of the República Argentina. F.G.S. and M.B.S. were supported by a fellowship from CONICET. W.G. and A.Z. are career members of CONICET.
We thank Juan Ignacio Ituarte for his assistance in alfalfa sampling procedures and S. Anderson for editing the manuscript.
Footnotes
Published ahead of print 6 April 2012
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bogino P, Banchio E, Bonfiglio C, Giordano W. 2008. Competitiveness of a Bradyrhizobium sp. strain in soils containing indigenous rhizobia. Curr. Microbiol. 56:66–72 [DOI] [PubMed] [Google Scholar]
- 3. Brosius J, Palmer JL, Kennedy HP, Noller HF. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 75:4801–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caetano-Anollés G, Favelukes G. 1986. Quantitation of adsorption of rhizobia in low numbers to small legume roots. Appl. Environ. Microbiol. 52:371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campbell GR, Reuhs BL, Walker GC. 2002. Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc. Natl. Acad. Sci. U. S. A. 99:3938–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cepeda C, Santos Y. 2000. Rapid and low-level toxic PCR-based method for routine identification of Flavobacterium psychrophilum. Int. Microbiol. 3:235–238 [PubMed] [Google Scholar]
- 7. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 8. Denarié J, Debellé F, Promé JC. 1996. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65:503–535 [DOI] [PubMed] [Google Scholar]
- 9. Ferguson GP, Roop RM, Walker GC. 2002. Deficiency of a Sinorhizobium meliloti bacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. J. Bacteriol. 184:5625–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Finan TM, et al. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fisher RF, Long SR. 1992. Rhizobium-plant signal exchange. Nature 357:655–660 [DOI] [PubMed] [Google Scholar]
- 12. Fraysse N, Couderc F, Poinsot V. 2003. Surface polysaccharide involvement in establishing the Rhizobium-legume symbiosis. Eur. J. Biochem. 270:1365–1380 [DOI] [PubMed] [Google Scholar]
- 13. Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM. 2006. Investigations of Rhizobium biofilm formation. FEMS Microbiol. Ecol. 56:195–206 [DOI] [PubMed] [Google Scholar]
- 14. Fujishige NA, Kapadia NN, Hirsch AM. 2006. A feeling for the microorganism: structure on a small scale. Biofilms on plant roots. Bot. J. Linn. Soc. 150:79–88 [Google Scholar]
- 15. Fujishige NA, et al. 2008. Rhizobium common nod genes are required for biofilm formation. Mol. Microbiol. 67:504–515 [DOI] [PubMed] [Google Scholar]
- 16. Galibert F, et al. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668–672 [DOI] [PubMed] [Google Scholar]
- 17. Glazebrook J, Walker GC. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661–672 [DOI] [PubMed] [Google Scholar]
- 18. Hirsch AM. 1999. Role of lectins (and rhizobial exopolysaccharides) in legume nodulation. Curr. Opin. Plant Biol. 2:320–326 [DOI] [PubMed] [Google Scholar]
- 19. Hirsch AM, Lum MR, Fujishige NA. 2009. Microbial encounters of a symbiotic kind—attaching to roots and other surfaces, p 295–314 In Emons AMC, Ketelaar T. (ed), Root hairs. Plant cell monographs, vol 12 Springer-Verlag, Berlin, Germany [Google Scholar]
- 20. Hotter GS, Scott B. 1991. Exopolysaccharide mutants of Rhizobium loti are fully effective on a determinate nodulating host but are ineffective on an indeterminate nodulating host. J. Bacteriol. 173:851–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jacobs A, Chenia HY. 2010. Biofilm-forming capacity, surface hydrophobicity and aggregation characteristics of Myroides odoratus isolated from South African Oreochromis mossambicus fish. J. Appl. Microbiol. 107:1957–1966 [DOI] [PubMed] [Google Scholar]
- 22. Janczarek M. 2011. Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int. J. Mol. Sci. 12:7898–7933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keller M, et al. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant Microbe Interact. 8:267–277 [DOI] [PubMed] [Google Scholar]
- 24. Krol E, Becker A. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Genet. Genomics 272:1–17 [DOI] [PubMed] [Google Scholar]
- 25. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrant E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics John Wiley & Sons, New York, NY [Google Scholar]
- 26. Leigh JA, Signer ER, Walker GC. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. U. S. A. 82:6231–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lindström K, Murwira M, Willems A, Altier N. 2010. The biodiversity of beneficial microbe-host mutualism: the case of rhizobia. Res. Microbiol. 161:453–463 [DOI] [PubMed] [Google Scholar]
- 28. Löbler M, Hirsch AM. 1993. A gene that encodes a proline-rich nodulin with limited homology to PsENOD12 is expressed in the invasion zone of Rhizobium meliloti-induced alfalfa nodules. Plant Physiol. 103:21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McInnes A, Thies JE, Abbott LK, Howieson JG. 2004. Structure and diversity among rhizobial strains, populations and communities—a review. Soil Biol. Biochem. 36:1295–1308 [Google Scholar]
- 30. Meade H, Long S, Ruvkun G, Brown S, Ausubel F. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morris CE, Monier JM. 2003. The ecological significance of biofilm formation by plant-associated bacteria. Annu. Rev. Phytopathol. 41:429–453 [DOI] [PubMed] [Google Scholar]
- 32. Nievas F, Bogino P, Nocelli N, Giordano W. 2012. Genotypic analysis of isolated peanut-nodulating rhizobial strains reveals differences among populations obtained from soils with different cropping histories. Appl. Soil Ecol. 53:74–82 [Google Scholar]
- 33. Nikitina VE, Ponomareva EG, Alen'kina SA, Konnova SA. 2001. The role of cell-surface lectins in the aggregation of Azospirilla. Microbiology 70:471–476 [PubMed] [Google Scholar]
- 34. O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 35. Pellock BJ, Teplitski M, Boinay RP, Bauer WD, Walker GC. 2002. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J. Bacteriol. 184:5067–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reuber TL, Walker GC. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269–280 [DOI] [PubMed] [Google Scholar]
- 37. Rinaudi L, et al. 2006. Effects of nutritional and environmental conditions on Sinorhizobium meliloti biofilm formation. Res. Microbiol. 157:867–875 [DOI] [PubMed] [Google Scholar]
- 38. Rinaudi L, Sorroche F, Zorreguieta A, Giordano W. 2010. Analysis of mucR gene regulating biosynthesis of exopolysaccharides: implications for biofilm formation in Sinorhizobium meliloti Rm1021. FEMS Microbiol. Lett. 302:15–21 [DOI] [PubMed] [Google Scholar]
- 39. Rinaudi LV, Giordano W. 2010. An integrated view of biofilm formation in rhizobia. FEMS Microbiol. Lett. 304:1–11 [DOI] [PubMed] [Google Scholar]
- 40. Rinaudi LV, Gonzalez JE. 2009. The low-molecular-weight fraction of exopolysaccharide II from Sinorhizobium meliloti is a crucial determinant of biofilm formation. J. Bacteriol. 191:7216–7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Russo DM, et al. 2006. Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J. Bacteriol. 188:4474–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 43. Santaella C, Schue M, Berge O, Heulin T, Achouak W. 2008. The exopolysaccharide of Rhizobium sp. YAS34 is not necessary for biofilm formation on Arabidopsis thaliana and Brassica napus roots but contributes to root colonization. Environ. Microbiol. 10:2150–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simon R, Hötte B, Klauke B, Kosier B. 1991. Isolation and characterization of insertion sequence elements from gram-negative bacteria by using new broad-host-range, positive selection vectors. J. Bacteriol. 173:1502–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Skorupska A, Janczarek M, Marczak M, Mazur A, Król J. 2006. Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb. Cell Fact. 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sorroche F, Rinaudi L, Zorreguieta A, Giordano W. 2010. EPS II-dependent autoaggregation of Sinorhizobium meliloti planktonic cells. Curr. Microbiol. 61:465–470 [DOI] [PubMed] [Google Scholar]
- 47. Spaink HP. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54:257–288 [DOI] [PubMed] [Google Scholar]
- 48. Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849 [Google Scholar]
- 49. Streeter JG. 1994. Plant-environment interactions symbiotic nitrogen fixation, p 245–262 In Wilkinson RE. (ed), Marcel Dekker Inc, New York, NY [Google Scholar]
- 50. Vincent JM. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications, Oxford, England [Google Scholar]
- 51. Wells DH, Chen EJ, Fisher RF, Long SR. 2007. ExoR is genetically coupled to the ExoS-ChvI two-component system and located in the periplasm of Sinorhizobium meliloti. Mol. Microbiol. 64:647–664 [DOI] [PubMed] [Google Scholar]
- 52. Williams A, et al. 2008. Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J. Bacteriol. 190:4706–4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Williams V, Fletcher M. 1996. Pseudomonas fluorescens adhesion and transport through porous media are affected by lipopolysaccharide composition. Appl. Environ. Microbiol. 62:100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhan HJ, Lee CC, Leigh JA. 1991. Induction of the second exopolysaccharide (EPSb) in Rhizobium meliloti SU47 by low phosphate concentrations. J. Bacteriol. 173:7391–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]