Abstract
Marine actinomycetes in the genus Salinispora fail to grow when seawater is replaced with deionized (DI) water in complex growth media. While bioinformatic analyses have led to the identification of a number of candidate marine adaptation genes, there is currently no experimental evidence to support the genetic basis for the osmotic requirements associated with this taxon. One hypothesis is that the lineage-specific loss of mscL is responsible for the failure of strains to grow in media prepared with DI water. The mscL gene encodes a conserved transmembrane protein that reduces turgor pressure under conditions of acute osmotic downshock. In the present study, the mscL gene from a Micromonospora strain capable of growth on media prepared with DI water was transformed into S. tropica strain CNB-440. The single-copy, chromosomal genetic complementation yielded a recombinant Salinispora mscL+ strain that demonstrated an increased capacity to survive osmotic downshock. The enhanced survival of the S. tropica transformant provides experimental evidence that the loss of mscL is associated with the failure of Salinispora spp. to grow in low-osmotic-strength media.
INTRODUCTION
The obligate marine actinomycete genus Salinispora is comprised of the formally described species S. tropica and S. arenicola (14) and a third species for which the name “S. pacifica” has been proposed (10). The genus is broadly distributed in tropical and subtropical marine sediments (10) and is the source of a large number of structurally diverse secondary metabolites (5), including the proteasome inhibitor salinosporamde A, which is in clinical trials as an anticancer agent (6). Salinispora species produce a dense, nonfragmenting mycelium and nonmotile spores that blacken the colony surface, as is typical of the closely related genus Micromonospora. One of the unique characteristics of Salinispora spp., however, is that strains fail to grow when seawater is replaced with deionized (DI) water in complex growth media that lack added salts (14, 16).
Among Gram-negative marine bacteria, the requirement of seawater for growth has been linked to a specific sodium ion requirement (18). While a sodium requirement was originally reported for Salinispora spp., growth has subsequently been demonstrated with as little as 5 mM Na+ if an appropriate osmotic environment is provided by the addition of alternative salts (25). In addition, it was reported that Salinispora cells lyse in low-ionic-strength media (25), suggesting they have poor tolerance for osmotic downshock. While the genetic basis for the failure of Salinispora strains to grow in low-osmotic-strength media has not been established, comparative genomics revealed a large family of highly duplicated polymorphic membrane proteins (PMPs), which were proposed to render cells unable to survive osmotic downshock (19). A more comprehensive bioinformatic analysis identified a larger pool of candidate marine adaptation genes and the lineage-specific loss of mscL (20), the product of which is a mechanosensitive channel that has been shown to alleviate cell lysis following osmotic downshock (17).
Free-living microorganisms have developed robust mechanisms to maintain cell volume and integrity in response to changes in osmotic stress (28). These mechanisms include the accumulation of compatible solutes and mechanisms to release osmolytes under hypo-osmotic conditions. Mechanosensitive channels are present in a large variety of bacteria and are thought to function as primary osmolyte release valves that reduce turgor pressure under conditions of osmotic downshock (8). The mechanosensitive channel of large conductance (MscL) is nonselective in the ions and small molecules it transports and has been shown to open following osmotic downshock (1). Cells lacking MscL are thus unable to tolerate the rapid transition from high- to low-osmolarity conditions (12), as might be experienced in the transition from a marine to a nonmarine environment.
The Escherichia coli mscL gene was the first mechanosensitive channel gene to be cloned (23). Subsequent genetic experiments with the marine bacterium Vibrio alginolyticus revealed that the introduction of this gene alleviates cell lysis following osmotic downshock (17). Similar functions were also demonstrated in the Gram-positive bacteria Lactococcus lactis (7) and Bacillus subtilis (8). Evidence that Salinispora spp. lack mscL coupled with the role of its protein product in relieving cell turgor pressure (24) led to the hypothesis that the loss of this gene may contribute to the inability of Salinispora spp. to grow on complex media that lack added salts (20). The aims of the present study were to test this hypothesis using a series of genetic complementation experiments and growth assays.
MATERIALS AND METHODS
Microorganisms.
The type strain S. tropica CNB-440T (accession number CP000667) (14) was chosen for complementation experiments based on an analysis of the genome sequence (27), which did not contain the mscL gene (20). Micromonospora sp. strain CNB-512 was used to complement CNB-440. It was isolated from a marine sediment sample and did not require seawater for growth (9). Three exconjugants were generated from S. tropica CNB-440; two contained the recombinant plasmid pSET152::mscL (c1, CNY-369; c2, CNY-370) and the third an empty plasmid (CNY-372). Strains CNB-440 and CNB-512 were grown in medium A1 (10 g starch, 4 g yeast extract, 2 g peptone, 1 liter natural seawater). E. coli was grown in Luria-Bertani (LB) medium (10 g Bacto tryptone, 5 g Bacto yeast extract, 5 g NaCl, 1 liter DI water). Descriptions of all Salinispora, Micromonospora, and E. coli strains and plasmids used are presented in Table 1.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Species and genotype | Reference or sourcea |
|---|---|---|
| Strains | ||
| CNB-440b | S. tropica | Bahamas (CP000667) |
| CNB-536 | S. tropica | Bahamas (AY040618) |
| CNH-898 | S. tropica | Bahamas (AY040622) |
| CNS-205b | S. arenicola | Palau (NC_009953) |
| CNH-665 | S. arenicola | Bahamas |
| CNS-325 | S. arenicola | Palau (GU593973) |
| CNH-662 | S. arenicola | Bahamas |
| CNT-133b | S. pacifica | Fiji (HQ218996) |
| CNS-844 | S. pacifica | Fiji (HQ642897) |
| CNT-131 | S. pacifica | Fiji (HQ642896) |
| CNY-369 | S. tropica carrying pSET152::mscL | This work |
| CNY-370 | S. tropica carrying pSET152::mscL | This work |
| CNY-372 | S. tropica carrying pSET152 empty | This work |
| CNB-512 | Micromonospora sp. | Bahamas (AY040624) |
| CNB-394 | Micromonospora sp. | Bahamas (AY040625) |
| CNX-434 | Micromonospora sp. | Palmyra |
| DH5α | E. coli, endA1hsdR17(r− m−) supE44 thi-1 recA1 gyrA(Nalr) relA1 Δ(lacZYA-argF)U169 deoR [ϕ80Δ(lacZ)M15] | |
| S17-1 | E. coli, recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 | 22 |
| Plasmids | ||
| pTOPO | Invitrogen | |
| pSET152 | 3 | |
| pSET152::mscL | This work |
16S rRNA accession numbers are in parentheses.
The genome sequence is available.
PCR analysis.
Two sets of PCR primers were designed based on the mscL gene sequence (accession number NC_014815) obtained from the Micromonospora sp. strain L5 genome (accession number CP002399). One set, mscL-int-F (5′-TGACCTCCTCGCTGGGAGCC-3′) and mscL-int-R (5′-CGCGGTCGGCGTCGTCATC-3′), amplifies a 320-bp internal fragment, and the second set, mscL-ext-F (5′-GCCATCCGCGCCGGCGACCCG-3′) and mscL-ext-R (5′-GTCAGCGCGCGGCCGGGGGCTCC-3′), amplifies 580 bp that includes the complete mscL gene and upstream flanking sequence, which includes the promoter region. These primers were used to test for the presence of the mscL gene in a total of nine Salinispora strains (Table 1) and to amplify the mscL gene sequence from Micromonospora strain CNB-512. S. tropica CNB-440, S. arenicola CNS-205, and S. pacifica CNT-133 have genome sequences available and were used as negative controls to verify that the primers were specific to mscL. Amplification was performed for 30 cycles (denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min, followed by a 7-min extension at 72°C).
Cloning of the Micromonospora mscL gene.
The mscL gene and flanking sequence (580 bp) was PCR amplified as described above from genomic DNA prepared from CNB-512 using the mscL-ext primers with restriction sites (in bold) at the 5′ ends, EcoRI-mscL-ext-F (5′-CTTGAATTCAGCCGGTGCTTTTCTCGAAG-3′) and XbaI-mscL-ext-R (5′-ATTCTAGAGTCAGCGCGCGGCCGGGGGCTCC-3′). The PCR product was purified, digested with the endonucleases EcoRI and XbaI, and ligated to the same sites of the Aprar conjugative plasmid vector pSET152 (3). The ligation mixture was electroporated into the E. coli host strain DH5α and plated on LB containing 50 μM apramycin, 0.5 mM isopropyl-d-thiogalactopyranoside (IPTG), and 40 μg/ml 5-bromo-4chloro-3-indolyl-d-galactopyranoside (X-Gal) at 37°C, and recombinants (white colonies) were screened by PCR using the same primers listed above. Plasmid DNA purified from one clone yielded an insert of the predicted size after digestion with EcoRI and XbaI and was subsequently sequence verified. This plasmid, pSET152::mscL, was electroporated into the conjugative helper E. coli S17-1 (22), producing the strain E. coli S17-1/pSET152::mscL. Similar procedures were followed to generate a control plasmid that lacked the insert (pSET::empty).
Conjugation assays.
To conduct E. coli/S. tropica CNB-440 crosses, overnight LB cultures of the donor strains E. coli S17-1/pSET152::mscL and S17-1/pSET::empty were grown for 4 h in 10 ml LB with 50 μg/ml apramycin. In parallel, S. tropica CNB-440 was grown in 30 ml A1 medium (70% seawater) for 2 days. The E. coli suspensions (0.5 ml) were then mixed with the S. tropica culture (0.5 ml) and the mixture spread onto A1 agar plates. After 20 h of incubation at 33°C, the plates were overlaid with 1 ml of 2-mg/ml nalidixic acid to eliminate the E. coli donor strain and 1 ml of 4-mg/ml apramycin to select for S. tropica CNB-440 exconjugants. Exconjugants were visible after 2 weeks of incubation at room temperature, and individual colonies were isolated onto A1 agar plates with 200 μg/ml apramycin and 100 μg/ml nalidixic acid (11). In control experiments, plasmid insertion was highly stable even after three passages under nonselective conditions.
RNA isolation and mscL-specific RT-PCR.
To isolate RNA, bacteria were grown for 5 days in medium A1 (70% seawater) at 27°C. Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA) and treated with amplification-grade, RNase-free DNase I (Gibco-BRL). Reverse transcription-PCR (RT-PCR) was performed with 200 ng of DNase-treated RNA using a single-tube RT-PCR kit (Gibco-BRL). PCR amplification of the mscL gene was performed as previously described using the internal primer set. Genomic DNA served as a positive control, and DNase-treated RNA that had not been reverse transcribed was used as a negative control. Twenty-microliter aliquots were removed after 30 PCR cycles, stained with SYBR green, electrophoresed on a 1% agarose Tris-borate-EDTA (TBE) gel, and analyzed using a Digital Science 120 system (Kodak).
Western blot MscL analysis.
Membrane preparations followed previously described methods (21) with slight modifications. Bacteria were grown for 5 days in 30 ml medium A1 with shaking (230 rpm, 27°C), chilled on ice, pelleted by centrifugation (7,500 × g, 15 min, 4°C), resuspended in lysis buffer (10 mM Tris-HCl [pH 8], 10 mM MgCl2), sonicated, and supplemented with 2 mM phenylmethylsulfonyl fluoride. Whole cells and debris were removed by low-speed centrifugation (5,000 × g, 10 min), and total membrane fractions were obtained after 45 min of centrifugation at 13,000 × g at 4°C. Total membrane fractions were solubilized in 50 μl of Tris-HCl buffer (100 mM, pH 8) and 1% SDS. Proteins (10 μg/lane) were separated by electrophoresis (12% SDS polyacrylamide gels), transferred to polyvinylidene difluoride (PVDF) membranes, and blocked for 1 h in blocking buffer (phosphate-buffered saline [PBS] and 5% nonfat milk with 3% bovine serum albumin) at room temperature. A rabbit polyclonal IgG antibody designed by Agent Inc. (San Diego, CA) based on the Micromonospora L5 MscL immunogenic motif LDDVLGRRQEPPAPRC was then diluted 1:500 in blocking buffer and incubated overnight with the membrane at 4°C. Membranes were then incubated for 1 h at room temperature with a 1:5,000 dilution of IRDye-conjugated goat anti-rabbit IgG (Li-Cor Biosciences) as a secondary antibody. Fluorescence was detected with a Li-Cor Odyssey kit (Li-Cor Biosciences) and the membrane scanned using an Odyssey CLx infrared imaging system (Li-Cor Biosciences) operated in the 700/800-nm channel. The bands were analyzed using Odyssey imaging software to quantify pixel intensity.
Growth estimates based on protein content.
S. tropica CNB-440 and CNB-440 mscL+ were grown in triplicate for 5 days in medium A1 (70% seawater) with apramycin (200 μg/ml), pelleted by centrifugation (7,000 × g), washed twice with phosphate-buffered saline, and diluted 1:100 in PBS. Aliquots (200 μl) containing approximately 2 × 106 CFU/ml were inoculated into 100 ml medium A1 (70% seawater) and A1 prepared with DI water and allowed to grow for 1 week at 27°C while shaking at 230 rpm. Duplicate 1-ml subsamples were taken every 24 h throughout the growth curve and assayed for total protein content using previously described methods (13) and modifications (15). In brief, the samples were centrifuged (13,800 × g) for 5 min. The pellets were washed by vortexing with 1 ml PBS (pH 7.0), centrifuged again as described above, and frozen (−20°C). For analysis, the pellets were resuspended in 0.1 ml of 1 M NaOH, placed in boiling water for 10 min, and neutralized by adding 0.02 ml of 5 M HCl, and the volumes were adjusted to 1 ml by adding PBS. The samples were then centrifuged for 30 min and the absorbance of 0.8 ml measured at 230 and 260 nm using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). The protein concentration (μg/ml) was determined from the equation [protein] = (183 × A230) − (75.8 × A260) (13). The assay is linear over the range of 6 to 225 μg protein/ml (13), and extracts from heavily turbid cultures were diluted in PBS to ensure that measurements remained within the linear range.
Effects of exposure to DI water on growth.
S. tropica CNB-440 and CNB-440 mscL+ were grown in triplicate for 5 days in 30 ml A1 (70% seawater). The cells were pelleted, washed twice with DI water, and resuspended in 20 ml DI water without shaking at room temperature for various times from 1 to 72 h. Aliquots (300 μl) were then spread plated onto medium A1 (70% seawater) and incubated at 30°C for 2 weeks. Growth was visually assessed.
Viability estimates.
S. tropica CNB-440 and CNB-440 mscL+ were grown in triplicate and exposed to DI water as described above. Live versus dead cells were distinguished using the BacLight LIVE/DEAD bacterial viability kit (L7012; Life Technologies, Grand Island, NY) following the manufacturer's instructions. In brief, equal volumes of dye components A and B were combined in a microcentrifuge tube and mixed, and 3 μl was added for each 1 ml of bacterial suspension analyzed. The suspensions were thoroughly mixed and incubated at room temperature in the dark for 15 min, and 5 μl was placed between a glass microscope slide and an 18-mm square coverslip. The samples were observed at a magnification of ×40 using an Olympus MVX10 fluorescence microscope (Olympus, Center Valley, PA) equipped with filter cube U-MCFPHQ/XL. Fluorescence associated with viable (green) and nonviable (red) cells was measured at 510 to 540 and 620 to 650 nm, respectively. Images of 10 different fields were captured for each treatment using an Olympus DC71 camera operated by DP Manager software. The experiment was repeated three times for each strain.
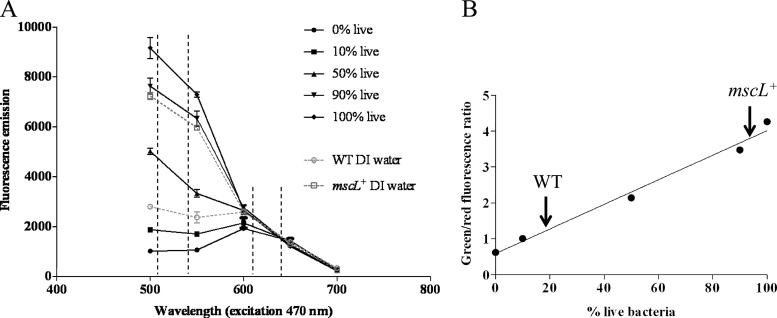
In an effort to quantify cell viability, S. tropica CNB-440 and CNB-440 mscL+ were cultured in triplicate for 5 days in 30 ml A1 (70% seawater). One half of each culture was heat killed by boiling for 20 min and confirmed to be nonviable by plating on A1 agar (70% seawater). The suspensions of live and heat-killed cells were adjusted to an optical density at 600 nm (OD600) of 0.30 using a spectrophotometer (BioPhotometer; Eppendorf). Live and dead bacterial suspensions (2 ml) were then prepared in ratios of 0:100, 10:90, 50:50, 90:10, and 100:0 and stained as described above using the BacLight LIVE/DEAD bacterial viability kit to generate a standard curve of fluorescence versus percent viable cells.
In parallel, S. tropica CNB-440 and CNB-440 mscL+ were grown in triplicate for 5 days in 30 ml A1 (70% seawater). Cells were pelleted and washed as described above and soaked in DI water for 24 h. The cell suspensions were then adjusted to an OD600 of ca. 0.30 in a 2-ml total volume and stained as described above, and 100 μl was pipetted into separate wells of a 96-well, flat-bottom microtiter plate. The plate was incubated at room temperature in the dark for 15 min, after which the fluorescence emission at 500 to 700 nm was measured using a microtiter plate reader (SpectraMax M2; Molecular Devices, Inc., Sunnyvale, CA) with the excitation wavelength set to 470 nm. The data were analyzed for each bacterial suspension by calculating the ratio of the integrated intensity of the green (510 to 540 nm) and red (620 to 650 nm) fluorescence emissions and plotting these values against the standard curved described above to estimate the percentage of live cells in the suspension.
Gadolinium experiments.
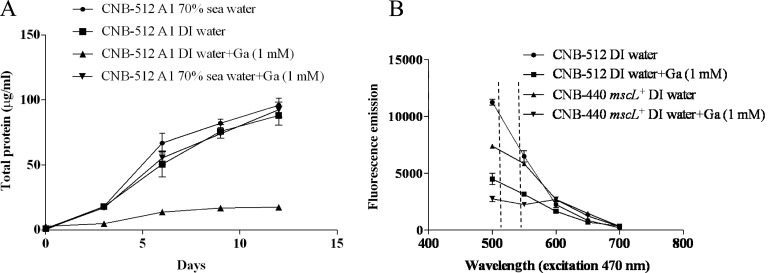
Micromonospora strain CNB-512 was grown in triplicate for 5 days in 30 ml medium A1 (70% seawater), pelleted by centrifugation (7,000 × g), washed twice with PBS, and resuspended in 10 ml DI water. Aliquots (200 μl) were inoculated into 100 ml medium A1 (70% seawater) with and without 1 mM gadolinium chloride and into A1 (DI water) with and without 1 mM gadolinium chloride and allowed to grow for 2 weeks at 27°C with shaking at 230 rpm. Duplicate 1-ml subsamples were taken every 24 h throughout the growth curve and assayed for total protein content using the method described above. In parallel, the viabilities of Micromonospora strain CNB-512 and S. tropica CNB-440 mscL+ in media prepared with 100% DI water with and without 1 mM gadolinium were measured using the BacLight bacterial viability kit as described above.
RESULTS
PCR probing for the mscL gene.
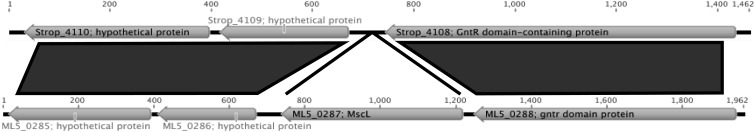
The mscL gene was not observed in the complete genome sequence of S. tropica strain CNB-440 (accession number CP000667) (Fig. 1) or S. arenicola strain CNS-205 (CP000850). It was also not present in the draft genomes of S. arenicola strain CNH-643 (PRJNA84391), S. arenicola strain CNT-088 (PRJNA84269), S. pacifica strain CNS-143 (PRJNA84389), and S. pacifica strain CNT-133 (PRJNA84271). To further explore the absence of this gene in the genus, we PCR probed for a 320-bp internal region and a 580-bp region that included the upstream mscL flaking sequence in six additional Salinispora strains (Table 1). No PCR products were obtained from any of these strains, while products of the predicted size and sequence were consistently amplified using both sets of primers and DNA templates prepared from three Micromonospora strains (Table 2).
Fig 1.
Regional synteny plot of the Micromonospora L5 and S. tropica CNB-440 genomes. Dark gray indicates syntenic regions. Gene numbers (locus tags) and GenBank annotations are listed. Tick marks represent base pairs.
Table 2.
PCR amplification of the mscL gene
| Species | Strain | Growth in: |
PCR product |
||
|---|---|---|---|---|---|
| Seawater | DI water | 320 bp | 580 bp | ||
| S. tropica | CNB-440a | +++ | − | No | No |
| S. tropica | CNB-536 | +++ | − | No | No |
| S. tropica | CNH-898 | +++ | − | No | No |
| S. arenicola | CNS205a | +++ | − | No | No |
| S. arenicola | CNH-665 | +++ | − | No | No |
| S. arenicola | CNS-325 | +++ | − | No | No |
| S. pacifica | CNT-133a | +++ | − | No | No |
| S. pacifica | CNS-844 | +++ | − | No | No |
| S. pacifica | CNT-131 | +++ | − | No | No |
| Micromonospora sp. | CNB-394 | +++ | +++ | Yes | Yes |
| Micromonospora sp. | CNB-512 | +++ | +++ | Yes | Yes |
| Micromonospora sp. | CNX-434 | +++ | +++ | Yes | Yes |
The genome sequence is available.
Genetic complementation and expression of mscL in S. tropica.
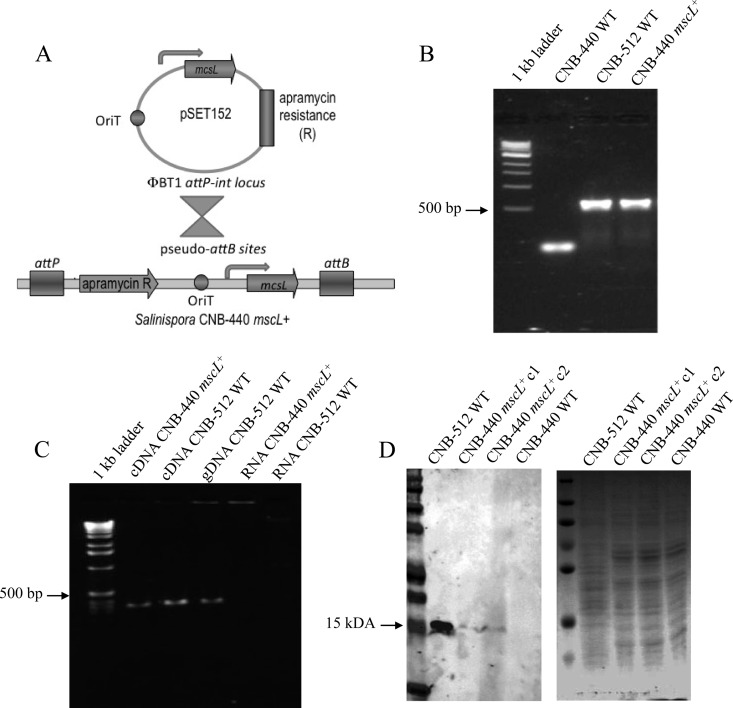
The genera Micromonospora and Salinispora are closely related within the family Micromonosporaceae. Nonetheless, sequence differences even among closely related taxa can present formidable barriers to the construction of interspecific hybrids. To construct a Salinispora interspecies recombinant, we PCR amplified the mscL gene from Micromonospora strain CNB-512 using primers designed to amplify the complete gene and upstream promoter region (580 bp). This PCR product was then successfully ligated into the pSET152 conjugative plasmid and introduced into E. coli S17-1 as a donor strain (E. coli S17-1/pSET152::mscL) (Fig. 2A). Retrosequencing revealed that pSET152 integration occurred at strop_0483, one of three previously identified S. tropica pseudointegration sites (11). Following transformation and the selection of an apramycin-resistant S. tropica exconjugant (S. tropica mscL+), PCR amplification yielded a 580-bp product that was sequence verified as mscL (Fig. 2B). Furthermore, RT-PCR experiments revealed that mscL was expressed in the CNB-440 exconjugant (Fig. 2C). Thus, a Salinispora interspecies genetic hybrid has been successfully constructed, and the native Micromonospora promoter is active in a Salinispora genetic background.
Fig 2.
Complementation experiments. (A) Diagram of the conjugation assay in which an E. coli donor strain harboring the Micromonospora CNB-512 mscL gene (S17-1/ pSET152::mscL) was used to introduce mscL into the recipient S. tropica CNB-440 strain. (B) PCR amplification of the mscL genes from S. tropica CNB-440 mscL+ and Micromonospora CNB-512, using the primer set EcoRI-mscL-ext-F/R (580-bp product). No appropriately sized product was observed from the CNB-440 WT strain. (C) PCR amplification of the mscL gene from cDNA generated from the CNB-440 mscL+ transformant and both cDNA and genomic DNA (gDNA) generated from Micromonospora strain CNB-512, using the primer set mscL-int-F/R (320-bp product). No products were observed from RNA controls. (D) Western blot analysis reveals the association of MscL with a membrane-enriched subcellular fraction as detected using an MscL-specific polyclonal antibody. The arrow shows the expected size of the protein, which was detected in relatively low quantities in two CNB-440 mscL+ transformants relative to the CNB-512 WT strain. The SDS-PAGE profile of the same samples stained with Coomassie blue shows that they contain similar protein concentrations.
Western blot analysis and MscL protein detection.
To determine if the mscL transcripts were translated and the resulting protein incorporated into the cell membrane of S. tropica mscL+, a polyclonal antibody targeting the Micromonospora CNB-512 MscL sequence was developed. Western blot analysis of membrane preparations derived from cultures of S. tropica mscL+ revealed a specific, 15-kDa band that corresponds to MscL (Fig. 2D). This band was present in both the wild-type (WT) Micromonospora strain CNB-512 and S. tropica mscL+; however, it was not observed in membrane preparations generated from the wild-type S. tropica CNB-440 strain. MscL production in Micromonospora CNB-512 was standardized to 100% (16.66 pixels) and compared with that in two recombinant S. tropica mscL+ strains. The fluorescence intensities of the hybridized probe were 6.89 and 6.75 pixels, corresponding to 41.5% and 40.66% of the positive control. These results demonstrate that MscL is incorporated into the S. tropica cell membrane, albeit at what appear to be reduced levels relative to those in the native Micromonospora strain.
Effect of osmotic downshock on Salinispora survival.
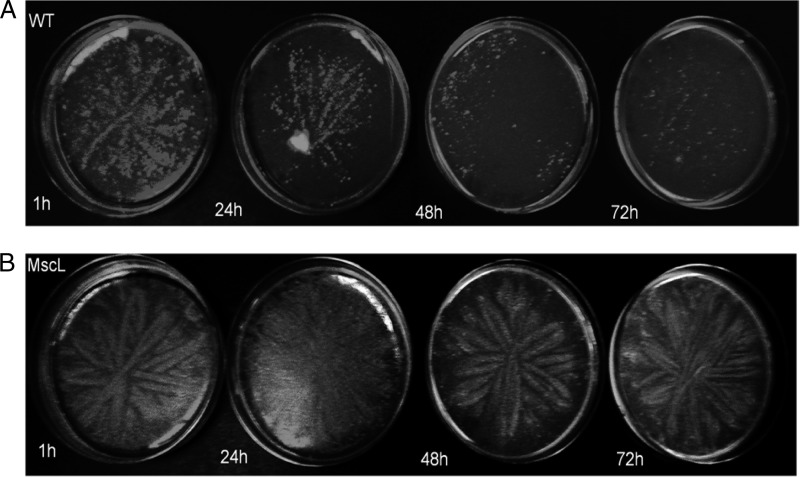
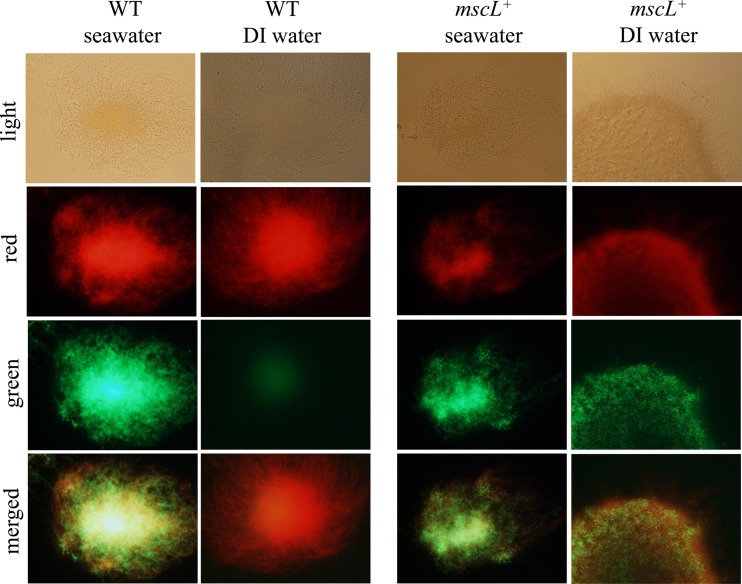
Initial efforts to cultivate S. tropica CNB-440 mscL+ revealed that this otherwise isogenic exconjugant, like the CNB-440 wild-type (WT) strain, failed to grow in complex medium prepared with DI water (data not shown). Consequently, we used two different approaches to test for the effects of exposure to DI water on cell viability. The first test involved a visual examination of growth on A1 medium prepared with seawater following exposure to DI water for 1 to 72 h. The results provide clear and reproducible evidence that growth was reduced in a time-dependent fashion in the WT strain yet remained largely unchanged in the S. tropica CNB-440 mscL+ exconjugant (Fig. 3). Given that Salinispora strains produce branching filaments, it was difficult to measure growth using traditional optical density or colony-counting methods. For this reason, the effect of exposure to DI water on cell viability was further explored using the BacLight LIVE/DEAD bacterial viability kit. When grown in medium prepared with seawater, WT and mscL+ strains were dominated by viable cells (Fig. 4). However, following a 24-h exposure to DI water, green fluorescence was dramatically reduced in the WT strain, indicating a lack of intact cell membranes (Fig. 4). The intense red emission from the same sample indicates that most cellular membranes had been disrupted and supports prior observations that Salinispora strains lyse in low-osmotic-strength media (26). Considerable green fluorescence is maintained in the mscL+ strain following exposure to DI water (Fig. 4), suggesting that the introduction of this gene has made the cells less susceptible to lysis. In an effort to quantify viability using the BacLight kit, the fluorescence emissions corresponding to various ratios of live and dead cells were measured (Fig. 5A). When plotted as the percentage of viable bacteria versus the ratio of green to red fluorescence, a linear relationship was observed (Fig. 5B). Following a 24-hour exposure to DI water, the green/red fluorescence ratio for the wild-type S. tropica CNB-440 strain corresponded to ca. 20% viable bacteria, while that for the mscL+ exconjugant was greater than 80%. Thus, it can be estimated that the introduction of the mscL gene increased viability by ca. 60%.
Fig 3.
Growth of the S. tropica strain CNB-440 wild-type (WT) strain and mscL+ transformant after exposure to DI water. (A) The WT showed a negative visual growth response in relation to increased exposure to DI water from 1 to 72 h prior to plating on medium prepared with seawater. (B) The otherwise isogenic mscL+ transformant grew considerably better following DI exposure. Results from a representative of three replicate experiments are shown.
Fig 4.
Viability of the S. tropica CNB-440 wild-type (WT) strain and the mscL+ transformant as measured using the BacLight bacterial viability kit following exposure to seawater (control) or DI water for 24 h. Mycelial masses were viewed at a magnification of ×40 using bright field and red (620- to 650-nm) and green (510- to 540-nm) filters; merged images are also shown.
Fig 5.
Viability quantification of S. tropica CNB-440 using the BacLight bacterial viability kit. (A) Fluorescence emissions in the viable (510- to 540-nm) and dead (620- to 650-nm) wavelengths for different ratios of live and dead cells along with the WT strain and the mscL+ transformant following 24 h of exposure to DI water. (B) The integrated 510- to 540-nm and 620- to 650-nm fluorescence ratio for the WT following a 24-hour exposure to DI water corresponds to ca. 20% viable bacteria, while that for the mscL+ exconjugant corresponds to greater than 80% viable bacteria. Averages ± standard deviations for three replicate experiments are plotted.
MscL chemical knockout.
Gadolinium chloride is a specific inhibitor of MscL function (2). To test the hypothesis that MscL provides resistance to osmotic downshock, the marine-derived Micromonospora strain CNB-512 was tested for growth on medium prepared with DI water supplemented with 1 mM GaCl2. While this strain grew equally well on media prepared with seawater, DI water, and seawater supplemented with GaCl2, growth as measured by total protein content was dramatically reduced when this compound was added to a medium prepared with DI water (Fig. 6A). Viability as measured using the BacLight kit was also reduced dramatically when GaCl2 was added to a medium prepared with DI water (Fig. 6B). Similar results were observed for the S. tropica CNB-440 mscL+ strain. Micromonospora strain CNB-512 was capable of growth on GaCl2 concentrations of as high as 5 mM, suggesting that compound toxicity was not a factor in the results. These experiments were repeated with two additional Micromonospora strains (Table 1), and similar results were obtained (data not shown).
Fig 6.
Chemical knockout of mscL function. (A) Growth as measured by protein content was equal in Micromonospora strain CNB-512 grown in media prepared with 70% seawater, 100% DI water, and 100% DI water plus 1 mM gadolinium, while growth in DI water with gadolinium was dramatically reduced. (B) Viability of Micromonospora strain CNB-512 and S. tropica CNB-440 mscL+ in media prepared with 100% DI water with and without 1 mM gadolinium as measured using the BacLight bacterial viability kit. Dashed lines indicate wavelengths of viable fluorescence emissions.
DISCUSSION
The genus Salinispora is unique among marine-derived actinomycetes in that all species cultured to date fail to grow in low-osmotic-strength media. While comparative genomics has been used to identify a pool of candidate marine adaptation genes that may be associated with this phenotype, it has been proposed that the lineage-specific loss of mscL plays a major role in the failure of Salinispora spp. to survive osmotic downshock (20). The present study provides experimental evidence in support of this hypothesis.
The recently released Micromonospora L5 genome sequence (accession number CP002399) facilitated the design of two mscL-specific primer sets that were used to successfully amplify this gene and the upstream promoter region from the marine-derived but non-seawater-requiring Micromonospora strain CNB-512 (9). mscL was not detected using either primer set in nine Salinispora strains representing all three currently recognized species; however, it remains possible that this negative PCR result is due to a lack primer specificity. In support of the lineage-specific loss of mscL, this gene was not detected in a total of six Salinispora genome sequences. Ongoing genome sequencing efforts will be used to further test this hypothesis, which if supported suggests that a PCR assay targeting mscL may represent a quick approach to distinguish between Salinispora and Micromonospora strains, which are not readily resolved based on morphological features.
While a recently developed genetic system has been used to inactive (4) and reintroduce (11) genes in Salinispora spp., the results presented here represent the first use of the pSET152 conjugative plasmid to introduce a non-Salinispora gene into a Salinispora genetic background. Remarkably, only a small genetic cassette harboring the mscL open reading frame and the 100-bp native promoter region was sufficient for the subsequent expression of this gene in S. tropica CNB-440, indicating that no additional species-specific factors are required. More importantly, a polyclonal antibody revealed that the gene product was associated with a membrane fraction of the CNB-440 mscL+ exconjugant, providing evidence that it was incorporated into the cytoplasmic membrane as has been shown in similar experiments with E. coli (24).
Although the recombinant Salinispora mscL+ strain expressed the MscL protein and it appears to have been incorporated into the cytoplasmic membrane, this in itself was not sufficient to facilitate growth in complex media prepared without added salts. There are a number of possible explanations for this, including preliminary evidence for low levels of MscL expression relative to those in the parent Micromonospora strain CNB-512 (Fig. 2). Alternatively, other marine adaptation genes, such as a highly duplicated family of polymorphic membrane proteins that appears to have been acquired from marine bacteria (19), may contribute to the inhibitory effects of a low-osmotic-strength environment. Nevertheless, the introduction of the single-copy mscL gene into S. tropica CNB-440 enhanced survival following osmotic downshock and provides yet another example of the role of MscL in osmoadaptation. The reduced viability observed following the chemical knockout of MscL function using gadolinium in both Micromonospora CNB-512 and the CNB-440 mscL+ exconjugant further supports the role of this protein in surviving osmotic downshock. It is also of interest to note that mscS homologs detected in both Salinispora genomes do not appear to complement mscL function as has been observed in E. coli (12). These results provide experimental evidence that the loss of mscL is associated with the inability of Salinispora spp. to grow in complex media that lack added salts. Although there are no known benefits associated with mscL loss in Salinispora, it may be an important factor that contributes to their reported requirement for seawater for growth.
The Salinispora 16S rRNA phylogeny reveals that it is closely related to a large number of nonmarine actinomycete genera. Thus, it can be proposed that this lineage is the result of a secondary introduction into the marine environment. Given the consistent salinity of seawater, it would not be surprising if the loss of mscL had no effect on the ability of an ancestral Salinispora strain to survive in the marine environment. This loss appears to have occurred prior to speciation within the genus and may account for the fact that Salinispora strains have yet to be reported outside the marine environment. It is of interest to note that no other marine-derived actinobacteria for which genome sequences are available lack mscL, although many other marine bacteria are missing this gene. This may be due to the possibility that Salinispora spp. have been in the marine environment longer than other marine actinobacteria or may simply reflect the stochastic nature of selectively neutral evolutionary events. It is intriguing to speculate that the random loss of a single gene may have resulted in the obligate marine distribution of this actinomycete lineage.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grant R01-GM086261 to P.R.J. S.A.B. was supported by a STIPAS postdoctoral fellowship from MECESUP 2 and the University of Chile.
We thank Brad Moore and Anna Lechner for providing the pSET152 plasmid, transformation protocols, and E. coli strains.
Footnotes
Published ahead of print 6 April 2012
REFERENCES
- 1. Ajouz B, Berrier C, Garrigues A, Besnard M, Ghazi A. 1998. Release of thioredoxin via the mechanosensitive channel MscL during osmotic downshock of Escherichia coli cells. J. Biol. Chem. 273:26670–26674 [DOI] [PubMed] [Google Scholar]
- 2. Berrier C, Coulombe A, Szabo I, Zoratti M, Ghazi A. 1992. Gadolinium ion inhibits loss of metabolites induced by osmotic shock and large stretch-activated channels in bacteria. Eur. J. Biochem. 206:559–565 [DOI] [PubMed] [Google Scholar]
- 3. Bierman M, et al. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49 [DOI] [PubMed] [Google Scholar]
- 4. Eustaquio AS, Pojer F, Noe JP, Moore BS. 2008. Discovery and characterization of a marine bacterial SAM-dependent chlorinase. Nat. Chem. Biol. 4:69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fenical W, Jensen PR. 2006. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat. Chem. Biol. 2:666–673 [DOI] [PubMed] [Google Scholar]
- 6. Fenical W, et al. 2009. Discovery and development of the anticancer agent salinosporamide A (NPI-0052). Bioorg. Med. Chem. 17:2175–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Folgering JH, Moe PC, Schuurman-Wolters GK, Blount P, Poolman B. 2005. Lactococcus lactis uses MscL as its principal mechanosensitive channel. J. Biol. Chem. 280:8784–8792 [DOI] [PubMed] [Google Scholar]
- 8. Hoffmann T, Boiangiu C, Moses S, Bremer E. 2008. Responses of Bacillus subtilis to hypotonic challenges: physiological contributions of mechanosensitive channels to cellular survival. Appl. Environ. Microbiol. 74:2454–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jensen P, Dwight R, Fenical W. 1991. Distribution of actinomycetes in near-shore tropical marine sediments. Appl. Environ. Microbiol. 57:1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jensen PR, Mafnas C. 2006. Biogeography of the marine actinomycete Salinispora. Environ. Microbiol. 8:1881–1888 [DOI] [PubMed] [Google Scholar]
- 11. Lechner A, Eustaquio AS, Gulder TAM, Hafner M, Moore BS. 2011. Selective overproduction of the proteasome inhibitor salinosporamide A via precursor pathway regulation. Chem. Biol. 18:1527–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Levina N, et al. 1999. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 18:1730–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Makkar HP, Sharma OP, Dawra RK, Negi SS. 1982. Simple determination of microbial protein in rumen liquor. J. Dairy Sci. 65:2170–2173 [DOI] [PubMed] [Google Scholar]
- 14. Maldonado LA, et al. 2005. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 55:1759–1766 [DOI] [PubMed] [Google Scholar]
- 15. Meyers PR, et al. 1998. Novel method for rapid measurement of growth of mycobacteria in detergent-free media. J. Clin. Microbiol. 36:2752–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mincer TJ, Jensen PR, Kauffman CA, Fenical W. 2002. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 68:5005–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakamaru Y, Takahashi Y, Unemoto T, Nakamura T. 1999. Mechanosensitive channel functions to alleviate the cell lysis of marine bacterium, Vibrio alginolyticus, by osmotic downshock. FEBS Lett. 444:170–172 [DOI] [PubMed] [Google Scholar]
- 18. Oh S, Kogure K, Ohwada K, Simidu U. 1991. Correlation between possession of a respiration-dependent Na+ pump and Na+ requirement for growth of marine bacteria. Appl. Environ. Microbiol. 57:1844–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Penn K, et al. 2009. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 3:1193–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Penn K, Jensen P. 2012. Comparative genomics reveals evidence of marine adaptation in Salinispora species. BMC genomics. 13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schnaitman CA. 1971. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J. Bacteriol. 108:545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 23. Sukharev SI, Blount P, Martinac B, Blattner FR, Kung C. 1994. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature 368:265–268 [DOI] [PubMed] [Google Scholar]
- 24. Sukharev SI, Blount P, Martinac B, Kung C. 1997. Mechanosensitive channels of Escherichia coli: the MscL gene, protein, and activities. Annu. Rev. Physiol. 59:633–657 [DOI] [PubMed] [Google Scholar]
- 25. Tsueng G, Lam K. 2008. A low-sodium-salt formulation for the fermentation of salinosporamides by Salinispora tropica strain NPS21184. Appl. Microbiol. Biotechnol. 78:821–826 [DOI] [PubMed] [Google Scholar]
- 26. Tsueng G, Lam KS. 2008. Growth of Salinispora tropica strains CNB440, CNB476, and NPS21184 in nonsaline, low-sodium media. Appl. Microbiol. Biotechnol. 80:873–880 [DOI] [PubMed] [Google Scholar]
- 27. Udwary DW, et al. 2007. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U. S. A. 104:10376–10381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wood JM, et al. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:437–460 [DOI] [PubMed] [Google Scholar]