Abstract
The intensification of human activities within the habitats of wild animals is increasing the risk of interspecies disease transmission. This risk is particularly important for great apes, given their close phylogenetic relationship with humans. Areas of high human density or intense research and ecotourism activities expose apes to a high risk of disease spillover from humans. Is this risk lower in areas of low human density? We determined the prevalence of Escherichia coli antibiotic-resistant isolates in a population of the critically endangered western lowland gorilla (Gorilla gorilla gorilla) and other wild mammals in Lopé National Park (LNP), Gabon, and we tested whether the observed pattern could be explained by bacterial transmission from humans and domestic animals into wildlife populations. Our results show a high prevalence of antibiotic-resistant bacterial isolates in humans and low levels in gorillas and other wildlife. The significant differences in the genetic background of the resistant bacteria isolated from humans and gorillas suggest that transmission is low or does not occur between these two species. These findings indicate that the presence of antibiotic-resistant strains in wildlife do not imply direct bacteria transmission from humans. Thus, in areas of low human density, human-wildlife E. coli transmission seems to be low. The presence of antibiotic-resistant isolates in gorillas may be better explained by other mechanisms for resistance acquisition, such as horizontal gene exchange among bacteria or naturally acquired resistance.
INTRODUCTION
The intensification of human activities within habitats of previously isolated wild animals is a key factor in the emergence of infectious diseases (7, 18). Although major focus has been given to the spread of zoonotic diseases into human populations (23, 43, 45), anthropogenic activities also cause the emergence of disease in wildlife populations (7, 8). In particular, the close phylogenetic relationship between great apes and humans exposes apes to a high risk of disease spillover from humans (44). In the last 2 decades, bushmeat hunting, forest encroachment, ecotourism, and research activities are increasing the levels of contact between humans and great apes. This in turn has resulted in several confirmed cases of human pathogen transmission to apes (e.g., human respiratory virus in chimpanzees [19] and mountain gorillas [28], among other examples [12]). Escherichia coli exchanges between humans, domestic animals, and great apes have been reported in densely human-populated areas of western Uganda. Within Uganda, habituated groups of wild apes are visited daily by researchers and tourists (e.g., chimpanzees at Kibale [13] and mountain gorillas at Bwindi Impenetrable National Park [33]). However, there has been little attention directed toward the quantification of bacterial transmission in areas with low human impact, which could serve to elucidate whether the pattern of pathogen prevalence observed in wildlife is explained by transmission from humans.
Ubiquitous and commensal bacteria such as E. coli represent a convenient model to study patterns of bacterial transmission between humans and domestic and wild animals (13, 33). In particular, the presence of antibiotic-resistant strains in untreated wild animals has been suggested to reflect bacterial exchange with humans or domestic animals, in which treatment with antibiotics actively selects antibiotic-resistant strains (1, 6, 9, 22, 32, 34). Furthermore, several molecular techniques are now available to compare the genetic structure of E. coli strains from different populations (4, 37), thus contributing to the understanding of the subjacent transmission process.
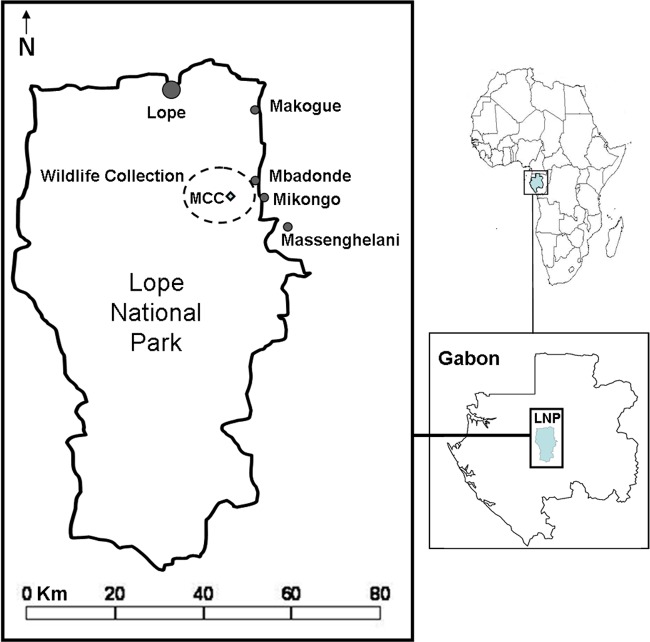
In this study, we determined the pattern of E. coli antibiotic-resistant isolates derived from feces of a population of the critically endangered western lowland gorilla (Gorilla gorilla gorilla) and other wild mammals, and we assessed if the observed pattern occurred as a result of bacterial transmission from humans. This study was conducted in Lopé National Park (LNP) in central Gabon (Fig. 1), which hosts important populations of western lowland gorillas and other wild mammals (38, 39). Lopé is surrounded by a human and domestic population density that is one of the lowest around the vicinity of any African protected area (17). In addition, the use of antibiotics in Lopé is almost entirely restricted to the treatment of human infectious diseases. Thus, the frequency of human-wildlife transmission of resistant strains is expected to be low.
Fig 1.
Study site. (Template map from About.com.)
Our results show high levels of antibiotic resistance in E. coli isolates from human feces and low levels in those from domestic animals, gorillas, and other wildlife. However, the significant difference in the genetic structure of E. coli-resistant isolates derived from humans and domestic animals and those from gorillas does not support direct bacterial transmission between these two populations. These findings suggest that the presence of antibiotic-resistant strains in wildlife do not imply direct bacterial transmission from humans, and that in areas of low human density, human-wildlife E. coli transmission is low.
MATERIALS AND METHODS
Sample collection. (i) Wild animals.
The collection of wildlife samples was conducted in an area of 42 km2 around the Mikongo Conservation Centre (MCC), located in the northeast region of LNP (0°18′23′′N, 11°42′06′′E) (Fig. 1). Since 2001, the MCC has been carrying out research and tourism activities on western lowland gorillas but without full gorilla habituation. Fresh fecal samples (less than 24 h after defecation) were collected during a 2-week census designed to study gorilla population density surrounding MCC in February 2010. Each day, four teams composed of three experienced trackers and one researcher walked predetermined forest transects designed to optimize the sampling area and to avoid duplicating the sampling of the same gorilla group (16). When a gorilla trace was found, the team tracked the traces to locate the nest built by the gorilla group the previous night. A total of 119 gorilla fecal samples were collected from 14 different gorilla nest sites. Only one fecal sample was collected per nest in each site. Samples from other mammals were also collected opportunistically, including 6 chimpanzees (Pan troglodytes troglodytes), 5 mandrills (Mandrillus sphinx), 5 monkeys (including black colobus, Colobus satanas, and the gray-cheeked mangabey, Lophocebus albigena), 12 duikers of several species, 5 river hogs (Potamochoerus porcus), 8 forest buffalos (Syncerus caffer nanus), and 7 African elephants (Loxodonta africana cyclotis) (Table 1). Approximately 2 g of fecal material sample was collected from the interior of the fresh fecal stool and placed in a 1.5-ml Eppendorf tube containing 1 ml of sterile and isotonic buffered solution composed of NaCl at 0.9%.
Table 1.
Summary of the distribution of resistant E. coli isolates according to each antibiotic and collection source
| Antibiotic | Isolate distributiona by sample source |
% of resistance from total fecal isolates (n = 226) | ||||
|---|---|---|---|---|---|---|
| Gorillas (n = 119) | Other wildlife (n = 48) | Humans (n = 25) | Domestic animals (n = 34) | Water (n = 10) | ||
| Ampicillinb | 4 (3.4) | 4 (8.3) | 9 (36.0) | 1 (2.9) | 0 | 8 |
| Ceftiofure | 0 | 0 | 0 | 0 | 0 | 0 |
| Chloramphenicolb | 0 | 1 (2.1) | 1 (4.0) | 0 | 0 | 0.9 |
| Tetracyclineb | 3 (2.5) | 2 (4.2) | 8 (32.0) | 0 | 0 | 5.8 |
| Ciprofloxacinc | 0 | 0 | 0 | 0 | 0 | 0 |
| Doxycyclined | 0 | 1 (2.1) | 6 (24.0) | 0 | 0 | 3 |
| Neomycinc | 0 | 0 | 0 | 0 | 0 | 0 |
| Rifampind | 2 (1.7) | 1 (2.1) | 2 (8.0) | 2 (5.9) | 0 | 3.1 |
| Streptomycind | 3 (2.5) | 1 (2.1) | 9 (36.0) | 1 (2.9) | 0 | 6.2 |
| Sulfamethoxazoled | 1 (0.8) | 0 | 11 (44.0) | 3 (8.8) | 0 | 6.6 |
The number (n) of resistant isolates to each antibiotic is given for each host (percentages are in parentheses).
Most commonly used (in Lopé).
Rarely used.
Not used in Lope but available in Gabon.
Not used in Lopé and very rarely used in Gabon.
(ii) Humans and domestic animals.
Samples from 25 adult humans that agreed to participate in this study and 34 domestic animals were collected in the five villages bordering the park located close to the area of wildlife sampling: Lopé (number of inhabitants [n] ≈ 300), Makoghé (n ≈ 50), Mbadondé (n ≈ 50), Mikongo (n ≈ 100), and Massenghelani (n ≈ 70) (Fig. 1 shows the locations). Each village had a small number of livestock (between 10 and 50 animals) composed essentially of sheep and goat, and several households possessed companion animals (i.e., cats and dogs). The incursion of wild animals, such as elephants, buffalos, or monkeys, into crop fields and streams used by the villagers for water consumption or bathing was common in the villages. Humans were given a 150-ml sterile Falcon (BD) tube and were asked to put a small fecal swab in the tube using provided sterile gloves and/or a sterile cocktail stick. Tubes containing samples were collected the next day and filled with buffer solution. None of the samples presented signs of diarrhea. Domestic animal samples included 21 sheep (Ovis aries), 4 goats (Capra aegagrus hircus), 8 dogs (Canis lupus familiaris), and 1 cat (Felis catus). Livestock animals were followed until defecation occurred, whereas samples from companion animals were collected rectally. The collection of human (and domestic animal) samples was approved by a written statement obtained from volunteers (or livestock owners), the village chief, and the Centre National de Recherche Scientifique et Technologique du Gabon (CENAREST; permit number AR0026/09).
(iii) Water and soil samples.
Twenty water samples were collected from five different streams encountered in the forest during the surveys and from the 15 water sources available within the five villages. These sources included 2 main rivers (Offoué and Ogooué), 12 streams, and 1 open well. Water samples were collected in a 50-ml syringe and were systematically filtered with a high-flow syringe containing a 0.45-μm polyether sulfone membrane filter (Sartorius Stedim Biotech), which retained bacteria (29). The membrane filter was then placed on top of a MacConkey (MC) agar plate (VWR) no more than 12 h after collection. Ten soil samples were collected in proximity to 10 gorilla nests sampled. Soil samples were stored in buffer solution for further bacterial isolation.
Bacterial isolation.
Bacteria from fecal samples were isolated in MC selective medium by following the methods described by Quinn (29) as soon as possible within the 48 h following their collection. Previous studies have shown that the short-term storage of samples (up to 2 weeks) does not affect the phylogenetic structure, abundance, and diversity of microbial communities (20). After collection, each fecal, soil, and water membrane sample was streaked on an MC agar plate (VWR) and incubated at 37°C for 24 to 48 h. Five isolated pink lactose-positive colonies, visually resembling E. coli, were transferred into a 200-μl, 96-well microtiter plate containing MC agar. Microtiter plates were kept at 4°C and shipped to France for further analyses. Once in France, colonies were reisolated and identified by biochemical testing (API 20E gallery or Vitek [bioMérieux]). Only one identified E. coli organism isolated per fecal sample was saved in 20% glycerol and stored at −80°C for further analysis. Unbalanced sampling per individual and pseudoreplication therefore were avoided. For descriptive purposes, all different morphotypes present within the five isolates belonging to water samples were identified (Table 2).
Table 2.
Bacteria species recovered from water samplesa
| Origin | No. of water sources | No. of Lac+ isolates | Species found (n) | No. of Lac− isolates | Species found (n) |
|---|---|---|---|---|---|
| Forest streams | 5 | 14 | Serratia marcescens (7), Escherichia coli (5), Klebsiella pneumonia (1), Klebsiella oxytoca (1) | 3 | Proteus mirabilis (2), Proteus vulgaris (1) |
| Villages | 15 | 49 | Serratia marcescens (33), Escherichia coli (5), Enterobacter sp. (5), Klebsiella pneumonia (3), Klebsiella oxytoca (2), Klebsiella terrigena (1) | 8 | Proteus mirabilis (3), Morganella morganii (3), Proteus vulgaris (2) |
Each morphologically distinct isolate (out of five isolates per sample) was identified to the species level for each water sample. The number of isolates belonging to each bacterial species is given in parentheses.
Analyses. (i) Antibiotic resistance.
Antibiotic susceptibility was tested using the agar dilution method recommended by the Clinical and Laboratory Standards Institute (5). Ten antibiotics were selected according to their availability in Lop é and Gabon. Ampicillin, chloramphenicol, and tetracycline represent the most common antibiotics used in Gabon (27), and they were available in the only local pharmacy of Lopé. Ciprofloxacin and neomycin were rarely available in the Lopé area. Doxycycline, sulfamethoxazole, rifampin, and streptomycin were not available at Lopé but are used in Gabon for the treatment of malaria (26), Shigella (27), and tuberculosis (25). Ceftiofur, an expanded-spectrum cephalosporin antibiotic, was never available at Lopé, and it is very rarely used in Gabon. Two-fold dilutions of the antibiotics were added to obtain final concentrations superior to the critical concentrations according to breakpoints provided by the Clinical and Laboratory Standards Institute guidelines. Resistance to antibiotics was tested at the following concentrations: ampicillin (>8 μg/ml), ceftiofur (>8 μg/ml), ciprofloxacin (>2 μg/ml), tetracycline (>8 μg/ml), chloramphenicol (>8 μg/ml), doxycycline (>8 μg/ml), neomycin (>4 μg/ml), rifampin (>16 μg/ml), streptomycin (>16 μg/ml), and sulfamethoxazole (>256 μg/ml). Inocula were prepared in saline solution to achieve 108 CFU/ml (0.5 McFarland standard) and then diluted 1 in 10 using sterile saline solution. A multipoint inoculator was used to dispense 1 μl of diluted inoculum to achieve 104 CFU/ml per plate, with and without antibiotics. Plates were incubated in air at 37°C for 18 h. Fully susceptible E. coli ATCC 25922 was used as a control. An isolate was considered resistant if it grew in the presence of the antibiotic and formed a homogenized colony similar to the control colony grown in medium without antibiotic. A replicate was systematically run to confirm each resistant case. To evaluate the reproducibility of the method, independent tests were performed for E. coli ATCC 25922 and 10 clinical strains (two tests each).
(ii) E. coli phylogenetic groups.
Each E. coli isolate was assigned to one of the four major phylogenetic groups (A, B1, B2, and D) (21) using a multiplex PCR assay that creates the simultaneous amplification of the chuA and yjaA genes and DNA fragment TSPE4.C2 (4). The presence of the three markers in the same strain denotes the subgroup B23. This method has been widely used for the genetic description of different E. coli populations given its rapidity and simplicity (4, 10, 37) and its equivalence to other more sophisticated methods, such as multilocus sequence typing (14). Primers used in this study were those from Clermont et al. (4).
(iii) Statistical analyses.
Fisher exact tests where used to compare the prevalence of resistant isolates and the proportion of the different phylogenetic groups of the E. coli isolates derived from different host populations. This test is recommend for the analysis of contingency tables with small samples sizes, including the distribution of genetic characteristics (11, 30). The number of isolates resistant to each antibiotic was compared by means of a Spearman correlation test, which is a nonparametric test that is suitable for a small number of data points (10 in our case) (35).
RESULTS
Patterns of antibiotic resistance.
A total of 236 E. coli isolates were considered in this study, of which 226 came from fecal samples (1 isolate per stool sample from the 25 humans, the 34 domestic animals, the 119 gorillas, and the other 48 wild animals sampled) and 10 from water (Table 1). Although all water samples harbored a variety of enterobacterial species (Table 2), E. coli was present in only 10 of the 20 samples. No enterobacteria were isolated from the 10 soil samples. Thirty of the 226 feces-derived isolates were resistant to at least one antibiotic. In humans, 48.0% (12 out of 25) of the E. coli isolates were resistant to at least one antibiotic, compared to 14.7% (5 out of 34) in domestic animals, 10.4% (5 out of 48) in wild animals, and 6.7% (8 out of 119) in gorillas (Fig. 2). Resistance to at least one antibiotic was significantly higher in E. coli isolates from stools collected in the villages (i.e., humans and domestic animals) than those collected in the forest (i.e., gorillas and other wildlife) (2-by-2 Fisher's exact test; odds ratio, 4.79; P < 0.001). However, this difference seems to be driven by the high prevalence of antibiotic-resistant isolates in humans, because no significant differences were found for isolates from domestic animals, gorillas, and other wildlife (2-by-3 Fisher's exact test; P = 0.69). E. coli isolates derived from human stools were resistant to 7 of the 10 antibiotics tested, whereas those from wild animals, gorillas, and domestic animals were resistant to 6, 5, and 4 antibiotics, respectively (Table 1). None of the E. coli isolates from water were resistant to any of the antibiotics tested in this study (Fig. 2).
Fig 2.
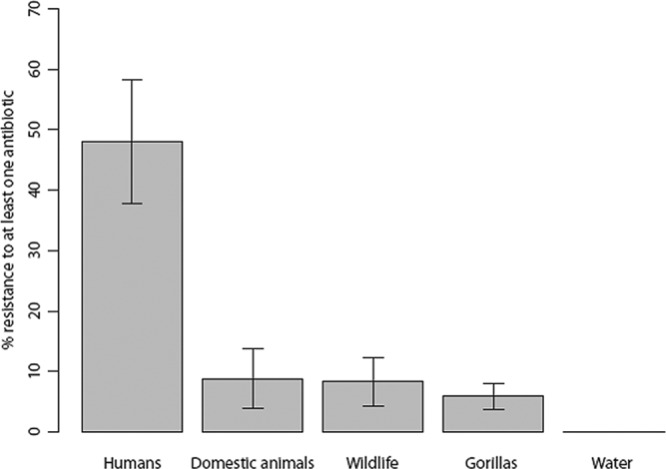
Antibiotic resistance per sample origin. Bars represent the proportion of isolates resistant to at least one antibiotic (given in percentages) for each source. Error bars were calculated as standard errors (SE) for a proportion, given by SE = p × (1 − p)/n, where p is the proportion of isolates resistant and n is the total number of isolates. SE were multiplied by 100 to scale bars given in percentages.
Overall, 7.6% of all isolates were resistant to ampicillin, 6.4% to sulfamethoxazole, 5.9% to streptomycin, and 5.5% to tetracycline. No resistance was observed for neomycin, ciprofloxacin, and ceftiofur. Multidrug resistance was detected in 17 (56%) of the 30 resistant isolates. Multidrug resistance averaged (means ± standard deviations [SD]) 3.8 ± 1.8 antibiotics in isolates from humans, 1.4 ± 0.9 in isolates from domestic animals, 2.0 ± 1.4 in isolates from wild animals, and 1.6 ± 0.7 in isolates from gorillas. In addition, 88.2% of multidrug-resistant combinations included ampicillin and 70.6% included tetracycline, streptomycin, or sulfamethoxazole. There was a large diversity of antibiotic phenotypes, with 18 isolates having a unique phenotype (Table 3).
Table 3.
Antibiotic-resistant phenotype and phylogenetic group for each resistant E. coli isolatea
| No. of isolates | Antibiotic-resistant phenotype | E. coli phylogenetic group |
|---|---|---|
| Gorillas | ||
| 1 | Tet, Amp, Rif | B23 |
| 1 | Amp, Str | B23 |
| 1 | Tet, Amp | D |
| 1 | Amp, Sul | B23 |
| 2 | Str | B23, B23 |
| 1 | Rif | D |
| 1 | Tet | B23 |
| Wildlife | ||
| 1 | Tet, Chl, Dox, Amp | D |
| 1 | Tet, Amp, Str | A |
| 2 | Amp | D, D |
| 1 | Rif | B2 |
| Domestic animals | ||
| 1 | Amp, Str, Sul | B1 |
| 2 | Rif | D, A |
| 2 | Sul | A, D |
| Humans | ||
| 1 | Tet, Chl, Dox, Amp, Str, Sul | D |
| 1 | Tet, Dox, Amp, Str, Sul, Rif | A |
| 3 | Tet, Dox, Amp, Str, Sul | D, B1, A |
| 1 | Dox, Amp, Str, Sul, Rif | A |
| 1 | Tet, Amp, Str, Sul | A |
| 1 | Tet, Dox, Str, Sul | D |
| 1 | Amp, Sul | B1 |
| 1 | Str, Sul | D |
| 1 | Amp | B1 |
| 1 | Sul | D |
Antibiotic-resistant phenotypes represent the antibiotics to which an isolate was resistant. Codes for antibiotics are the following: Amp, ampicillin; Tet, tetracycline; Chl, chloramphenicol; Dox, doxycycline; Rif, rifampin; Set, streptomycin; and Sul, sulfamethoxazole.
The number of strains resistant to each antibiotic was correlated between isolates from the stools of humans and gorillas (Spearman correlation test, rho = 0.75; P = 0.01). Thus, when the number of isolates resistant to an antibiotic was high in humans, it was also high in gorillas, and if the number was low in humans, it was also low in gorillas. This correlation was also observed between gorillas and other wildlife (Spearman's rho = 0.71; P = 0.02) but not between gorillas and domestic animals (Spearman's rho = 0.61; P > 0.05).
E. coli population structure.
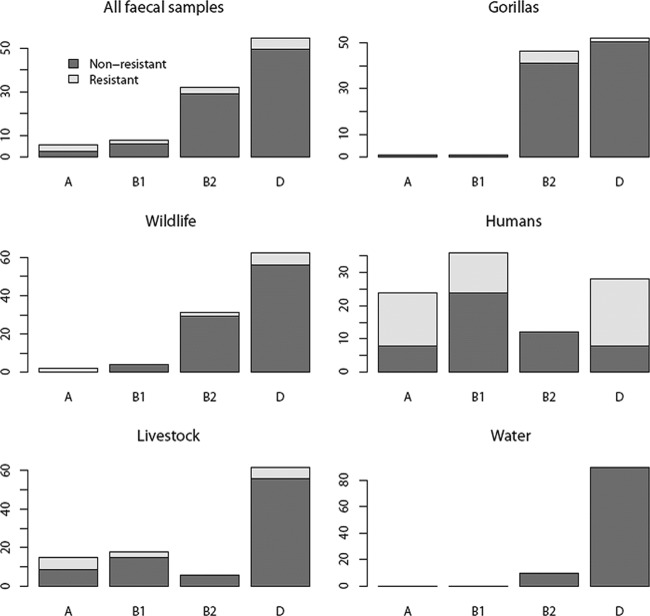
The majority of E. coli isolates belonged to groups D (54.7%) and B2 (32.2%), and only a few belonged to groups B1 (7.6%) and A (5.5%) (Fig. 3). Isolates collected from water samples belonged mostly to groups D and B2. The stools of gorillas and other wild animals exhibited similar E. coli composition (2-by-4 Fisher exact test, P > 0.05), with high prevalence in isolates from groups D and B2. The stools of humans and domestic animals harbored E. coli composition (2-by-4 Fisher exact test, P > 0.05) similar to that of E. coli isolates from all phylogenetic groups (Fig. 3). However, the E. coli composition in gorilla (or other wild animals) and human (or domestic animals) stool samples were significantly different (2-by-4 Fisher exact test, P < 0.001). The patterns of phylogenetic groups of resistant isolates reflected that of nonresistant isolates (Fisher exact test, P > 0.1) (Fig. 3). Thus, all antibiotic-resistant isolates found in gorillas belonged to group B2 or D, which are also the most prevalent groups among nonresistant isolates. Combining both genetic and antibiotic phenotype profiles revealed that resistant E. coli isolates present among gorillas or other wildlife do not match the ones observed among humans or domestic animals (Table 3). Moreover, 6 out of the 8 antibiotic-resistant isolates found in gorillas belong to the subgroup B23, which was not found in any of the antibiotic-resistant samples detected in humans or domestic animals.
Fig 3.
E. coli phylogenetic group pattern for each sample origin. Each bar represents the percentage of E. coli isolates identified in each group. Within a bar, each color represents the percentage of resistant and nonresistant strains.
DISCUSSION
Humans as reservoirs of antibiotic-resistant E. coli.
A high percentage of E. coli isolates found in human stools were resistant to at least one antibiotic (Fig. 1). This result confirms that central African regions share the worldwide trend in increasing antimicrobial resistance (41), and it suggests that human populations are the main reservoir for antibiotic-resistant strains in the study area. Resistance was observed particularly for antibiotics commonly used in Lopé, such as ampicillin and tetracycline (Table 1). The lack of resistance to ceftiofur, which is not available in the area, and to ciprofloxacin and neomycin, which are rarely used in Lopé, suggests that the high number of resistant isolates is mostly generated by the selective pressures of antibiotics prescribed to humans within the study area. However, the presence of resistance to other antibiotics, such as sulfamethoxazole and streptomycin (available in Gabon but not in Lopé), also suggests that humans in Lopé are receiving resistant strains from humans living in other areas of Gabon where these antibiotics are used. Levels of antibiotic resistance were low in isolates from domestic animals, which are almost never treated with antibiotics in this area. However, similarities in E. coli phylogenetic groups of resistant and nonresistant isolates in humans and domestic animals suggest that transmission between these two populations is occurring.
Transmission of antibiotic-resistant E. coli from humans to wildlife appears very low.
If E. coli antibiotic-resistant isolates were found in the stools of gorillas and other wild animals at a low level compared to that for humans, where are these resistant E. coli isolates coming from? Previous studies have proposed that contact and subsequent transmission of antibiotic-resistant bacterial strains from highly resistant sources, such as humans or livestock, could account for the presence of antibiotic resistance in wild animals (9, 22, 32, 34). In particular, the presence of multiresistant strains among gorillas has been used previously as evidence of resistant bacterial transmission from a source such as humans, which are subject to high selection pressure from antibiotics (e.g., see reference 33). However, in our study, we found significant differences in the genetic background of resistant and nonresistant E. coli isolates derived from humans-domestic animals and gorillas-other wildlife (Fig. 3). In particular, E. coli-resistant isolates in gorillas mainly belong to group B23, which was not a phylogenetic group observed for any resistant isolates from humans. Furthermore, the combination of both antibiotic-resistant phenotypes and phylogenetic group information showed that no isolates from gorillas have characteristics identical to those of human isolates (Table 3). All of these results favor the idea that the direct transmission of E. coli-resistant strains from humans to wild animals is not occurring. Furthermore, although water may be acting as an indirect transmission pathway for resistant strains, we did not find resistant strains in water samples. However, a larger sample size is required to draw definitive conclusions. This could be achieved by the repeated sampling of the same source, higher volumes of filtered water, or by using an enrichment medium prior to bacterial isolation.
Other mechanisms explaining the observed resistance in wildlife.
Mechanisms other than the direct transmission of strains may account for the low prevalence of antibiotic-resistant E. coli isolates found in gorillas and other wildlife stools. These include both the genetic exchange of resistant material by horizontal gene transfer between unrelated strains (e.g., tetracycline [31] and ampicillin [36]) or the independent selection of resistance in wild animals that confers protection, for example, against naturally occurring antibiotics and heavy metals (1). Although specific antibiotic-resistant phenotypes of gorillas and humans did not match in our sample, our results did show a significant positive correlation in the specific prevalence of resistance to each antibiotic between these two species. This could have been generated from high transmission rates of resistant material rather than the bacteria itself. The physiological cost of resistance in the absence of antibiotic pressure (2, 24) may have subsequently caused the loss of some of this genetic material. This loss could explain the low prevalence of resistance (and multiresistance) observed in wild animals and the differences in antibiotic-resistant phenotypes between the isolates coming from humans and wild animals. Further analysis of the genetic component of antibiotic phenotypes reported here, such as the amplification of transposons, integrons, or plasmids, could help elucidate the origin and mechanisms of antibiotic-resistant spread among wild animals.
Microbial exchanges through common food sources.
This study reveals that E. coli strains from wild gorillas and other wildlife are genetically similar and belong almost exclusively to groups B2 and D. The high prevalence of group B2 strains has been previously associated with herbivorous and omnivorous animals (15) and suggests that diet is a major determinant of the composition of E. coli flora in animals. In our study, this trend is reinforced, as we sampled during a short dry season where ripe fruit is available but is less abundant than in the wet season, and wild animals tend to forage within the same available trees (40, 42). Interestingly, E. coli strains from groups B2 and D are not associated with the presence of virulent factors responsible for enteropathogenic E. coli causing diarrhea (10). Thus, it is unlikely that wild animals are the reservoirs for this disease in Lopé.
Conclusions.
The purpose of this study was to infer patterns of E. coli antibiotic resistance in wild animals in a region where human density is low and to assess whether the observed pattern can be explained by transmission from humans or domestic animals. Direct transmission can be suspected if the genotype of the transmitted bacteria to a receiving host is a subset of the genotypes within the transmitting host (3). The genetic background of resistant strains from both gorillas and other wildlife was different from the background observed in humans and domestic animals. Thus, our study indicates that the observed antibiotic resistance in wild animals is not caused by the direct acquisition of human bacterial strains. Rather, among wild animals, sharing the same local environment, such as common foraging places, may be the major determinant of the similarities in population structure of E. coli. Therefore, our study predicts that in areas of low human density where contact opportunities with wild animals are rare, the potential for E. coli transmission is low.
ACKNOWLEDGMENTS
We are very grateful to the Government of Gabon, especially the CENAREST and the Agence Nationale des Parcs Nationaux (ANPN), for authorizing this study. We also thank all of the Mikongo Conservation Centre staff, ZSL volunteers, inhabitants of each sampled village, and the authorities of Lopé National Park for their support in data collection. We particularly thank Alice, Pierre, Jean-Claude, Aimé, ZB, Didier, Bob, Manix, Yve, Olivier, and all Moukoungama trackers for their support in the field. We also thank the staff of the Hospital Arnaud de Villeneuve (Arnaud, Patrick, Claire, and Mylène) for their substantial help with bacterial analyses. We are grateful to Gaston B. and Valérie Durande for their support. We thank the three anonymous reviewers for their comments and manuscript improvements.
This work received financial support from the Ministère Français de l'Enseignement Supérieur and a CONICYT scholarship from the Chilean Government to J.A.B.
Footnotes
Published ahead of print 6 April 2012
This is contribution ISEM 2012-044.
REFERENCES
- 1. Allen HK, et al. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8:251–259 [DOI] [PubMed] [Google Scholar]
- 2. Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489–493 [DOI] [PubMed] [Google Scholar]
- 3. Archie EA, Luikart G, Ezenwa VO. 2009. Infecting epidemiology with genetics: a new frontier in disease ecology. Trends Ecol. Evol. 24:21–30 [DOI] [PubMed] [Google Scholar]
- 4. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2006. Performance standards for antimicrobial disc susceptibility tests; approved standard, 9th ed Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Cole D, et al. 2005. Free-living Canada geese and antimicrobial resistance. Emerg. Infect. Dis. 11:935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daszak P, Cunningham AA, Hyatt AD. 2001. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop. 78:103–116 [DOI] [PubMed] [Google Scholar]
- 8. Dobson A, Foufopoulos J. 2001. Emerging infectious pathogens of wildlife. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 356:1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dolejska M, Cizek A, Literak I. 2007. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J. Appl. Microbiol. 103:11–19 [DOI] [PubMed] [Google Scholar]
- 10. Escobar-Páramo P, et al. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085–1094 [DOI] [PubMed] [Google Scholar]
- 11. Fisher RA. 1925. Statistical methods for research workers. Oliver and Boyd, Edinburgh, Scotland [Google Scholar]
- 12. Gillespie TR, Nunn CL, Leendertz FH. 2008. Integrative approaches to the study of primate infectious disease: implications for biodiversity conservation and global health. Am. J. Phys. Anthropol. Suppl. 47:53–69 [DOI] [PubMed] [Google Scholar]
- 13. Goldberg TL, et al. 2007. Patterns of gastrointestinal bacterial exchange between chimpanzees and humans involved in research and tourism in western Uganda. Biol. Conservation 135:511–517 [Google Scholar]
- 14. Gordon DM, Clermont O, Tolley H, Denamur E. 2008. Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ. Microbiol. 10:2484–2496 [DOI] [PubMed] [Google Scholar]
- 15. Gordon DM, Cowling A. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575–3586 [DOI] [PubMed] [Google Scholar]
- 16. Guschanski K, et al. 2009. Counting elusive animals: comparing field and genetic census of the entire mountain gorilla population of Bwindi Impenetrable National Park, Uganda. Biol. Conservation 142:290–300 [Google Scholar]
- 17. Harcourt AH, Parks SA, Woodroffe R. 2001. Human density as an influence on species/area relationships: double jeopardy for small African reserves? Biodiversity Conservation 10:1011–1026 [Google Scholar]
- 18. Jones KE, et al. 2008. Global trends in emerging infectious diseases. Nature 451:990–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kondgen S, et al. 2008. Pandemic human viruses cause decline of endangered great apes. Curr. Biol. 18:260–264 [DOI] [PubMed] [Google Scholar]
- 20. Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol. Lett. 307:80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lecointre G, Rachdi L, Darlu P, Denamur E. 1998. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 15:1685–1695 [DOI] [PubMed] [Google Scholar]
- 22. Literak I, et al. 2010. Antimicrobial-resistant faecal Escherichia coli in wild mammals in central Europe: multiresistant Escherichia coli producing extended-spectrum beta-lactamases in wild boars. J. Appl. Microbiol. 108:1702–1711 [DOI] [PubMed] [Google Scholar]
- 23. Lloyd-Smith JO, et al. 2009. Epidemic dynamics at the human-animal interface. Science 326:1362–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martinez JL, et al. 2009. A global view of antibiotic resistance. FEMS Microbiol. Rev. 33:44–65 [DOI] [PubMed] [Google Scholar]
- 25. Mefane C, Guerch M. 1986. Antibiotic sensitivity of 85 strains of Mycobacterium tuberculosis at Libreville (Gabon). Méd. d'Afrique Noire 33:5 [Google Scholar]
- 26. Metzger W, Mordmüller B, Graninger W, Bienzle U, Kremsner PG. 1995. High efficacy of short-term quinine-antibiotic combinations for treating adult malaria patients in an area in which malaria is hyperendemic. Antimicrob. Agents Chemother. 39:245–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okome-Kouakou M, Nkana JAE, Kombila M. 1999. Shigellosis epidemiology in Gabonese adults. Med. Maladies Infect. 29:516–519 [Google Scholar]
- 28. Palacios G, et al. Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerg. Infect. Dis. 17:711–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quinn PJ. 1994. Clinical veterinary microbiology. Wolfe, London, United Kingdom [Google Scholar]
- 30. Raymond M, Rousset F. 1995. An exact test for population differentiation. Evolution 49:4. [DOI] [PubMed] [Google Scholar]
- 31. Roberts ML, Buchanan KL, Evans MR. 2004. Testing the immunocompetence handicap hypothesis: a review of the evidence. Anim. Behav. 68:227–239 [Google Scholar]
- 32. Rolland RM, Hausfater G, Marshall B, Levy SB. 1985. Antibiotic-resistant bacteria in wild primates: increased prevalence in baboons feeding on human refuse. Appl. Environ. Microbiol. 49:791–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rwego IB, Isabirye-Basuta G, Gillespie TR, Goldberg TL. 2008. Gastrointestinal bacterial transmission among humans, mountain gorillas, and livestock in Bwindi Impenetrable National Park, Uganda. Conservation Biol. 22:1600–1607 [DOI] [PubMed] [Google Scholar]
- 34. Skurnik D, et al. 2006. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 57:1215–1219 [DOI] [PubMed] [Google Scholar]
- 35. Spearman C. 1904. The proof and measurement of association between two things. Am. J. Psychol. 15:72–101 [PubMed] [Google Scholar]
- 36. Sutcliffe JG. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. U. S. A. 75:3737–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207–217 [DOI] [PubMed] [Google Scholar]
- 38. Tutin CEG, Fernandez M. 1993. Composition of the diet of chimpanzees and comparisons with that of sympatric lowland gorillas in the Lope Reserve, Gabon. Am. J. Primatol. 30:195–211 [DOI] [PubMed] [Google Scholar]
- 39. Tutin CEG, Ham RM, White LJT, Harrison MJS. 1997. The primate community of the Lope Reserve, Gabon: diets, responses to fruit scarcity, and effects on biomass. Am. J. Primatol. 42:1–24 [DOI] [PubMed] [Google Scholar]
- 40. Tutin CEG, White LJT. 1998. Primates, phenology and frugivory: present, past and future patterns in the Lope Reserve, Gabon. In Prins DM, Brown HT. (ed), Dynamics of tropical communities: the 37th symposium of the British Ecological Society, Cambridge University, 1996, Newbery Blackwell Science Ltd, Oxford, United Kingdom [Google Scholar]
- 41. Vlieghe E, Phoba MF, Tamfun JJM, Jacobs J. 2009. Antibiotic resistance among bacterial pathogens in central Africa: a review of the published literature between 1955 and 2008. Int. J. Antimicrob. Agents 34:295–303 [DOI] [PubMed] [Google Scholar]
- 42. White LJT. 1994. Patterns of fruit-fall phenology in the Lope Reserve, Gabon. J. Trop. Ecol. 10:289–312 [Google Scholar]
- 43. Wolfe ND, Dunavan CP, Diamond J. 2007. Origins of major human infectious diseases. Nature 447:279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolfe ND, et al. 1998. Wild primate populations in emerging infectious disease research: the missing link? Emerg. Infect. Dis. 4:149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Woolhouse MEJ, Gowtage-Sequeria S. 2005. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11:1842–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]