Abstract
While the microbial water quality in the Platte River is seasonally impacted by excreta from migrating cranes, there are no methods available to study crane fecal contamination. Here we characterized microbial populations in crane feces using phylogenetic analysis of 16S rRNA gene fecal clone libraries. Using these sequences, a novel crane quantitative PCR (Crane1) assay was developed, and its applicability as a microbial source tracking (MST) assay was evaluated by determining its host specificity and detection ability in environmental waters. Bacteria from crane excreta were dominated by bacilli and proteobacteria, with a notable paucity of sequences homologous to Bacteroidetes and Clostridia. The Crane1 marker targeted a dominant clade of unclassified Lactobacillales sequences closely related to Catellicoccus marimammalium. The host distribution of the Crane1 marker was relatively high, being positive for 69% (66/96) of the crane excreta samples tested. The assay also showed high host specificity, with 95% of the nontarget fecal samples (i.e., n = 553; 20 different free-range hosts) being negative. Of the presumed crane-impacted water samples (n = 16), 88% were positive for the Crane1 assay, whereas none of the water samples not impacted by cranes were positive (n = 165). Bayesian statistical models of the Crane1 MST marker demonstrated high confidence in detecting true-positive signals and a low probability of false-negative signals from environmental water samples. Altogether, these data suggest that the newly developed marker could be used in environmental monitoring studies to study crane fecal pollution dynamics.
INTRODUCTION
The Platte River is a major river in the state of Nebraska in the United States and has many beneficial uses to humans, such as groundwater recharge for the drinking water supply, irrigation, and recreation activities. This river is the preferred transient roosting habitat of various migratory birds, including the endangered whooping crane, sandhill cranes, snow geese, Canada geese, white-fronted geese, mallard ducks, and pintail ducks, among others. Sandhill cranes are among the most numerous migratory birds in the region, with an estimated 500,000 birds seasonally in the area (http://www.nwf.org/Wildlife/Wild-Places/Platte-River.aspx). Sandhill cranes primarily use the Central Flyway migration route, coming into the Platte River in early spring, which coincides with resuming human recreational activities in the river basin. This is important, as migratory birds have the potential to be seasonal sources of waterborne fecal bacteria, including pathogens (9, 11, 29). In fact, sandhill cranes were identified as the fecal source in a recent campylobacteriosis outbreak in Alaska associated with the consumption of peas (8), underscoring the importance of monitoring microbial water quality in waterfowl-impacted waters.
Fecal indicator bacteria (FIB) such as Escherichia coli and enterococci are commonly used to determine the microbial quality of environmental waters. While measuring FIB can be used to establish the overall level of fecal pollution, traditional FIB detection methods are uninformative for determining the primary fecal source(s) in polluted waters (30). Specifically, in the Platte River there are other potential fecal contributors, such as cattle and wildlife. Several avian markers have been developed (7, 19, 20, 21), but to our knowledge, there is no crane-specific assay. This is in part due to the lack of data on the microbial composition of crane excreta. To address this, 16S rRNA gene fecal clone sequences from sandhill crane were analyzed to characterize microbial populations and to identify potentially crane-specific markers for use in fecal source tracking. Additionally, we investigated the applicability of a putative crane-specific assay to environmental water samples collected from different geographic locations. We also studied the microbial composition of snow geese, as this waterfowl species uses the Platte River as a roosting site during its winter migration.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Most sandhill crane (Grus canadensis, n = 95) and all snow goose (Chen caerulescens, n = 22) excreta were collected as fresh droppings on sand bars in a Platte River watershed (latitude, 40.78°N; longitude, 98.48°W). The sandhill crane samples were collected on five different sampling dates: 15 March 2010 (n = 6), 18 March 2010 (n = 16), 25 March 2010 (n = 16), 5 April 2010 (n = 26), and 21 March 2012 (n = 31). A sandhill crane sample and 11 whooping crane (G. americana) samples collected in Texas (1 March 2012) were also processed in this study. In addition, 71 samples from 12 captive cranes (collected in March 2012) were analyzed: black-crowned crane (Balearica pavonina, n = 1), black-necked crane (G. nigricollis, n = 5), Brolga crane (G. rubicunda, n = 3), Eurasian crane (G. grus, n = 1), gray-crowned crane (Balearica regulorum, n = 1), hooded crane (G. monacha, n = 4), red-crowned crane (G. japonensis, n = 4), sandhill crane (G. canadensis, n = 11), Siberian crane (G. leucogeranus, n = 10), wattled crane (Bugeranus carunculatus, n = 1), white-naped crane (G. vipio, n = 3), and whooping crane (n = 27). Five individual samples from sandhill cranes and snow geese were used to develop 16S rRNA gene clone libraries. Crane fecal clone sequences were subsequently used to develop a crane-specific assay (the Crane1 assay). A total of 553 nontargeted fecal samples from 20 different hosts were collected and used to test host specificity. To evaluate the applicability of the developed assay with environmental water samples, a total of 43 water samples were collected in 1-liter sterilized plastic bottles from three locations (upstream, within, and downstream of the crane roosting area) in the Central Platte River, Nebraska, during the sandhill crane migratory season (i.e., between January 2010 and May 2010). Water samples were collected on Sunday night or Monday morning, when effluents from power plants were not discharged. A total of 138 water samples were also collected from the Arecibo watershed (Puerto Rico) presumed to be primarily impacted by cattle, human, and wildlife (but not impacted by migratory birds) and used as putative negative-control samples. Water and fecal samples were shipped overnight on ice packs or dry ice to the EPA laboratory (Cincinnati, OH). Triplicates of 100-ml water samples were filtered onto polycarbonate membranes (0.4-μm pore size, 47-mm diameter; GE Water and Process Technologies, Trevose, PA) immediately upon receipt. Filters and fecal samples were stored at −80°C until DNA extraction. DNA extractions from membrane filters and fecal samples were performed using MoBio PowerSoil kits according to the manufacturer's protocol (MoBio Laboratories, Carlsbad, CA). DNA extracts were stored at −20°C until further processing.
Cloning and sequencing analyses.
DNA was amplified using primer set 8F-787R, targeting the 16S rRNA gene of the domain Bacteria as described elsewhere (31), with some minor modifications. Briefly, PCR amplifications were performed in 25 μl using the polymerase TaKaRa Ex Taq (TaKaRa Bio Inc.) in a Bio-Rad Tetrad2 Peltier thermal cycler (Bio-Rad, Hercules, CA) under the following cycling conditions: one initial denaturation step at 95°C for 5 min and 30 cycles of 1 min at 95°C, 1 min at 56°C, and 1 min at 72°C. PCR products were visualized in 1.5% agarose gels using GelStar nucleic acid gel stain (Lonza, Rockland, ME). PCR products were cloned into the pCR4 TOPO vector and transformed to TOPO10 chemically competent E. coli cells as described by the manufacturer (Invitrogen, Carlsbad, CA). 16S rRNA gene clone libraries were developed from PCR products generated using crane fecal DNA extracts. Individual E. coli clones were then subcultured into 300 μl of Luria broth containing 50 μg/ml ampicillin and screened for inserts using M13 PCR. The PCR products were submitted to the Children's Hospital DNA Core Facility (Cincinnati, OH) for sequencing using an Applied Biosystems Prism 3730XL DNA analyzer. Raw sequences were processed using Sequencher, version 4.9, software (Gene Codes, Ann Arbor, MI). Chimeric sequences detected using the Bellerophon program (12) were not included in further analyses. Sequences were submitted to Greengenes for alignment using the Nearest Alignment Space Termination algorithm (4, 5). The clone libraries were compared using the Naive Bayesian rRNA Classifier, version 2.0, of the Ribosomal Database Project (RDP) with a 95% confidence threshold (3). For 16S rRNA gene sequences, homology searches of DNA sequences in the GenBank (NR) database were undertaken with the National Center for Biotechnology Information (NCBI) BLASTn (http://www.ncbi.nlm.nih.gov/BLAST/) program (1).
New marker development and qPCR assay.
A phylogenetic tree was created using a neighbor-joining algorithm within the ARB software (23), 16S rRNA gene sequences within the Silva database, and the sequences from crane and snow goose fecal libraries (Fig. 1). In order to design host-specific primers, unique clades were identified and candidate primers were designed using ARB. In silico testing of primer specificity was performed in ARB against a database that included published and unpublished sequences. Forward and reverse primers (i.e., targeting Catellicoccus-like species and Bacteroidetes; Table 1) were optimized through temperature gradient PCR with eight individual crane excreta DNA templates. The assays were also tested for sensitivity (host distribution) and specificity against individual DNA from fecal samples of different animals (Table 2).
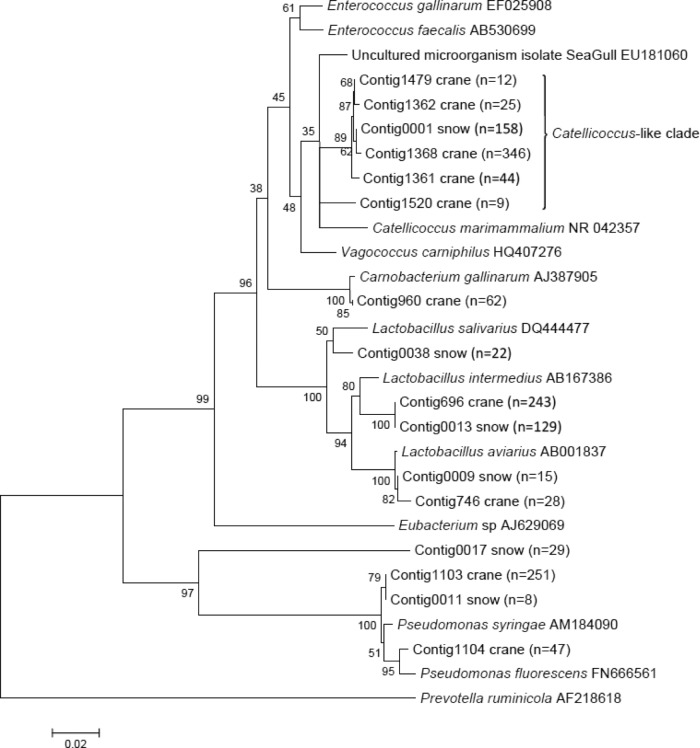
Fig 1.
Neighbor-joining tree of 16S rRNA gene sequences obtained from crane and snow goose clone libraries. GenBank accession numbers are provided for bacterial sequences used as references. Bootstrap values are provided for 1,000 replicated trees.
Table 1.
Summary of candidate primers for crane-specific PCR assays
| Target bacteria | Primer direction | Primer namea | Primer sequence (5′ to 3′) |
|---|---|---|---|
| Catellicoccus-like spp. | Forward | 76F | GGTGCTTGCACCGACYTAAG |
| 183F | ACAAGCGCATGCTTGTGA | ||
| Reverse | 446R | CTCTCACACGTGTTCTTC | |
| 471R | GATACCGTCAAGGAGAAG | ||
| Bacteroidetes | Forward | 153F | GGTATGGTGAAGTTGCATGA |
| Reverse | 466R | GTACATGCAAAAACCTAC | |
| 659R | ATTCCGCCTACCTCGACC |
Numbers represent positions within the 16S rRNA gene of E. coli. Catellicoccus-like forward (F) primers were tested against the Catellicoccus-like reverse (R) primers (i.e., four combinations), while the forward Bacteroidetes primer was tested against both Bacteroidetes reverse primers (two combinations). The crane-specific, or Crane1, assay consisted of primers 76F and 471R.
Table 2.
Prevalence of the Crane1 marker in animal fecal samples
| Animal | Species | Location of samples | No. of samples |
|
|---|---|---|---|---|
| Total | SYBR green qPCR positive | |||
| Crane | Sandhill crane (Grus canadensis) | Nebraska, Texasa | 96 | 66 |
| Sandhill crane (Grus canadensis) | Wisconsinb | 11 | 1 | |
| Whooping crane (G. americana) | Texas | 11 | 7 | |
| Whooping crane (G. americana) | Wisconsinb | 27 | 0 | |
| Black-crowned crane (Balearica pavonina) | Wisconsinb | 1 | 0 | |
| Black-necked crane (G. nigricollis) | Wisconsinb | 5 | 1 | |
| Brolga crane (G. rubicunda) | Wisconsinb | 3 | 0 | |
| Eurasian crane (G. grus) | Wisconsinb | 1 | 1 | |
| Gray-crowned crane (B. regulorum) | Wisconsinb | 1 | 0 | |
| Hooded crane (G. monacha) | Wisconsinb | 4 | 2 | |
| Red-crowned crane (G. japonensis) | Wisconsinb | 4 | 0 | |
| Siberian crane (G. leucogeranus) | Wisconsinb | 10 | 1 | |
| Wattled crane (Bugeranus carunculatus) | Wisconsinb | 1 | 0 | |
| White-naped crane (G. vipio) | Wisconsinb | 3 | 1 | |
| Noncrane species | Snow geese | Nebraska | 22 | 12 |
| Canada geese | Alaska | 25 | 0 | |
| Gull | California | 64 | 0 | |
| Gull | Alaska | 60 | 0 | |
| Duck | Puerto Rico | 16 | 0 | |
| Mallard ducks | Alaska | 6 | 0 | |
| Red Knot | Delaware | 17 | 2 | |
| Ruddy Turnstone | Delaware | 5 | 1 | |
| Pelican | California | 10 | 0 | |
| Swan | Puerto Rico | 22 | 0 | |
| Guineafowl | Puerto Rico | 11 | 0 | |
| Pigeon | Puerto Rico | 11 | 0 | |
| Heron | Puerto Rico | 1 | 0 | |
| Chicken | Puerto Rico | 98 | 10 | |
| Turkey | Puerto Rico | 5 | 0 | |
| Total noncrane species | 373 | 25 | ||
| Nonavian species | Cattle | Puerto Rico | 66 | 0 |
| Goat | Puerto Rico | 32 | 0 | |
| Monkey | Puerto Rico | 9 | 0 | |
| Fish | Puerto Rico | 13 | 0 | |
| Horse | Puerto Rico | 30 | 0 | |
| Pig | Puerto Rico | 30 | 0 | |
| Total nonavian species | 180 | 0 | ||
Fecal samples were collected during five sampling events within the study area (Nebraska) (15 March to 5 April 2010, and 21 March 2012) and one event in Texas (1 March 2012).
Samples collected from captive crane species.
Quantitative PCR (qPCR) assays were conducted against fecal and water samples and aliquots from respective DNA extracts (2 μl) as the templates. Reaction mixtures (25 μl) contained 1× Power SYBR green master mix (Applied Biosystems, Foster City, CA), 0.2 μg/μl bovine serum albumin, and 0.2 μM (final concentration) primers. The amplification protocol involved incubation at 50°C for 2 min, followed by 95°C for 10 min and then 40 cycles of 95°C for 15 s and 61°C for 1 min with a fluorescence read, followed by a melt-curve analysis from 60 to 90°C in 0.1-degree increments. The qPCR assays were performed using a 7900 HT Fast real-time sequence detector (Applied Biosystems, Foster City, CA). All reaction mixtures were prepared in triplicate in MicroAmp Optical 96-well reaction plates with MicroAmp Optical caps (Applied Biosystems, Foster City, CA). PCR data were analyzed using ABI's Sequence Detector software (version 2.2.2). PCR signals were recorded as presence/absence of data and signal quantity values. Duplicate serial dilutions of crane fecal DNA (10−8 to 10−12 g/reaction) were used to generate standard curves. Percent amplification efficiencies were calculated by the instrument manufacturer's instructions (Applied Biosystems). No-template controls were used to check for cross contamination (two per PCR plate). Assays were performed with a range of from 0.5 to 2 ng/μl fecal DNA extracts. Tenfold dilutions of each DNA extract were used to test for PCR inhibition. Dissociation curves were examined to determine the presence of potential primer dimers and other nonspecific reaction products. Signal intensity values were recorded for those reactions showing one corresponding amplification peak within the disassociation curves. PCR products were also visualized in 1.5% agarose gels using GelStar nucleic acid gel stain (Lonza, Rockland, ME) to confirm the size of amplification products.
Bayesian statistics.
Bayes's theorem was used to estimate the confidence of each assay at detecting crane excreta in environmental waters (16), using the following formula:
| (1) |
This involved calculating the posterior probability [P(A|B)] by determining the ratio of true positives [P(B|A)] and false positives [P(B|A′)] in fecal samples and the ratio of water samples that tested positive [i.e., the prior probability, or P(A)]. The posterior probability was also calculated by varying the prior probability from the worst-case scenario (i.e., negative signals in all samples, or 0) to the best-case scenario (i.e., positive signals in all samples, or 1) as described by Lamendella et al. (18). Specifically, the conditional probability [i.e., posterior probability, or P(AB) in the Bayesian formula] was calculated from Equation 1. The sensitivity was calculated as the ratio of positive signals in crane excreta samples, or P(BA). The specificity was calculated as the ratio of negative signals in noncrane excreta samples or 1 − P(BA′). The prevailing rate was calculated as the ratio of positive signals in water samples, or P(A).
Nucleotide sequence accession numbers.
Sequences of different representative taxa obtained in this study were deposited in GenBank under the following accession numbers: JQ015167 to JQ015182.
RESULTS AND DISCUSSION
Taxonomic analysis of 16S rRNA gene sequences.
A total of 1,151 clone sequences were analyzed to determine the identities of crane excreta bacteria. Excluding sequences unclassified or classified as unknowns, 18 bacterial genera were represented in the crane clone libraries (Table 3). The crane bacterial community was mostly composed of populations closely related to bacilli (57.0%), Gammaproteobacteria (28.0%), Bacteroidetes (3.2%), and Clostridia (1.7%). Within the bacilli, 239 sequences were classified as Lactobacillus (20.8%) and 9 sequences were classified as Catellicoccus spp. (0.8%), while 147 and 227 sequences formed clades of unclassified Enterococcaceae (12.8%) and unclassified Lactobacillales (19.7%), respectively. Most unclassified Enterococcaceae in the crane clone library were closely related to Catellicoccus marimammalium, showing 94% sequence identity to the latter species. Since these Catellicoccus-like sequences are novel, we considered them potential targets for developing host-specific markers. Lactobacillales sequences pertaining to unclassified genera were also numerous. Representatives of these unique clades within the bacilli were used to design new markers.
Table 3.
Distribution of 16S rRNA genes in the clone library of crane excreta from Nebraska
| Class (% clones of total) | Genus | No. of clones |
|---|---|---|
| Fusobacteria (2.0) | Fusobacterium | 18 |
| Unclassified Fusobacteriales | 5 | |
| Bacilli (57.0) | Lactobacillus | 239 |
| Carnobacterium | 21 | |
| Catellicoccus | 9 | |
| Unclassified Lactobacillaceae | 11 | |
| Unclassified Enterococcaceae | 147 | |
| Unclassified Lactobacillales | 227 | |
| Clostridia (1.7) | Clostridium | 2 |
| Sporacetigenium | 8 | |
| Unclassified Peptostreptococcaceae | 3 | |
| Unclassified Veillonellaceae | 4 | |
| Alphaproteobacteria (0.7) | Erythromicrobium | 2 |
| Novosphingobium | 1 | |
| Unclassified Sphingomonadaceae | 2 | |
| Betaproteobacteria (3.4) | Janthinobacterium | 19 |
| Sutterella | 3 | |
| Deltaproteobacteria (28.0) | Pseudomonas | 266 |
| Anaerobiospirillum | 4 | |
| Escherichia/Shigella | 6 | |
| Yersinia | 4 | |
| Epsilonproteobacteria (0.3) | Campylobacter | 4 |
| Bacteroidetes (3.2) | Bacteroides | 21 |
| Prevotella | 11 | |
| Unclassified Prevotellaceae | 5 | |
| Flavobacteria (0.4) | Flavobacterium | 5 |
| Unknown (3.3) | Unknown | 38 |
A small number of sequences from genera known to contain species considered human pathogens (i.e., Campylobacter, Shigella, and Yersinia) were identified in this study (Table 3). Bacterial and viral pathogens have been isolated from different species of birds, and examples of their zoonotic transmission have been documented (14, 15, 22, 24, 26, 28, 35, 36), although the overall frequency of zoonotic transmission of human pathogens by avian species is still poorly understood. For example, while a high prevalence of campylobacters in gull excreta (i.e., 45% positive in 159 fecal samples) was previously observed (22), the low occurrence of pathogenic campylobacter species is considered to represent a relatively low level of risk for human infection (32). However, Pacha et al. (27) reported that human-pathogenic Campylobacter jejuni isolates were detected in more than 70% of sandhill crane and duck excreta samples. Other studies have also documented the presence of Campylobacter spp. in crane excreta (11, 13). In this study, a few C. jejuni sequences (i.e., 0.3%, 99% identity) were obtained from crane fecal clone libraries. Due to the relatively low sequencing coverage and number of clone libraries processed in this study, methods that directly detect the presence of this pathogen should be used to better estimate the importance of sandhill cranes as vectors of C. jejuni. On the other hand, the presence of C. jejuni in our study and the implication of sandhill cranes as the source of a C. jejuni food-borne outbreak suggest that these migratory birds may carry human-pathogenic bacteria for long distances (8). Besides Campylobacter, pathogenic species of Salmonella and Mycobacterium have been isolated from cranes (17, 34). Altogether, cranes seem to be a reservoir of many bacterial pathogens and potentially contribute a significant level of human health risk. Future studies should focus on studying the correlation of avian-specific assays with microbial water quality indicators and pathogens to develop better microbial risk assessment models for waterfowl.
Crane-specific marker development.
Bacteroidetes assays based on 16S rRNA sequences have been used to generate several host-specific assays (2, 7, 25). However, the abundance of Bacteroidetes in avian excreta is relatively low (20), and for some avian species, Bacteroidetes 16S rRNA gene sequences do not appear to form host-specific clusters (6). These previous findings and the low number of sequences closely related to Bacteroidetes found in this study (i.e., 3.2% of crane clone sequences) suggested that this bacterial group is not an ideal target for waterfowl host-specific assay development. Nonetheless, we identified a potentially unique clade within Bacteroidetes and used it to develop one forward and two reverse primers for possible crane-specific markers (Table 1). Additionally, we identified two forward and two reverse primers targeting sequences closely related to the C. marimammalium 16S rRNA gene (i.e., 94% identity) (Table 1). Sequences specific to C. marimammalium have been used to develop gull markers (20, 33).
Among the four combinations of Catellicoccus-like primers and two combinations targeting Bacteroidetes, only one combination (i.e., the 76F and 471R primers, or Crane1) amplified excreta DNA from free-range cranes, whereas the other combinations failed to amplify their targeted regions. The Crane1 assay was further optimized using temperature gradients with various concentrations of free-range crane excreta DNA templates and used in validation studies using DNA extracts from various fecal samples and environmental water samples.
Evaluation of the crane-specific assay and its environmental application.
Relative quantification of SYBR green assays was performed using serial 10-fold dilutions of genomic DNA from crane excreta. The detection limit was 0.1 pg, and the range of quantification (ROQ) for qPCR was from 10−8 to 10−12 g of genomic DNA per reaction. The qPCR amplification efficiency ranged from 81.7 to 90.3%, with R2 values of ≥0.980. The PCR product size (i.e., 414 bp) might contribute to the relatively low amplification efficiency of the assay. PCR inhibition tests were performed with 10-fold dilutions of each crane DNA extract. None of the latter samples showed increases of signal intensity compared to the undiluted DNA templates, suggesting that PCR inhibition did not interfere with the amplification efficiency. No-template controls indicated the absence of contamination in the qPCR experiments. The Crane1 assay was tested with 96 individual sandhill crane excreta samples collected from the wild (Table 2), and 69% were positive with the assay. These results are similar to those of other source-tracking methods targeting avian hosts (7, 20).
The sandhill cranes present in the Platte River primarily come from southwest states (e.g., Arizona, New Mexico, Texas, and Louisiana) and northern Mexico. At any given time during their winter migration (i.e., primarily during March and April), different crane flocks leave and arrive at the Platte River. The sandhill crane samples were collected within the Platte River watershed on four different dates in 2010 (n = 64) and one date in 2012 (n = 31). Therefore, it is very likely that the fecal samples collected in this study are from birds that originate from different flocks migrating from different regions. Additionally, 64% of fecal samples from free-range whooping cranes (i.e., n = 11) were positive with the sandhill crane marker. These samples were collected in regions inhabited by sandhill cranes, suggesting potential cross contamination between these two crane species. Whooping cranes are considered an endangered species, with only a few hundred birds living in the wild. Thus, from a source-tracking standpoint, cross amplification with whooping crane fecal samples is not as significant as that with samples from other waterfowl.
The Crane1 marker was not detected in any of the nonavian fecal samples (n = 180), whereas 25 of 373 noncrane avian fecal samples (6.7%) were positive with the assay. Snow geese, chickens, and shore birds (i.e., Red Knot and Ruddy Turnstone) excreta samples showed the highest levels of cross amplification with the Crane1 marker. Cross-amplification signals with snow goose samples can be explained due to the fact that this group coinhabits the Platte River with sandhill cranes for some periods of time. Crane1 signals with the shore birds and chicken samples analyzed in this study are intriguing, as no sizeable crane populations were suspected in the sampling areas for these nontargeted birds. Future studies should include the analysis of 16S rRNA gene clone libraries using universal primers, as well as the crane-specific primer, and fecal DNA from these nontargeted hosts to further elucidate the identity of these signals. The presence of double peaks with nontarget avian host samples (possibly due to nonspecific amplification) precluded us from assessing the relative strength of the signals in avian excreta. With the exception of the results obtained with the snow goose, chicken, and shore bird samples, the specificity results suggested that the Crane1 assay may provide reasonable crane host specificity.
We also studied samples from 12 captive crane species for the presence of the crane marker (Table 2). Less than 10% of the samples tested (n = 71) were positive. Interestingly, none of the sandhill or whooping crane samples tested were positive, suggesting that environmental conditions as well as diet may influence the relative prevalence of the marker in cranes. Thus, although the incidence of the marker in the captive birds was relatively low, the latter results from this study suggest that the Crane1 assay might be useful at detecting fecal pollution associated with free-range crane species. Moreover, the fact that free-range whooping cranes and snow geese were positive by the Crane1 assay further implies that sandhill cranes are potential primary vectors of the host-specific population.
To further determine the identity of the snow goose bacteria cross amplifying with the Crane1 assay, snow goose excreta clone libraries were analyzed (Table 4). Similar to crane, the snow goose bacterial community was mostly composed of populations closely related to bacilli (76.6%) and Proteobacteria (17.0%), whereas there were few sequences homologous to Bacteroidetes (0.4%) and Clostridia (0.8%). Within the bacilli, 194 sequences were classified as Lactobacillus (36.6%) and 21 sequences as Catellicoccus (4.0%), while 135 and 39 sequences formed clades of unclassified Enterococcaceae (25.5%) and unclassified Lactobacillales (7.4%). The bacterial composition of snow geese was somewhat different from the results reported for Canadian geese, in which Clostridia represented a third of all sequences examined. Turicibacter spp. were the most abundant bacilli, and Pseudomonas spp. represented <1% of all sequences (21). Additionally, three previously developed Canadian goose-specific assays (7, 21) were negative for all snow goose samples tested in the current study, further suggesting that these goose species may harbor different gut microbial communities (data not shown). The differences between the fecal microbiota of geese might be related to unknown environmental factors, dietary regime, and stress related to migration. These results show that the differences in the excreta microbiota between some avian species are significant, making the development of assays for a group of species (e.g., geese) relatively challenging. In contrast to the Canadian goose libraries, 59% (310/530) of the snow goose clones were related to the sequences (i.e., >99% identity) obtained in the crane libraries (Fig. 1). The similarities between these avian species might be due to the fact that they overlapped in the same habitat. Interestingly, 158 of 530 goose sequences clustered within the clade previously identified to be crane specific. In fact, the Crane1 primers showed no mismatches to these goose sequences, confirming the likelihood for cross amplification. These results explain the relatively high level of cross amplification with snow goose excreta and suggest that the Crane1 marker could be useful to determine the contribution of both avian species rather than being specific to cranes. However, since most studies have been conducted with a limited number of animals from an unknown number of different flocks, additional studies are needed to better document the differences between the fecal microbiota of waterfowl species. In spite of some of these limitations, these results indicate the difficulty in developing host-specific assays for avian species that share habitats, which is a relatively common occurrence among waterfowl during their migratory journeys. The data also underline the importance of molecular surveys in source-tracking studies.
Table 4.
Distribution of 16S rRNA genes in the clone library of snow goose excreta from Nebraska
| Class (% clones of total) | Genus | No. of clones |
|---|---|---|
| Fusobacteria (1.7) | Cetobacterium | 6 |
| Unclassified Fusobacteriaceae | 3 | |
| Bacilli (76.6) | Lactobacillus | 194 |
| Carnobacterium | 8 | |
| Catellicoccus | 21 | |
| Unclassified Lactobacillaceae | 3 | |
| Unclassified Enterococcaceae | 135 | |
| Unclassified Lactobacillales | 39 | |
| Clostridia (0.8) | Acetivibrio | 1 |
| Unclassified Clostridiales | 3 | |
| Alphaproteobacteria (0.6) | Devosia | 1 |
| Unclassified Sphingomonadaceae | 1 | |
| Betaproteobacteria (7.9) | Janthinobacterium | 32 |
| Unclassified Oxalobacteraceae | 8 | |
| Deltaproteobacteria (7.0) | Pseudomonas | 28 |
| Yersinia | 2 | |
| Citrobacter | 1 | |
| Epsilonproteobacteria (1.5) | Geobacter | 5 |
| Desulfobulbus | 1 | |
| Bacteroidetes (0.4) | Unclassified Bacteroidaceae | 2 |
| Flavobacteria (1.1) | Flavobacterium | 5 |
| Unknown (2.5) | Unknown | 13 |
As indicated above, the host specificity results suggested that the assay could be used to detect excreta from snow geese and cranes in the Platte River. Of the presumed crane- and snow goose-impacted water samples collected from the central Platte River between mid-March and April (n = 16), 88% were positive by the Crane1 assay (Table 5). On the contrary, none of the water samples collected before/after the widespread presence of cranes and snow geese in the watershed (n = 27) were positive. Normally, the watershed would be impacted by snow goose during winter; however, due to low temperatures during the winter of the sampling year, they did not arrive until approximately the same time that the crane arrived (i.e., mid-March). Hence, these waters can be considered negative controls for both cranes and snow geese, which is compatible with the marker results. Besides the Platte River water samples, the Crane1 assay was tested against tropical water samples (n = 138). The marker was not detected in any of the tropical water samples, which is not surprising, as these waters are not frequented by either goose or crane species. Since the Arecibo watershed is primarily contaminated by human (wastewater and septic tanks), domesticated animal (mostly cattle), and some avian (chicken and duck) sources, these results further shed light on the potential use of the Crane1 marker for environmental applications.
Table 5.
Detection of the Crane1 marker in water samples
| Sampling location | Sampling period | No. of water samplesa |
Presumed primary fecal contamination sourceb | |
|---|---|---|---|---|
| Total | Crane1 assay positive | |||
| Platte River, NE | March–April 2010c | 16 | 14 | Crane and snow goose |
| Platte River, NE | January–May 2010d | 27 | 0 | Cattle and wildlife |
| Puerto Rico | September 2010–January 2011 | 138 | 0 | Domesticated animals |
All samples were from freshwater.
There is historical knowledge that host animals are present during sampling periods.
Water samples were collected within and downstream of areas frequented by cranes and snow geese during their migration periods.
Water samples were collected upstream of the crane-roosting area before, during, and after crane migration.
Since the Crane1 assay showed some level of false positives (cross amplification with some of the noncrane avian samples) and false negatives (no signals with some of the free-range crane samples), we conducted a Bayesian analysis to further describe the reliability of the assay for environmental monitoring. The deterministic Bayesian values with free-range crane and snow goose-impacted water samples were estimated as described by Kildare et al. (16). The predictive positive value of the Crane1 assay was estimated to be 0.99, suggesting a very high confidence level for water samples that tested positive. This is in agreement with the relatively high sensitivity (i.e., host distribution) and specificity rates (0.69 and 0.95, respectively) exhibited by the Crane1 assay. The prevailing rate was 0.88. The Bayesian analysis also indicated a relatively high confidence in detecting true-positive signals and a low probability of detecting false-negative signals in water samples under a wide range of prior probabilities of crane fecal contamination (Fig. 2). Altogether, these data indicate that the Crane1 assay has desirable properties for its use as a microbial source tracking (MST) marker for environmental application.
Fig 2.
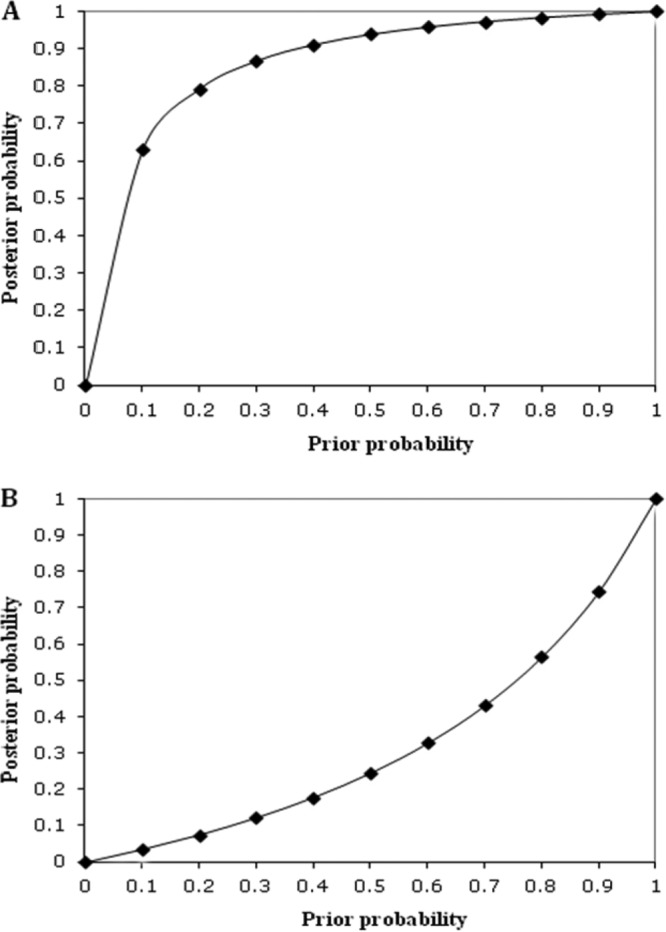
Probability of crane contamination of water using MST marker Crane1. Bayesian analysis of qPCR result with a posterior probability of contamination given a positive result (A) and a negative result (B) over a range of prior probabilities.
Various kinds of birds contribute different levels of fecal loadings to watersheds and beaches. In terms of risk management of areas impacted with different avian sources, accurate characterization of fecal contamination sources is important, as such data could be used to develop new guidance criteria and manage human risks. Unlike domesticated animal- and human-specific MST markers, to date few avian species-specific assays have been developed (7, 20, 21). Specifically, there are host-specific assays for only two waterfowl species: gulls and geese. In this study, we developed an assay for a third waterfowl group: cranes. This avian group, particularly the sandhill cranes, is considered an important fecal pollution source not only at the Nebraska study site but also in other regions in the United States that are impacted during their flyway migration. Indeed, sandhill cranes can migrate from southern regions to Alaska and even cross intercontinental lines. The assay was intended to target sandhill cranes, although we found that other crane species cross-reacted to the assay.
Most host-specific assays have been based on Bacteroidetes targets. Since Bacteroidetes are uncommon in bird feces, this study provides further evidence that alternative bacterial groups (i.e., lactobacilli) are good targets for waterfowl-specific assays. Moreover, our study shows that there is an untapped diversity of bacterial groups that might be physiologically relevant to the host and, from an MST standpoint, can be used for assay development. Since the cranes are found in many regions globally, the availability of this marker will allow other scientists to further validate the assay's host specificity and to test for the prevalence of this lactobacillus group in other water systems. However, in most cases the validation of avian-specific assays against environmental samples is not comprehensive. Since all bacterial host-specific assays show cross amplification against nontarget feces to some extent, the results from this and other studies indicate that it may not be realistic to develop either perfect avian-specific assays or general avian assays.
Avian MST assays could be useful by implying the waterfowl contributing to fecal indicator loadings. Unfortunately, the correlation between fecal indicator bacteria, pathogens, and avian MST targets is unknown or has been generated using a small number of fecal and water samples. Future research should address these gaps, particularly in environmental applications, in which there is a poor correlation between indicator bacteria, human source tracking, and bacterial pathogens (10). Moreover, while the focus on source-tracking activities has emphasized potential public health implications of waterfowl pollution, future studies are needed to better understand the significance of the waterfowl gut microbiota in migratory bird health.
ACKNOWLEDGMENTS
We thank Brandon Iker and Michael Henson for technical assistance. We gratefully acknowledge the contribution of Mark Peyton (Central Nebraska Public Power and Irrigation District) and Barry Hartup (International Crane Foundation) for providing crane fecal samples, Kirsten Grond for providing shore bird samples, and Carlos Hernandez-Toledo for providing water and fecal samples.
The U.S. Environmental Protection Agency, through its Office of Research and Development, funded and managed, or partially funded and collaborated in, the research described herein. This work has been subjected to the agency's administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print 6 April 2012
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeSantis TZ, et al. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394–W399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dick LK, et al. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fremaux B, Boa T, Yost CK. 2010. Quantitative real-time PCR assays for sensitive detection of Canada goose-specific fecal pollution in water sources. Appl. Environ. Microbiol. 76:4886–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gardner TJ, et al. 2011. Outbreak of campylobacteriosis associated with consumption of raw peas. Clin. Infect. Dis. 53:26–32 [DOI] [PubMed] [Google Scholar]
- 9. Graczyk TK, Majewska AC, Schwab KJ. 2008. The role of birds in dissemination of human waterborne enteropathogens. Trends Parasitol. 24:55–59 [DOI] [PubMed] [Google Scholar]
- 10. Hellein KN, et al. 2011. Culture-based indicators of fecal contamination and molecular microbial indicators rarely correlate with Campylobacter spp. in recreational waters. J. Water Health 9:695–707 [DOI] [PubMed] [Google Scholar]
- 11. Hoar BM, Whiteside DP, Ward L, Inglis GD, Morck DW. 2007. Evaluation of the enteric microflora of captive whooping cranes (Grus americana) and sandhill cranes (Grus canadensis). Zoo Biol. 26:141–153 [DOI] [PubMed] [Google Scholar]
- 12. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 13. Inglis GD, Hoar BM, Whiteside DP, Morck DW. 2007. Campylobacter canadensis sp. nov., from captive whooping cranes in Canada. Int. J. Syst. Evol. Microbiol. 57:2636–2644 [DOI] [PubMed] [Google Scholar]
- 14. Kapperud G, Rosef O. 1983. Avian wildlife reservoir of Campylobacter fetus subsp. jejuni, Yersinia spp., and Salmonella spp. in Norway. Appl. Environ. Microbiol. 45:375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keller JI, Shriver WG, Waldenstrom J, Griekspoor P, Olsen B. 2011. Prevalence of Campylobacter in wild birds of the mid-Atlantic region, USA. J. Wildl. Dis. 47:750–754 [DOI] [PubMed] [Google Scholar]
- 16. Kildare BJ, et al. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701–3715 [DOI] [PubMed] [Google Scholar]
- 17. Kitadai N, Ninomiya N, Murase T, Obi T, Takase K. 2010. Salmonella isolated from the feces of migrating cranes at the Izumi Plain (2002–2008): serotype, antibiotic sensitivity and PFGE type. J. Vet. Med. Sci. 72:939–942 [DOI] [PubMed] [Google Scholar]
- 18. Lamendella R, et al. 2009. Evaluation of swine-specific PCR assays used for fecal source tracking and analysis of molecular diversity of swine-specific Bacteroidales populations. Appl. Environ. Microbiol. 75:5787–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu J, Santo Domingo JW, Shanks OC. 2007. Identification of chicken-specific fecal microbial sequences using a metagenomic approach. Water Res. 41:3561–3574 [DOI] [PubMed] [Google Scholar]
- 20. Lu J, Santo Domingo JW, Lamendella R, Edge T, Hill S. 2008. Phylogenetic diversity and molecular detection of gull feces. Appl. Environ. Microbiol. 74:3969–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu J, Santo Domingo JW, Hill S, Edge TA. 2009. Microbial diversity and host-specific sequences of Canada goose feces. Appl. Environ. Microbiol. 75:5919–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu J, Ryu H, Santo Domingo JWJ, Griffith Ashbolt N. 2011. Molecular detection of Campylobacter spp. in California gull (Larus californicus) excreta. Appl. Environ. Microbiol. 77:5034–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moore JE, et al. 2002. Occurrence of Campylobacter spp. and Cryptosporidium spp. in seagulls (Larus spp.). Vector Borne Zoonotic Dis. 2:111–114 [DOI] [PubMed] [Google Scholar]
- 25. Okabe S, Okayama N, Savichtcheva O, Ito T. 2006. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 74:890–901 [DOI] [PubMed] [Google Scholar]
- 26. Olsen B, et al. 2006. Global patterns of influenza A virus in wild birds. Science 312:384–388 [DOI] [PubMed] [Google Scholar]
- 27. Pacha RE, Clark GW, Williams EA, Carter AM. 1988. Migratory birds of central Washington as reservoirs of Campylobacter jejuni. Can. J. Microbiol. 34:80–82 [DOI] [PubMed] [Google Scholar]
- 28. Quessy S, Messier S. 1992. Prevalence of Salmonella spp., Campylobacter spp. and Listeria spp. in ring-billed gulls (Larus delawarensis). J. Wildl. Dis. 28:526–531 [DOI] [PubMed] [Google Scholar]
- 29. Reed KD, Meece JK, Henkel JS, Shukla SK. 2003. Birds, migration and emerging zoonoses: West Nile virus, Lyme disease, influenza A and enteropathogens. Clin. Med. Res. 1:5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Santo Domingo JW, Bambic DG, Edge TA, Wuertz S. 2007. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res. 41:3539–3552 [DOI] [PubMed] [Google Scholar]
- 31. Santo Domingo JW, et al. 2011. Molecular survey of concrete sewer biofilm microbial communities. Biofouling 27:993–1001 [DOI] [PubMed] [Google Scholar]
- 32. Schoen ME, Ashbolt NJ. 2010. Assessing pathogen risk to swimmers at non-sewage impacted recreational beaches. Environ. Sci. Technol. 44:2286–2291 [DOI] [PubMed] [Google Scholar]
- 33. Sinigalliano CD, et al. 2010. Traditional and molecular analyses for fecal indicator bacteria in non-point source subtropical recreational marine waters. Water Res. 44:3763–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thoen CO, Himes EM, Barrett RE. 1977. Mycobacterium avium serotype 1 infection in a sandhill crane (Grus canadensis). J. Wildl. Dis. 13:40–42 [DOI] [PubMed] [Google Scholar]
- 35. Waldenström J, On SLW, Ottvall R, Hasselquist D, Olsen B. 2007. Species diversity of Campylobacter in a wild bird community in Sweden. J. Appl. Microbiol. 102:424–432 [DOI] [PubMed] [Google Scholar]
- 36. Whelan CD, Monaghan P, Girdwood RW, Fricker CR. 1988. The significance of wild birds (Larus sp.) in the epidemiology of Campylobacter infections in humans. Epidemiol. Infect. 101:259–267 [DOI] [PMC free article] [PubMed] [Google Scholar]