Abstract
Pantoea vagans C9-1 is a biocontrol strain that produces at least two antibiotics inhibiting the growth of Erwinia amylovora, the causal agent of fire blight disease of pear and apple. One antibiotic, herbicolin I, was purified from culture filtrates of P. vagans C9-1 and determined to be 2-amino-3-(oxirane-2,3-dicarboxamido)-propanoyl-valine, also known as Nß-epoxysuccinamoyl-DAP-valine. A plasposon library was screened for mutants that had lost the ability to produce herbicolin I. It was shown that mutants had reduced biocontrol efficacy in immature pear assays. The biosynthetic gene cluster in P. vagans C9-1 was identified by sequencing the flanking regions of the plasposon insertion sites. The herbicolin I biosynthetic gene cluster consists of 10 coding sequences (CDS) and is located on the 166-kb plasmid pPag2. Sequence comparisons identified orthologous gene clusters in Pantoea agglomerans CU0119 and Serratia proteamaculans 568. A low incidence of detection of the biosynthetic cluster in a collection of 45 Pantoea spp. from biocontrol, environmental, and clinical origins showed that this is a rare trait among the tested strains.
INTRODUCTION
Erwinia amylovora, the causal agent of the blight disease, is a major threat to pome fruit production. The pathogen colonizes flowers and, under favorable weather conditions, enters through the nectarthodes, kills tissues, and spreads throughout the plant (34, 44). Until now, no cure for infected plants has been known, and diseased tissues have to be removed by pruning. Measures to reduce disease incidence include antibiotic and biocontrol agent application during bloom. The emerging resistance of E. amylovora to the most effective antibiotic, streptomycin (6, 25), raises the need for further management measures. Application of biocontrol agents of the related enterobacterial genus Pantoea can remarkably reduce epiphytic growth of the pathogen (2, 11, 29, 40, 41). Site exclusion, nutrient competition, and antibiotic production contribute to the effectiveness of biocontrol agents (23, 24, 28, 45, 47, 48).
Pantoea vagans C9-1, recently reassigned from the closely related Pantoea agglomerans (31, 32), is an effective biocontrol agent inhibiting the growth of E. amylovora in vitro (18) and in orchard trials (41, 42). The biosynthesis of at least two antibiotics contributes to the suppression of E. amylovora in immature pear assays (18). The effectiveness of P. vagans C9-1 was evaluated in field trials, resulting in a significant reduction of fire blight incidence (41, 42). P. vagans C9-1 has been commercialized as BlightBan C9-1 (NuFarm Americas, Burr Ridge, IL) for biocontrol of fire blight of pear and apple.
Many Pantoea species produce one or multiple antibiotics that are effective against fire blight, including a phenazine antibiotic produced by P. agglomerans Eh1087 (12), pantocin A, produced by P. agglomerans strains P10c and Eh318 and Pantoea sp. Eh252 (21, 31, 40, 47), and pantocin B, produced by P. agglomerans Eh318 (3, 49). Although the production of antibiotics is observed for many Pantoea species, the biosynthesis genes of only a few have been identified (12, 15, 20, 21). The identification of antibiotic biosynthetic gene clusters allows the screen for potential antibiotic producing strains and the assessment of novel biocontrol agents.
The main objective of this study was to genetically and chemically characterize the antibiotic produced by P. vagans C9-1 given the common name herbicolin I. A recent study indicated that herbicolin I could be identical to 2-amino-3-(oxirane-2,3-dicarboxamido)-propanoyl-valine, produced by Pantoea agglomerans 48b/90 (35), of which the biosynthetic genes were to date unknown. This study confirms the chemical structure of herbicolin I, while the gene cluster was identified using plasposon mutants. The analysis of the biosynthetic genes revealed a similar gene cluster in P. agglomerans CU0119, which produces a family of dapdiamide antibiotics (dapdiamide A to E) (7, 16). Additionally, we evaluated the incidence of homologous biosynthetic genes across a wide range of Pantoea spp., including plant, environmental, and clinical isolates.
MATERIALS AND METHODS
Bacterial strains and plasmids.
P. vagans C9-1, originally isolated from apple (Malus × domestica ‘Jonathan’; Michigan) (18), was used throughout. Additional bacterial strains used are listed in Table 1. Escherichia coli S17-1 λpir was used for transformation of rescued pTnMod-RKm′ plasposons (R6K ori, TnMod, Kmr, RP4, oriT, Tn5tnp). All strains were stored at −80°C and routinely cultured in Luria-Bertani (LB) medium. E. coli was cultured at 37°C, and all other strains were cultured at 26°C. Kanamycin (Km) was used at a concentration of 50 μg ml−1 as appropriate.
Table 1.
Pantoea species collection used for prevalence screening of pantocin A and dapdiamide biosynthetic genes
| Strain | Origin (reference) | Host | Properties | PCR result |
|
|---|---|---|---|---|---|
| paaABCa | ddaD, ddaFb | ||||
| Pantoea vagans | |||||
| C9-1 | USA (18) | Malus × domestica (apple stem) | Biocontrol agent | + | + |
| C9-1W | Switzerland (38) | NAc | Nonpigmented, pPag3 plasmid-cured variant of C9-1 | + | + |
| LMG 24196 | Argentina (31) | Eucalyptus sp. | Phytopathogen | − | − |
| LMG 24199T | Uganda (31) | Eucalyptus sp. | Phytopathogen | − | − |
| Pantoea agglomerans | |||||
| CPA-2 | Spain (27) | Malus × domestica (fruit surface) | Biocontrol agent | − | − |
| E325 | USA (29) | Malus × domestica (flower) | Biocontrol agent | − | − |
| Eh1087 | New Zealand (24) | Malus × domestica (flower) | Biocontrol agent | − | − |
| Eh239 | USA (4) | Hordeum vulgare (barley, kernel) | Biocontrol agent | − | − |
| Eh318 | USA (49) | Malus × domestica (stem) | Biocontrol agent | + | − |
| Eh454 | USA (4) | Hordeum vulgare (kernel) | Biocontrol agent | − | − |
| Eh460 | USA (4) | Hordeum vulgare (kernel) | Biocontrol agent | − | − |
| P10c | New Zealand (46) | Malus × domestica (flower) | Biocontrol agent | + | − |
| EPS125 | Spain (5) | Pyrus communis (pear, fruit surface) | Biocontrol agent | − | − |
| ATCC 27987 (CDC 1400-74) | USA (31) | Human (ear) | Clinical isolate | − | − |
| ATCC 27998 (CDC 1741-71) | USA (31) | Human (trachea) | Clinical isolate | − | − |
| EM 21cb | Spain (31) | Human (bile) | Clinical isolate | − | − |
| EM 22cb | Spain (31) | Human (blood) | Clinical isolate | − | − |
| LMG 1286T | Zimbabwe (31) | Human (knee wound) | Clinical isolate | − | − |
| VA21971 | Spain (31) | Human (arm wound) | Clinical isolate | − | − |
| CIP 82.100 | Canada (31) | Triticum aestivum (wheat) | Environmental | − | − |
| LMG 2557 | United Kingdom (31) | Pyrus communis | Environmental | − | − |
| LMG 2595 | South Africa (31) | Allium cepa (onion, necrotic stalk/leaf) | Environmental | − | − |
| LMG 2941 | NA (31) | Malus sylvestris (leaf, epiphyte) | Environmental | − | − |
| P3SA | Australia (31) | Triticum aestivum (rhizosphere) | Environmental | − | − |
| Pantoea agglomerans pv. gypsophilae | |||||
| ATCC 43348 | USA (31) | Gypsophila paniculata (baby's breath, plant gall) | Phytopathogen | − | − |
| CFBP 4342 | The Netherlands (31) | Gypsophila paniculata (plant gall) | Phytopathogen | − | − |
| Pantoea sp. | |||||
| Eh252 | USA (44) | Malus pumila | Biocontrol agent | + | − |
| EPS486 | Spain (31) | M. × domestica (bud) | Environmental | − | − |
| EPS595 | Spain (31) | Pyrus communis (bud) | Environmental | − | − |
| Pantoea septica | |||||
| LMG 5345 | USA (31) | Human | Clinical isolate | − | − |
| Pantoea brenneri | |||||
| ATCC 29001 (CDC 164-75) | USA (31) | Human (prostate) | Clinical isolate | − | − |
| LMG 5343T | USA (31) | Human (urethra) | Clinical isolate | − | − |
| Pantoea dispersa | |||||
| CIP 102701 | France (31) | Human (ear) | Clinical isolate | − | − |
| LMG 2770 | USA (31) | Human (blood) | Clinical isolate | − | − |
| LMG 2603T | Japan (31) | Soil | Environmental | − | − |
| LMG 2605 | Tanzania (31) | Vigna unguiculata (cowpea, seed) | Environmental | − | − |
| Pantoea ananatis | |||||
| ATCC 27995 (CDC 4854-73) | USA (31) | Human | Clinical isolate | − | − |
| LMG 5342 | USA (31) | Human | Clinical isolate | − | − |
| ATCC 27996 | USA (31) | Insect | Environmental | − | − |
| LMG 20103 | South Africa (31) | Eucalyptus sp. | Phytopathogen | − | − |
| LMG 2665T | Brazil (31) | Ananas comosus (pineapple) | Phytopathogen | − | − |
| LMG 2676 | USA (31) | Puccinia graminis f. sp. tritici (cereal stem rust-black rust) | Phytopathogen | − | − |
| Pantoea conspicua | |||||
| EM 17cb | Spain (31) | Human (blood) | Clinical isolate | − | − |
| Pantoea stewartii subsp. indologenes | |||||
| CFBP 3614T | India (31) | Setaria italica (foxtail millet, leaf spot) | Phytopathogen | − | − |
| Pantoea stewartii subsp. stewartii | |||||
| CFBP 3517T | USA (31) | Zea mays var. rugosa (maize, corn) | Phytopathogen | − | − |
| Erwinia oleae | |||||
| CFBP 6632T | Spain (31) | Olea europaea (olive, plant gall) | Phytopathogen | − | − |
| Enterobacter sp. | |||||
| LMG 5339 | USA (31) | Gallus gallus (chicken, liver) | Veterinary isolate | − | − |
| Tatumella citrea (ex. Pantoea citrea) | |||||
| LMG 23359 | Philippines (31) | Ananas comosus | Environmental | − | − |
| Tatumella punctata (ex. Pantoea punctata) | |||||
| LMG 22097 | Japan (31) | Citrus × sinensis (orange) | Environmental | − | − |
paaABC, pantocin A biosynthesis genes used as a target in PCR to identify potential pantocin A-producing strains. Data are taken from reference 31.
ddaD, ddaF, dapdiamide biosynthesis genes used as a target in PCR to identify potential dapdiamide-producing strains.
NA, not applicable.
Antimicrobial production and activity assays.
Antibiotic production was examined in a double diffusion assay on MGA medium (morpholinopropanesulfonic acid [MOPS], gluconate, asparagine medium) at 26°C (18). Bacteria were grown for 18 to 24 h in LB medium or LB medium with Km (LB-Km) for mutants. A 10-μl cell suspension (0.1 optical density at 600 nm [OD600]) was spotted onto the basal MGA layer. The colonies were grown for 48 h, exposed to chloroform, and subsequently overlaid with molten MGA agar seeded with wild-type E. amylovora Ea110 and pantocin A (CIR555)- and herbicolin I (CIR550)-resistant mutants (18). The overlays were incubated for 48 h and visually inspected for zones of inhibition. Antimicrobial activities of purified antibiotics were assayed similarly, except that test samples were spotted on MGA agar and dried before being overlaid with an indicator strain.
Mutational analysis.
Plasposon mutants of P. vagans C9-1 were generated according to the method of Dennis et al. (8). Briefly, pTnMod-RKm′ was introduced into P. vagans C9-1 cells by electroporation. Mutants containing pTnMod-RKm′ were selected on LB agar containing Km. Antibiotic-deficient mutants were identified by their inability to form a zone of inhibition against the pantocin A-resistant E. amylovora strain CIR555. Genomic DNA of the herbicolin I mutants containing pTnMod-RKm′ was isolated using the method of Desomer et al. (9). The genomic DNA was digested with the restriction enzyme PstI, self ligated overnight, and transformed into E. coli S17-1 λpir. Replicating plasmids were recovered, and the flanking regions were sequenced using the primers TnMod_FP-1 and TnMod_RP-1 (Table 2). Sequences were identified in the published genome sequence of P. vagans C9-1 plasmid pPag2 (GenBank accession no. CP001894) (37).
Table 2.
Primers used for sequencing the flanking regions of TnMod-RKm′ and for prevalence screening
| Primer name | Sequence (5′–3′) | Size (bp) | Tannealb (°C) |
|---|---|---|---|
| TnMod_FP-1 | TCCCTCACTTTCTGGCTGGA | —a | 56 |
| TnMod_RP-1 | CCTCTCAAAGCAATTTTGAG | ||
| ddaD_F | GGATCTTGCATCGTTCGCAC | 807 | 58 |
| ddaD _R | CGATCGCCTGTGCGGTAGTA | ||
| ddaF_F | ATCCCTGCATTTCAGGCGCT | 853 | 63 |
| ddaF _R | ATGCCCCAGACACTCTTCGA | ||
| paaA_fw | CTCTTGCCAAAATGGATGGT | 2,398 | 55 |
| paaC_rev | TTGCAAATTCTGCACTCTCG |
—, different amplicon sizes.
Annealing temperature.
Immature pear fruit assay.
Inhibition of E. amylovora by P. vagans C9-1 and its antibiotic-deficient derivatives was tested in immature pear fruit (30). Fruit surfaces were disinfected with 70% ethanol, bisected longitudinally, and placed on sterile, moistened Whatman filters in petri dishes. A 50-μl suspension of P. vagans in 5 mM phosphate buffer (pH 6.5) at 5 × 105 CFU ml−1 or 5 × 106 CFU ml−1 or buffer alone was introduced into a 5-mm-deep-well in each pear half with a sterile pipette tip and left to absorb for 2 h. A 50-μl suspension of E. amylovora (5 × 105 CFU ml−1) in 5 mM phosphate buffer (pH 6.5) was introduced into the same well. Treatments consisted of six replicate pear fruit halves, and each experiment was repeated three times.
Disease symptoms (necrosis and/or bacterial ooze) were first observed 2 days after inoculation and recorded daily over 5 days after inoculation. The incidence of disease was calculated for each treatment and day. Disease incidence was transformed into the arcsine square root for normalization prior to analysis with the analysis-of-variance (ANOVA) procedure of the software program SAS (Statistical Analysis Systems, Cary, NC). Disease incidence data did not vary significantly among experiments and were pooled. Treatment means for each day were separated by Fisher's protected least-significant-difference test at a P value of 0.05.
Sequence analysis.
The BLASTN subroutine in the software program GenDB (26) was used to identify the insertion sites of the plasposons in the genome sequence of P. vagans C9-1 (37). Routine sequence manipulations were done using the subroutines of the Lasergene software package, version 8.1.5 (DNAstar, Madison, WI). Additional BLAST searches (1) were done at NCBI.
Antibiotic purification.
Herbicolin I was purified from culture supernatants of P. vagans C9-1 by a combination of cation exchange chromatography, reverse-phase high-pressure liquid chromatography (HPLC), and instant thin-layer chromatography (ITLC). Cells of C9-1 were grown until late log phase in 10 liters MGA. Samples of concentrated supernatants were adjusted to pH 2.5 with 5 N H2SO4, and quickly added to a 5.5-cm-by-10-cm column of Dowex 50W × 4 (200-400 mesh) equilibrated in 2.5 mM ammonium acetate buffer (pH 5). After washing with 2.5 mM ammonium acetate buffer, antibiotics were eluted with 20 mM ammonium acetate buffer (pH 5), concentrated to dryness under reduced pressure at 40°C, and then resuspended in distilled water. Preparations were kept frozen at −20°C. Active fractions were applied to octydecyl (C18) bonded-phase sorbent packed in a 100-ml flash chromatography column and equilibrated in 0.1% (vol/vol) trifluoroacetic acid (TFA) in water. The column was eluted with 0, 5, 10, 20, and 50% methanol in 0.1% TFA, and fractions were immediately adjusted to a pH of 3.5 to 4.0 with 2% ammonium hydroxide. Fractions with antibiotic activity were pooled and concentrated under reduced pressure. Following flash chromatography, the active component was further purified by HPLC on a C8 semipreparative column (25 cm by 9.4 mm) eluted with acetonitrile-water-TFA (5:94.9:0.1). Peaks at absorbance at 215 nm were collected and adjusted to pH 3.5 to 4.0. After concentration, the antibacterial activity of each sample was assayed. Samples with antibacterial activity were repeatedly lyophilized to remove salts. The active compound was finally purified on instant thin-layer chromatography (ITLC-SA) papers developed in acetonitrile-water (80:20).
To determine pH stability, aliquots of samples were pH adjusted with 0.5 N H2SO4 and 2% NH4OH and after 2 h were neutralized with 1 M potassium phosphate buffer (pH 7.0) and adjusted to a constant volume. Heat stability was determined by examining residual antibiotic activity in preparations heated at 95°C for 2 h in sealed vials. Beta-lactamase assays were carried out with penicillinase 1 (Sigma Chemical Co., St. Louis, MO) and were incubated for 1 h at 26°C.
The 1H nuclear magnetic resonance (1H-NMR) spectra in D2O were obtained on a Bruker WM-250 instrument (Bruker BioSpin Corporation, Billerica, MA) with suppression of the solvent HDO resonance signal. The molecular weight and chemical formula were obtained by fast atom bombardment mass spectrometry (FAB-MS) of intact, underivatized herbicolins (0.01 mg in 0.01 ml glycerol matrix). The amino acid composition and sequence of herbicolin I were determined by Edman degradations liberated as phenylthiohydantoin amino acids and analyzed by HPLC.
Sensitivities of orchard isolates of E. amylovora to antibiotics of P. vagans C9-1.
Thirty-four streptomycin-sensitive isolates and 25 streptomycin-resistant isolates of E. amylovora from commercial orchards in the U.S. Pacific Northwest (25) were evaluated for sensitivities to the antibiotics of P. vagans C9-1, using the double diffusion assay on MGA medium described above. l-Histidine (10 mM) was added to the overlay medium in replicate plates for each isolate to suppress antibiosis due to production of pantocin A. Additionally, isolates of Ea153 that were recovered from 20 diseased blossom clusters on pear trees treated twice with P. vagans C9-1 during bloom also were tested (42). Sensitivity assays were repeated twice.
RESULTS
Chemical identification of antibiotic compound.
The antibiotic referred to as herbicolin I was isolated from liquid cultures of P. vagans C9-1 and chemically characterized. The antibiotic was purified from culture supernatants of P. vagans C9-1 by a combination of cation exchange chromatography, reverse-phase HPLC, and instant thin-layer chromatography (ITLC). A different method than that of Sammer et al. (35) was required to exclude the copurification of pantocin A. The purified antibiotic was insensitive to beta-lactamase, base labile (pH > 10), and heat and acid stable. The observed [M+H]+ value of the antibiotic was 317.14461, corresponding to a calculated [M+H]+ value of 317.14610 and a chemical formula of C12H21N4O6. The antibiotic contained the amino acid valine. The proton NMR spectrum of herbicolin I was identical to that of the dapdiamide produced by P. agglomerans 48b/90: 2-amino-3-(oxirane-2,3-dicarboxamido)-propanoyl-valine (Fig. 1) (35). We previously proposed (37) that this compound would be identical to dapdiamide E, but now we are referring to the compound as herbicolin I to be consistent with earlier publications (7, 18) and because the stereochemistry for both compounds has not been determined.
Fig 1.
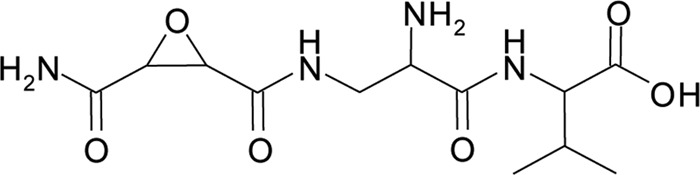
Chemical structure of herbicolin I [2-amino-3-(oxirane-2,3-dicarboxamido)-propanoyl-valine].
Identification of the herbicolin I gene cluster.
To identify the herbicolin I biosynthetic genes, a pTnMod-RKm′ plasposon mutant library of P. vagans C9-1 was constructed. This library, consisting of 3,800 plasposon mutants, was screened for the loss of antibiotic production in a double agar diffusion assay using the pantocin A-resistant and dapdiamide (herbicolin I)-sensitive biosensor strain CIR555. A total of 38 mutants were potentially deficient in dapdiamide biosynthesis. The flanking regions of all mutants were sequenced and analyzed using the BLASTN subroutine of the program GenDB (26) against the fully annotated genome sequence of P. vagans C9-1 (37). A total of 25 plasposon insertions were located in a single locus, which was identified as the biosynthetic operon of herbicolin I (Table 3). The operon consists of 10 genes (ddaA-ddaI) (Fig. 2) and is located on plasmid pPag2 in a region with a low G+C content compared to the overall G+C content of the plasmid. This indicates that P. vagans C9-1 acquired herbicolin I biosynthesis most probably by horizontal gene transfer. However, immediately flanking the operon, no direct evidence for transposition was found, so this cluster may have been acquired together with the complete plasmid pPag2, which is described as a biocontrol-specific feature (37). The other plasposon insertions are infrequently distributed on the chromosome and the other plasmids, pPag1 and pPag3, and are most likely not involved in the biosynthesis of herbicolin I.
Table 3.
Plasposon TnMod-RKm′ mutant insertion identification
| C9-1 plasposon mutant(s) | Locus tag | Gene | Putative function |
|---|---|---|---|
| CIR620, CIR621, CIR625, CIR635, CIR646 | Pvag_pPag20158 | ddaB | Ornithine cyclodeaminase |
| CIR613 | Pvag_pPag20159 | ddaC | Fe(II)/α-KG-dependent dioxygenase |
| CIR591, CIR603, CIR619, CIR633 | Pvag_pPag20162 | ddaF1 | Biotin carboxylase |
| CIR624 | Pvag_pPag20163 | ddaF2 | Biotin carboxylase |
| CIR592, CIR594, CIR596, CIR597, CIR600, CIR616, CIR617, CIR626 | Pvag_pPag20164 | ddaG | Phenylacetate-CoAa ligase |
| CIR599 | Pvag_pPag20165 | ddaH | Asparagine synthetase |
| CIR589, CIR614, CIR636, CIR641 | Pvag_pPag20166 | ddaI | Putative membrane protein |
CoA, coenzyme A.
Fig 2.
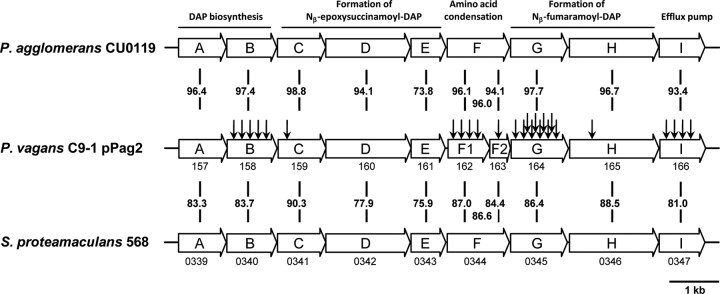
Dapdiamide operons of Pantoea vagans C9-1, P. agglomerans CU0119, and S. proteamaculans 568. The proposed biochemical functions encoded by the genes of P. agglomerans CU0119 (16) are indicated above. Plasposon insertion sites are indicated by arrows. Numbers between clusters indicate the identities at the amino acid level. Where available, locus tags are indicated directly below clusters: for P. vagans strain C9-1, the prefix is Pvag_pPag20, and for S. proteamaculans strain 568, it is Spro_. Note that the gene cluster of P. vagans C9-1 differs in the number of genes due to the naturally separated genes ddaF1 and ddaF2, which together constitute the biochemical function of ddaF in P. agglomerans CU0119.
The most frequent insertion sites of the plasposons were identified in ddaB, ddaF1, ddaG, and ddaI. DdaB is predicted to be involved in l-2,3-diaminopropionic acid (DAP) biosynthesis, one of the central monomers linked to valine and fumaramic acid via condensation reactions by DdaF and DdaG to form the dapdiamide (16). DdaI is predicted to be involved in export of the antibiotics. The gene products of ddaB, ddaF1, ddaG, and ddaI appear to be strictly required for herbicolin I biosynthesis. We identified only a few or no plasposon integration in the genes ddaCDE and ddaH. In studies of genes cloned from P. agglomerans CU0119 and expressed in E. coli, ddaH and ddaCDE are involved in the biosynthesis of the fumaramic acid moiety and formation of the epoxide precursor of Nß-epoxysuccinamoyl-DAP, respectively (17). One mutant identified as containing a plasposon insertion in the putative self-resistance gene ddaI produced less antibiotic than the wild type and remained viable on the antibiotic production medium, MGA.
P. vagans C9-1 significantly reduced the incidence of disease symptoms on immature pear fruits inoculated with E. amylovora Ea110 at each time point compared to results with the buffer treatment (Fig. 3). Biocontrol was significantly decreased with P. vagans C9-1 TnMod mutants lacking production of herbicolin I (CIR591) or pantocin A (CIR638) compared to that with the wild type (Fig. 3). The mutant lacking production of herbicolin I and pantocin A (CIR600) did not suppress symptoms, and the incidence of disease was similar to that with treatment with buffer (Fig. 3). Growth rates of all mutants were unaltered in compared to that of the wild type (data not shown), and each showed reduced biocontrol activity even when applied at 5 × 106 CFU ml−1 (Fig. 3A).
Fig 3.
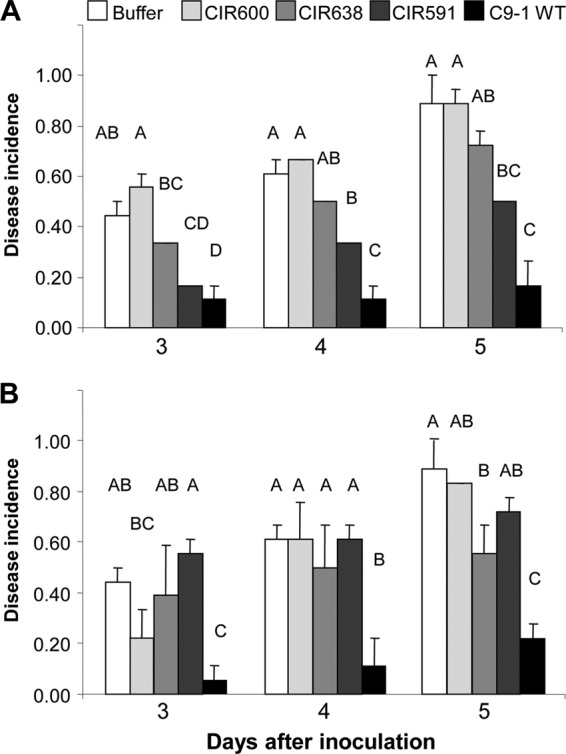
Reduction of incidence of disease symptoms (necrosis and/or bacterial ooze) in immature pear fruit by P. vagans C9-1 and antibiotic-deficient derivatives CIR591 (herbicolin I−/pantocin A+), CIR638 (herbicolin I+/pantocin A−), and CIR600 (herbicolin I−/pantocin A−). Immature pear fruits were treated with buffer or 5 × 106 CFU ml−1 wild-type C9-1 or derivatives (A) or buffer or 5 × 105 CFU ml−1 of wild-type C9-1 or antibiotic-deficient mutants (B) and subsequently challenged with 5 × 105 CFU ml−1 E. amylovora Ea110. Vertical lines indicate standard errors of the means. Similar letters above bars for a time point indicate that the transformed incidence of symptoms between treatments is not significantly different according to Fisher's protected least significant difference at P = 0.05.
Among 59 isolates of the pathogen E. amylovora from commercial orchards, 2 isolates were tolerant of herbicolin I in the double diffusion agar assay but sensitive to the histidine-reversible antibiotic pantocin A. These two orchard isolates had an inhibition pattern similar to that of the herbicolin I-resistant biosensor CIR550, with an inhibition zone distal from the P. vagans C9-1 colony, which was abolished when histidine was added to the overlay medium. One of the herbicolin I-tolerant isolates was resistant to 100 μg ml−1 streptomycin, and the other was sensitive; both caused necrosis and production of bacterial ooze in immature pear fruits. All of the isolates of strain Ea153 recovered from diseased blossom clusters on inoculated trees treated with P. vagans C9-1 were sensitive to both herbicolin I and pantocin A.
Sequence analysis.
BLAST search (NCBI) revealed the presence of highly similar gene clusters in the genomes of P. agglomerans CU0119 (7) and Serratia proteamaculans 568 (43) and a distantly related gene cluster in Vibrio caribbeanicus ATCC BAA-2122 (14). The sequence of the gene cluster of P. agglomerans 48b/90 is not available, but the operon structure of P. agglomerans CU0119 (7) is very similar to that for P. vagans C9-1 (Fig. 2). The gene cluster of P. agglomerans CU0119 has been cloned and expressed in E. coli and shown to produce a mixture of dapdiamides (7). Since the sequence of P. agglomerans CU0119 (7) comprises only the dapdiamide biosynthesis cluster, no evidence for horizontal transfer to P. agglomerans CU0119 is available.
In comparison to the 10 genes in P. vagans C9-1, the clusters of P. agglomerans CU0119 and S. proteamaculans 568 consist of 9 genes. Overall the gene cluster is highly conserved between P. vagans C9-1 and P. agglomerans CU0119 (above 90% amino acid identity), except for ddaE (Pvag_pPag20161) (73.8% identity). In the two other strains, the genes ddaF1 and ddaF2 are combined to only one open reading frame (ORF), a homolog of biotin carboxylase functioning in the condensation reaction between Nß-fumaramoyl-DAP and valine (17). A putative insertion event of 6 bp led to amino acid changes in Pvag_pPag20162. The insertion resulted in a leucine instead of a proline, an additional cysteine, and an early stop codon in Pvag_pPag20162 compared to the other two strains. The bases integrated directly in front of a methionine codon, which serves as a start codon for ddaF2 (Pvag_pPag20163).
Screening for dapdiamide biosynthetic genes in Pantoea species.
A broad collection of Pantoea spp. that includes biocontrol, clinical, and environmental isolates (31) was screened for the presence of the dapdiamide biosynthetic genes (Table 1). The collection was tested for the presence of dapdiamide biosynthetic genes using two different primer combinations targeting ddaD (Pvag_pPag20160) and ddaF1 and ddaF2 (Pvag_pPag162 and Pvag_pPag20163). The primers were generated by comparison of the three available sequences (Fig. 1). The dapdiamide genes were detected only in P. vagans C9-1 and its nonpigmented variant C9-1W cured of its megaplasmid pPag3 (38).
DISCUSSION
The establishment and antibiotic synthesis of antagonists in the floral court are critical for successful suppression of E. amylovora (13). Biocontrol strains that either naturally produce no antibiotic or are inactivated in biosynthesis of an antibiotic can still reduce the growth of E. amylovora (13, 40, 45). Antibiotic biosynthesis mutants inhibit the growth of the pathogen to a lesser extent than the producing strains, since site exclusion and competition for limited nutrients (e.g., nitrogen and iron) are still active. Recovered isolates of the pathogen inoculated onto trees treated with the biocontrol agent were sensitive to both antibiotics produced by P. vagans C9-1. Among isolates of the pathogen from commercial pear orchards, 3% were resistant to herbicolin I, and all were sensitive to pantocin A. None of the commercial orchards in the U.S. Pacific Northwest were exposed to P. vagans C9-1, which was isolated in Michigan, so the herbicolin I resistance was not correlated to exposure or selection pressure from this biocontrol agent. In double diffusion assays, spontaneous mutants of E. amylovora may arise over time that are resistant to herbicolin I or pantocin A; this was the source of the biosensor strains (18). We anticipate that a low incidence of spontaneous mutation may lead to isolates of the pathogen with resistance to herbicolin I. Nonetheless, we postulate that P. vagans C9-1 would continue to be an effective management tool for fire blight, even if some strains of the pathogen were resistant to herbicolin I. P. vagans C9-1 produces another antibiotic, pantocin A, and the isolates insensitive to herbicolin I were sensitive to pantocin A, an antibiotic demonstrated to contribute to biocontrol efficacy (40, 45, 49). Along with antibiosis, P. vagans C9-1 effectively competes with the pathogen for floral nutrients and colonization sites. The multiprong approach of P. vagans C9-1 to colonize and secure niches on flowers likely will mitigate a “breakdown” in biological control efficacy due to herbicolin I-resistant populations of the pathogen (10).
P. vagans C9-1 produces at least two antibiotics in culture that suppress growth of E. amylovora (18). One antibiotic of P. vagans C9-1 is suppressed by histidine and is presumed to be pantocin A based on the presence of a pantocin A gene cluster in the genome and preliminary characterization of the antibiotic (19, 37). In our Pantoea collection, the presence of pantocin A producers is much lower than that in a previous report indicating 61 of 88 P. agglomerans strains that produced a histidine-suppressible antibiotic were positive for pantocin A biosynthesis genes (22, 31). The other, herbicolin I, was shown here to belong to the dapdiamide family of antibiotics. The most abundant histidine-insensitive antibiotic isolated from P. vagans C9-1 is the epoxide of dapdiamide A, Nß-epoxysuccinamoyl-DAP-valine, which has been synthesized and shown to be biologically active against E. amylovora (17). The 1H-NMR spectrum of the purified antibiotic was identical to that of 2-amino-3-(oxirane-2,3-dicarboxamido)-propanoyl-valine, identified in P. agglomerans 48b/90, which has the same [M+H]+ value and biochemical properties as the isolated antibiotic (35). These findings are consistent with the detection of 2-amino-3-(oxirane-2,3-dicarboxamido)-propanoyl-valine in culture filtrates of P. vagans C9-1 (34) by liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS). Based on recent studies with dapdiamides and synthesized dapdiamide analogues, dapdiamides target cell wall biosynthesis by attacking glucosamine-6-phosphate synthase (17).
Given the similarities between the biosynthetic gene clusters in P. vagans C9-1 and P. agglomerans CU0119, the antibiotic produced by the gene cluster in P. vagans C9-1 most likely uses a similar biosynthetic pathway. The biosynthesis of the dapdiamide antibiotics comprises the linkage of l-2,3-diaminopropionic acid (DAP) to two variable units (e.g., amino acids, fumaramic acid and derivates) via amide bond formation (7, 15). Culture supernatants of E. coli expressing the dapdiamide biosynthetic cluster from P. agglomerans CU0119 contained dapdiamide A as well as the less-abundant variants dapdiamide B and C, having an isoleucine or leucine moiety instead of valine. Dapdiamide D and dapdiamide E also were present. These differ in linkage of DAP to fumaramic acid and an epoxide instead of the fumaramic acid double bond, respectively (7, 15). It is possible that P. vagans C9-1 produces other dapdiamides that were not detected or isolated by the protocols used for isolation of Nß-epoxysuccinamoyl-DAP-valine. For example, another histidine-insensitive antibiotic produced with the same antimicrobial activity spectrum as herbicolin I was detected in cultures of P. vagans C9-1 but due to low yield was not characterized further. Further studies of optimization of dapdiamide production and isolation from P. vagans C9-1 would be valuable in this respect and in future studies on the importance of Nß-epoxysuccinamoyl-DAP-valine or other dapdiamides in the biological control of fire blight by P. vagans C9-1.
DdaI, a putative membrane protein potentially involved in the export of the antibiotic, conferred resistance to dapdiamide A to sensitive E. coli (7). In our screen, mutants with an integrated plasposon in this gene resulted in viable cells, which formed a reduced zone of inhibition against E. amylovora. This suggests that different mechanisms of self-resistance exist within P. vagans C9-1, since the mutation in the putative exporter does not lead to lethality caused by the intracellular accumulation of dapdiamides. The putative target, glucosamine-6-phosphate synthase, might be insensitive to dapdiamide due to enzymatic or steric protection. Alternatively, the antibiotic might not be activated before its export, therefore displaying no toxic effect on the cells. However, additional mutagenesis and complementation studies would be required to determine the function of ddaI in P. vagans C9-1.
Biosynthesis of dapdiamide antibiotics is not a common trait in Pantoea spp. We identified dapdiamide biosynthetic genes only in P. vagans C9-1 and P. agglomerans CU0119 after screening a wide range of biocontrol, environmental, and clinical isolates. The rarity of dapdiamide biosynthesis within Pantoea spp. suggests that pathogen-inhibitory activity described for biocontrol Pantoea strains is likely due to production of other antibiotic compounds (28, 30). Analysis of the genes revealed that the operon, consisting of nine coding sequences (CDS) in P. agglomerans CU0119 (7), has 10 CDS due to base insertions in P. vagans C9-1. Besides this, the clusters have a similar gene organization, and the gene products display high similarity to S. proteamaculans 568 at the amino acid level (Fig. 2). Another member of this genus, Serratia plymuthica, also was found to produce a dapdiamide compound (35, 36). Notably, Nß-epoxysuccinamoyl-DAP-valine was not isolated from culture supernatants of E. coli expressing the dapdiamide genes from P. agglomerans CU0119. Although the genes and predicted proteins share a high degree of similarity, differences in dapdiamide production between P. agglomerans CU0119 and wild-type P. vagans C9-1 might reflect slight genetic differences between dapdiamide structural genes or its regulation. It is also possible that sequences outside the dapdiamide gene cluster affect the composition of dapdiamide antibiotics produced. We identified several plasposon mutants containing insertions outside the biosynthetic cluster that were deficient or reduced in herbicolin I. Methods used to purify dapdiamide antibiotics also may have influenced the types of dapdiamides detected.
The operon is most likely acquired by horizontal gene transfer, which is evidenced by the lower G+C content (47%) of the genes in both Pantoea strains than the G+C content of the chromosome (53.8%) (37). The dapdiamide biosynthesis operon is located on plasmid pPag2, which comprises diverse IS elements, which indicates that many important biocontrol attributes of P. vagans C9-1 might be acquired traits (37). Sorbitol metabolism, indole acetic acid biosynthesis from aldoximes, and tellurite resistance are located on this plasmid and might contribute to the ecological fitness of P. vagans C9-1. The biosynthesis of pantocin A and dapdiamide, as well as the overlapping nutrient utilization profile (37, 39, 41) of P. vagans C9-1, contribute to the effectiveness of this biocontrol agent at inhibiting the growth of E. amylovora.
Confirmation of the chemistry and genetics of this biocontrol trait in C9-1 resolves outstanding regulatory questions regarding active-ingredient mechanisms of action. Identification of the biosynthetic genes of dapdiamide will facilitate streamlining the screening process for new biocontrol agents by rapid selection of environmental isolates that produce dapdiamide antibiotics and have higher potential for effective pathogen suppression (33). The genetic characterization presented here also provides a foundation for discovery of novel compounds produced by similar pathways or by a subset of the biosynthetic genes using different substrates for synthesis.
ACKNOWLEDGMENTS
We thank C. Pelludat for insightful discussions and R. Hollingsworth and R. R. Brubaker for advice and assistance with antibiotic isolation and characterization. We thank C. Hardy for technical assistance with screening mutant libraries and S. Lindow for supplying immature pear fruits.
Funding was provided by the Swiss Federal Office of Agriculture, the EU Interreg IV project Gemeinsam gegen Feuerbrand, the U.S. Department of Agriculture, Agricultural Research Service, and the Colorado Agricultural Experiment Station. This work was conducted in part within the Swiss ProfiCrops and European COST Action 873 research networks.
Footnotes
Published ahead of print 13 April 2012
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Anderson LM, Stockwell VO, Loper JE. 2004. An extracellular protease of Pseudomonas fluorescens inactivates antibiotics of Pantoea agglomerans. Phytopathology 94:1228–1234 [DOI] [PubMed] [Google Scholar]
- 3. Brady SF, et al. 1999. Pantocin B, an antibiotic from Erwinia herbicola discovered by heterologous expression of cloned genes. J. Am. Chem. Soc. 121:11912–11913 [Google Scholar]
- 4. Braun-Kiewnick A, Jacobsen BJ, Sands DC. 2000. Biological control of Pseudomonas syringae pv. syringae, the causal agent of basal kernel blight of barley, by antagonistic Pantoea agglomerans. Phytopathology 90:368–375 [DOI] [PubMed] [Google Scholar]
- 5. Bonaterra A, Camps J, Montesinos E. 2005. Osmotically induced trehalose and glycine betaine accumulation improves tolerance to desiccation, survival and efficacy of the postharvest biocontrol agent Pantoea agglomerans EPS125. FEMS Microbiol. Lett. 250:1–8 [DOI] [PubMed] [Google Scholar]
- 6. Chiou CS, Jones AL. 1995. Molecular analysis of high-level streptomycin resistance in Erwinia amylovora. Phytopathology 85:324–328 [Google Scholar]
- 7. Dawlaty J, Zhang X, Fischbach MA, Clardy J. 2010. Dapdiamides, tripeptide antibiotics formed by unconventional amide ligases. J. Nat. Prod. 73:441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dennis JJ, Zylstra GJ. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of Gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desomer J, Crespi M, Van Montagu M. 1991. Illegitimate integration of non-replicative vectors in the genome of Rhodococcus fascians upon electrotransformation as an insertional mutagenesis system. Mol. Microbiol. 5:2115–2124 [DOI] [PubMed] [Google Scholar]
- 10. Duffy B, Schouten A, Raaijmakers JM. 2003. Pathogen self-defense: mechanisms to counteract microbial antagonism. Annu. Rev. Phytopathol. 41:501–538 [DOI] [PubMed] [Google Scholar]
- 11. Francés J, et al. 2006. Pathogen aggressiveness and postharvest biocontrol efficiency in Pantoea agglomerans. Postharvest Biol. Technol. 39:299–307 [Google Scholar]
- 12. Giddens SR, Feng Y, Mahanty HK. 2002. Characterization of a novel phenazine antibiotic gene cluster in Erwinia herbicola Eh1087. Mol. Microbiol. 45:769–783 [DOI] [PubMed] [Google Scholar]
- 13. Giddens SR, Houliston GJ, Mahanty HK. 2003. The influence of antibiotic production and pre-emptive colonization on the population dynamics of Pantoea agglomerans (Erwinia herbicola) Eh1087 and Erwinia amylovora in planta. Environ. Microbiol. 5:1016–1021 [DOI] [PubMed] [Google Scholar]
- 14. Hoffmann M, et al. 19 September 2011. Vibrio caribbeanicus sp. nov., isolated from marine sponge Scleritoderma cyanea. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] doi:10.1099/ijs.0.032375-0 [DOI] [PubMed] [Google Scholar]
- 15. Hollenhorst MA, et al. 2010. The nonribosomal peptide synthetase enzyme DdaD tethers N(β)-fumaramoyl-L-2,3-diaminopropionate for Fe(II)/α-ketoglutarate-dependent epoxidation by DdaC during dapdiamide antibiotic biosynthesis. J. Am. Chem. Soc. 132:15773–15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hollenhorst MA, Clardy J, Walsh CT. 2009. The ATP-dependent amide ligases DdaG and DdaF assemble the fumaramoyl-dipeptide scaffold of the dapdiamide antibiotics. Biochemistry 48:10467–10472 (Erratum, 49:6296, 2010.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hollenhorst MA, Ntai I, Badet B, Kelleher NL, Walsh CT. 2011. A head-to-head comparison of eneamide and epoxyamide inhibitors of glucosamine-6-phosphate synthase from the dapdiamide biosynthetic pathway. Biochemistry 50:3859–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishimaru CA, Klos EJ, Brubaker RR. 1988. Multiple antibiotic production by Erwinia herbicola. Phytopathology 78:746–750 [Google Scholar]
- 19. Ishimaru CA, et al. 2010. Pantocin A antibiotic produced by Pantoea vagans C9-1: chemical and genetic characterization. Phytopathology 100:S54 [Google Scholar]
- 20. Jin M, Fischbach MA, Clardy J. 2006. A biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J. Am. Chem. Soc. 128:10660–10661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jin M, Liu L, Wright SA, Beer SV, Clardy J. 2003. Structural and functional analysis of pantocin A: an antibiotic from Pantoea agglomerans discovered by heterologous expression of cloned genes. Angew. Chem. Int. Ed. Engl. 42:2898–2901 [DOI] [PubMed] [Google Scholar]
- 22. Jin M, Wright SAI, Beer SV, Clardy J. 2003. The biosynthetic gene cluster of pantocin A provides insights into biosynthesis and a tool for screening. Angew. Chem. Int. Ed. Engl. 42:2902–2905 [DOI] [PubMed] [Google Scholar]
- 23. Johnson KB, Stockwell VO. 1998. Management of fire blight: a case study in microbial ecology. Annu. Rev. Phytopathol. 38:227–248 [DOI] [PubMed] [Google Scholar]
- 24. Kearns LP, Mahanty HK. 1998. Antibiotic production by Erwinia herbicola Eh1087: its role in inhibition of Erwinia amylovora and partial characterization of antibiotic biosynthesis genes. Appl. Environ. Microbiol. 64:1837–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loper JE, et al. 1991. Evaluation of streptomycin and oxytetracycline and copper resistance of Erwinia amylovora isolated from pear orchards in Washington state. Plant Dis. 75:287–290 [Google Scholar]
- 26. Meyer F, et al. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nunes C, Usall J, Teixidó N, Fons E, Viñas I. 2002. Post-harvest biological control by Pantoea agglomerans (CPA-2) on Golden Delicious apples. J. Appl. Microbiol. 92:247–255 [DOI] [PubMed] [Google Scholar]
- 28. Pusey PL, Stockwell VO, Reardon C, Smits THM, Duffy B. 2011. Antibiotic production by Pantoea agglomerans biocontrol strain E325 and activity against Erwinia amylovora on apple flower stigmas. Phytopathology 101:1234–1241 [DOI] [PubMed] [Google Scholar]
- 29. Pusey PL, Stockwell VO, Rudell DR. 2008. Antibiosis and acidification by Pantoea agglomerans strain E325 may contribute to suppression of Erwinia amylovora. Phytopathology 98:1136–1143 [DOI] [PubMed] [Google Scholar]
- 30. Rezzonico F, Duffy B. 2007. The role of luxS in the fire blight pathogen Erwinia amylovora is limited to metabolism and does not involve quorum sensing. Mol. Plant Microbe Interact. 20:1284–1297 [DOI] [PubMed] [Google Scholar]
- 31. Rezzonico F, Smits THM, Montesinos E, Frey JE, Duffy B. 2009. Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol. 9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rezzonico F, Vogel G, Duffy B, Tonolla M. 2010. Application of whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid identification and clustering analysis of Pantoea species. Appl. Environ. Microbiol. 76:4497–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rezzonico F, et al. 2007. Is the ability of biocontrol fluorescent pseudomonads to produce the antifungal metabolite 2,4-diacetylphloroglucinol really synonymous with higher plant protection? New Phytol. 173:861–872 [DOI] [PubMed] [Google Scholar]
- 34. Rosen HR. 1935. The mode of penetration of pear and apple blossoms by the fire-blight pathogen. Science 81:26. [DOI] [PubMed] [Google Scholar]
- 35. Sammer UF, et al. 2009. 2-Amino-3-(oxirane-2,3-dicarboxamido)-propanoyl-valine, an effective peptide antibiotic from the epiphyte Pantoea agglomerans 48b/90. Appl. Environ. Microbiol. 75:7710–7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shoji J, et al. 1989. Isolation of CB-25-I, an antifungal antibiotic, from Serratia plymuthica. J. Antibiot. 42:869–874 [DOI] [PubMed] [Google Scholar]
- 37. Smits THM, et al. 2011. Metabolic versatility and antibacterial metabolite biosynthesis are distinguishing genomic features of the fire blight antagonist Pantoea vagans C9-1. PLoS One 6(7):e22247 doi:10.1371/journal.pone.0022247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smits THM, et al. 2010. Genomic and phenotypic characterization of a nonpigmented variant of Pantoea vagans biocontrol strain C9-1 lacking the 530-kb megaplasmid pPag3. FEMS Microbiol. Lett. 308:48–54 [DOI] [PubMed] [Google Scholar]
- 39. Smits THM, et al. 2010. Complete genome sequence of the fire blight bacterium Erwinia amylovora CFBP 1430 and comparison to other Erwinia strains. Mol. Plant Microbe Interact. 23:384–393 [DOI] [PubMed] [Google Scholar]
- 40. Stockwell VO, Johnson KB, Sugar D, Loper JE. 2002. Antibiosis contributes to biological control of fire blight by Pantoea agglomerans strain Eh252 in orchards. Phytopathology 92:1202–1209 [DOI] [PubMed] [Google Scholar]
- 41. Stockwell VO, Johnson KB, Sugar D, Loper JE. 2010. Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 applied as single strains and mixed inocula. Phytopathology 100:1330–1339 [DOI] [PubMed] [Google Scholar]
- 42. Stockwell VO, Johnson KB, Sugar D, Loper JE. 2011. Mechanistically compatible mixtures of bacterial antagonists improve biological control of fire blight of pear. Phytopathology 101:113–123 [DOI] [PubMed] [Google Scholar]
- 43. Taghavi S, et al. 2009. Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl. Environ. Microbiol. 75:748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomson SV. 1986. The role of the stigma in fire blight infections. Phytopathology 76:476–482 [Google Scholar]
- 45. Vanneste JL, Yu J, Beer SV. 1992. Role of antibiotic production by Erwinia herbicola Eh252 in biological control of Erwinia amylovora. J. Bacteriol. 174:2785–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vanneste JL, Cornish DC, Yu J, Voyle MD. 2002. P10c: a new biological control agent for control of fire blight which can be sprayed or distributed using honey bees. Acta Hortic. 590:231–235 [Google Scholar]
- 47. Vanneste JL, Yu J, Cornish DA. 2008. Presence of genes homologous to those necessary for synthesis of microcin MccEh252 in strains of Pantoea agglomerans. Acta Hortic. 793:391–396 [Google Scholar]
- 48. Wodzinski RS, Umholtz TE, Rundle JR, Beer SV. 1994. Mechanisms of inhibition of Erwinia amylovora by Erw. herbicola in vitro and in vivo. J. Appl. Bacteriol. 76:22–29 [Google Scholar]
- 49. Wright SAI, Zumoff CH, Schneider L, Beer SV. 2001. Pantoea agglomerans strain EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro. Appl. Environ. Microbiol. 67:284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]


