Abstract
The complete nucleotide sequences of two large, low-copy-number plasmids of 229.6 kb (pBSC2-1) and 143.5 kb (pBSC2-2) were determined during assembly of the whole-genome shotgun sequences of the methane-oxidizing bacterium Methylocystis sp. strain SC2. The physical existence of the two plasmids in strain SC2 was confirmed by pulsed-field gel electrophoresis followed by Southern hybridization. Both plasmids have a conserved replication module of the repABC system and carry genes involved in their faithful maintenance and conjugation. In addition, they contain genes that might be involved in essential metabolic processes. These include several heavy metal resistance genes and copper transport genes in pBSC2-1 and a complete nitrous oxide reductase operon and a pmoC singleton in pBSC2-2, the latter encoding the PmoC subunit of particulate methane monooxygenase.
INTRODUCTION
Methanotrophic bacteria, or methanotrophs, are able to oxidize the greenhouse gas methane via the enzyme methane monooxygenase (MMO), which is present in the particulate form (pMMO) in most of them (29). Methanotrophs have been shown to contain plasmids of considerable sizes (24), but no function has been ascribed to any of them yet. Earlier surveys have reported plasmids to be present in both type I and type II methanotrophs, ranging in size from 8 to 186 kb (23, 24, 40). However, no detectable homology was found among the plasmids isolated from different methanotrophs by using DNA-DNA hybridization and restriction pattern analysis (24). Given a genome size of approximately 4 Mb for methanotrophic bacteria, the collective size of the plasmids reported to be present in a single organism would account for approximately 5 to 10% of their total gene content. This calls for sequencing these large plasmids, in order to characterize the genes located on them and eventually understand their putative function in methanotrophs. For a long time, Methylococcus capsulatus Bath was the only methanotrophic bacterium whose complete genome sequence was available (39). However, it was known not to contain any plasmid (24). With the availability of high-throughput sequencing facilities, an increasing number of methanotroph genomes are being sequenced. This includes the type I methanotrophs Methylobacter tundripaludum SV96 (36), Methylomonas methanica MC09 (5), and Methylomicrobium alcaliphilum strain 20Z (38), the type II methanotrophs Methylosinus trichosporium OB3b (33) and Methylocystis sp. strain Rockwell (ATCC 49242) (31), the facultative Methylocella silvestris BL2 (12), and the acidophilic “Candidatus Methylacidiphilum infernorum” V4 (22). However, apart from the very recently announced genome of the type I methanotroph Mm. alcaliphilum strain 20Z, no plasmid sequence has been reported for any of these methanotrophic bacteria.
The present study reports the complete sequences of two novel, large plasmids, pBSC2-1 and pBSC2-2, identified in Methylocystis sp. strain SC2. Basic plasmid-related features and the putative functions of genes present in the two plasmids, including a singleton pmoC located on pBSC2-2, are discussed.
MATERIALS AND METHODS
PFGE and Southern hybridization.
Strain SC2 was grown in NH4Cl mineral salt (AMS) medium with 20% methane in the headspace (2). Pulsed-field gel electrophoresis (PFGE) was performed to detect the presence of plasmids in strain SC2. Agarose plugs were prepared from cultures of strain SC2 harvested at different growth phases (log and stationary phases) or grown to log phase at different methane concentrations (0.2% and 20%) following standard methods (4). To allow migration of plasmid DNA based on size, plugs were digested with S1 nuclease. Single gel plugs were treated with 1 unit of S1 nuclease (Invitrogen, Carlsbad, CA) for 20 min (4). The undigested or S1-digested DNA was analyzed on 1% PFGE-grade low-melting-temperature agarose (Bio-Rad, Hercules, CA), and electrophoresis was performed in 0.5× TBE (44.5 mM Tris-borate, 12.5 mM EDTA [pH 8.0]) at 14°C for 48 h. The settings were 6 V/cm at a pulse switch time ramped from 50 to 90 s in a contour-clamped homogenous electric field apparatus (CHEF DRIII apparatus; Bio-Rad). Gels were stained with Gel-Red (Biotium, Inc., Hayward, CA) and visualized on a Typhoon scanner (GE Healthcare Life Sciences). Southern hybridization probes were prepared from PCR products obtained with primer pairs that specifically target regions of the genome and the two plasmids (see Table S1 in the supplemental material). These included a 1,629-bp product (with primers pmo2F and pmo2R) from the genome, a 1,074-bp product (with primers P1-F and Pl-R) from pBSC2-1, and a 1,168-bp product (with primers P2-F and P2-R) from pBSC2-2. PCR products were gel purified and checked by sequencing before labeling using the DIG High Prime DNA labeling kit (Roche Diagnostics GmbH, Mannheim, Germany). Following PFGE, the DNA was blotted onto positively charged nylon membranes (Zeta-Probe GT membrane; Bio-Rad) by overnight capillary transfer with 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) as recommended in the DIG application manual for filter hybridization (Roche). After transfer, DNA was UV cross-linked and the membrane was hybridized with one of the probes at 45°C for 16 h in DIG Easy Hyb solution (Roche). A chemiluminescence-based method (using the CSPD chemiluminescent substrate) was used to detect probe-target hybrids, according to the manufacturer's instructions (Roche). Membrane stripping and rehybridization were performed following standard methods (Roche).
Plasmid curing.
Attempts to cure the plasmids by using ethidium bromide or acridine orange were made. Prior to curing, the sublethal concentrations of the curing agents were determined for strain SC2. At 30°C, these were around 100 μg ml−1 and 200 μg ml−1 for ethidium bromide and acridine orange, respectively. Cells were grown with the above concentrations of DNA-intercalating agents (initial optical density at 600 nm [OD600] of 0.04) for 15 days. Cell broth was then suitably diluted and spread on AMS agar plates and incubated. Similar trials were also done at 37°C. However, due to very poor growth, the plates were not analyzed further. Single colonies that appeared on plates incubated at 30°C were randomly selected (100 colonies for each treatment) and checked for loss of plasmids by PCR using the same primers as used for probe generation in Southern hybridization (see Table S1 in the supplemental material). For colony PCR, cells were lysed using Lyse and Go PCR reagent (Thermo Scientific, Rockford, IL). The cell lysates contained both genomic and plasmid DNA templates.
Sequencing strategy and assembly.
Two plasmid sequences were assembled from a whole-genome shotgun project for Methylocystis sp. strain SC2. Whole-genome shotgun data were obtained by pyrosequencing using the GS FLX Titanium platform (454 Life Sciences, Branford, CT). In addition, a fosmid library with 37-kb inserts (CopyControl fosmid library production kit; Epicentre, Madison, WI) was constructed. End sequencing of 4,000 fosmid inserts was performed using BigDye 3.1 chemistry and 3730XL capillary sequencers (ABI, Darmstadt, Germany). Sequence reads obtained from 454 and Sanger sequencing were assembled by the MIRA assembler (13). A preliminary data analysis indicated the presence of two plasmids in strain SC2. Corresponding contigs were finished by primer walking and manually curated in Consed (17). The obtained plasmid sequences for pBSC2-1 and pBSC2-2 show sequencing coverages of 52- and 56-fold, respectively, on average. The potential open reading frames (ORFs) were established with GLIMMER 2.1 (15). The predicted ORFs and putative intergenic sequences were further examined manually using the Artemis platform (6). BLAST searches (1) against the NCBI nonredundant protein database and Swiss-Prot protein database were performed to determine the ORFs. Circular plasmid maps were drawn using DNA plotter (7). The deduced amino acid sequences of the pmoC product were aligned in MEGA (v4.0.2) using CLUSTALW, and a phylogenetic tree was constructed using the p-distance matrix of neighbor-joining algorithms. Bootstrap analyses were performed with 1,000 replications. Similar tree topologies were obtained with neighbor-joining and maximum-likelihood methods.
Nucleotide sequence accession numbers.
The complete sequences of pBSC2-1 and pBSC2-2 have been deposited in the EMBL, GenBank, and DDBJ databases under the accession numbers FO000001 and FO000002, respectively.
RESULTS AND DISCUSSION
Two large, low-copy-number plasmids are present in strain SC2.
Complete nucleotide sequences of two novel plasmids were obtained during assembly of the genome sequence of strain SC2 and were found to be of 229.6 kb (pBSC2-1) and 143.5 kb (pBSC2-2) with average G+C contents of 60.7 and 60.4 mol%, respectively (Fig. 1). Sequences of both plasmids were obtained together with the genome sequence from a whole-genome shotgun approach. This allowed us to estimate the copy numbers of the plasmids by comparing their average sequence coverages (27). As pBSC2-1 and pBSC2-2 were obtained with sequence coverages (52- and 56-fold, respectively) similar to that of the chromosome (53-fold), we assume that the plasmids are present in single copy. PFGE followed by Southern blot analysis clearly showed the presence of two bands corresponding to the expected sizes (based on sequence assembly) of the plasmids, in addition to the genome (Fig. 2). Distinct bands representing the two plasmids were always detected in PFGE, regardless of whether log- or stationary-phase cells (Fig. 2) or cells grown at different methane concentrations (0.2% and 20% methane; data not shown) were analyzed. The banding patterns in PFGE and Southern hybridization from S1 nuclease-treated and untreated plugs were similar. These results strongly support the independent existence of two plasmids in strain SC2, ruling out the rare possibility that the assembled contigs are integrative conjugative elements or mobile excisable elements.
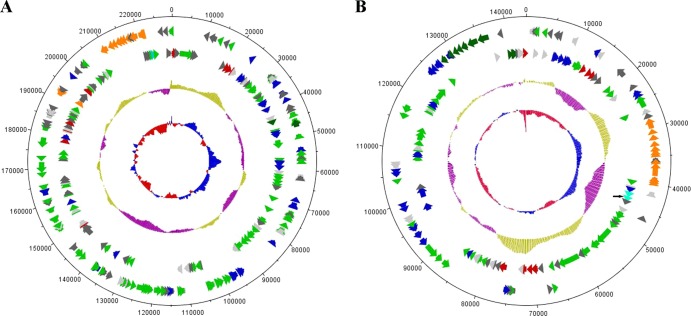
Fig 1.
Circular representation of pBSC2-1 (A) and pBSC2-2 (B). Features depicted by the circles from outside (1) to inside (5) are as follows: circle 1, scale marked in 5-kb intervals; circle 2, ORFs present in the reverse strand; circle 3, ORFs present in the forward strand; circle 4, G+C content (sea green and magenta indicate values greater than and less than the average G+C content, respectively); circle 5, G+C skew bias ([G − C]/[G + C]) (red indicates values below average and blue indicates values above average). ORFs are colored according to their function: red, plasmid replication and maintenance; orange, conjugation; light green, ORFs having homology to some functional proteins; dark green, nitrogen metabolism; blue, transposase-like proteins; light gray, hypothetical proteins; dark gray, conserved hypothetical proteins. The singleton pmoC (marked with an arrow) and its upstream regulatory gene in pBSC2-2 are colored sky blue.
Fig 2.
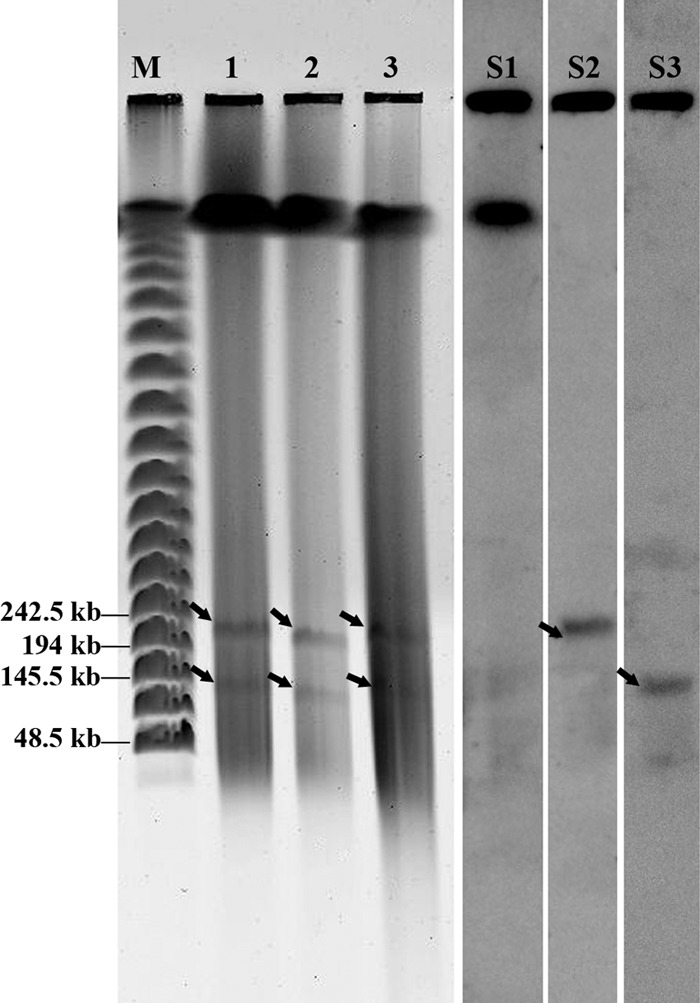
PFGE followed by Southern hybridization shows the presence of two plasmids in strain SC2. Lane M shows Lambda PFG marker (New England BioLabs, Ipswich, MA) containing successively larger concatemers of lambda DNA, with sizes of representative bands shown on the left. Lanes 1 and 2 contain plugs of strain SC2 cells harvested in log phase and stationary phase, respectively. Lane 3 contains a plug of strain SC2 cells harvested in log phase followed by S1 nuclease treatment. The two plasmids are marked by arrows. The plasmids showed similar migration patterns, regardless of whether or not SC2 cells had been treated with S1 nuclease. Therefore, Southern hybridization results are shown only for the electrophoresis pattern in lane 1, using probes specific to genomic DNA (lane S1), pBSC2-1 (lane S2), and pBSC2-2 (lane S3).
Of the 240 and 152 predicted ORFs in pBSC2-1 and pBSC2-2, respectively, 121 (50%) and 72 (47%) were assigned putative functions, 58 (24%) and 25 (16%) encoded conserved hypothetical proteins, and 28 (12%) and 23 (15%) ORFs were putative novel. Moreover, 33 (14%) ORFs in pBSC2-1 and 32 (21%) in pBSC2-2 encode transposase-like proteins, thereby suggesting possible DNA rearrangement events in both plasmids. The ORFs present in the two plasmids and their annotations are described in Tables S2 and S3 in the supplemental material. For the unique ORF identifiers, P1 or P2 refers to the respective plasmid pBSC2-1 or pBSC2-2, and the subsequent number corresponds to the ORF number.
The plasmids contain a repABC replication module.
Dot plot analysis showed that the two plasmids share only a few conserved regions (see Fig. S1 in the supplemental material), in particular, the replication module consisting of repABC genes (P1_12 to P1_14 and P2_74 to P2_76). The repABC family of replicons is reported to be present in large, low-copy-number plasmids and on some secondary chromosomes in at least 19 alphaproteobacterial genera, mainly belonging to the Rhizobiales, Rhodobacterales, and Rhodospirallales (8, 10). It has also been reported that more than one plasmid replicating via the repABC module can coexist in a single organism (25). The replicon consists of three clustered genes (repA, repB, and repC) that are transcribed in the same direction. The first two genes are involved in partition with the partition site (parS) located at different positions throughout the replicon in different plasmids (10). The third gene, repC, is involved in replication, with the replication origin present in an AT-rich stretch of the gene itself (9, 10). A short, AT-rich, highly conserved intergenic sequence (igs) is located between repB and repC (9, 10). From the igs region, a small nontranslated antisense RNA (ctRNA) is transcribed in the direction opposite that of repABC. The ctRNA is found in all repABC operons and controls replication of the plasmid (10, 11). We could identify complete repABC replication modules in both plasmids having all essential features, including the conserved igs region and the consensus promoter sequence for the ctRNA (Fig. 3). A putative parS-like consensus sequence was also identified within repB in both plasmids (Fig. 3). In addition, several copies of genes encoding single-strand binding proteins were identified in both of them (P1_149, P1_180, P1_186, P1_192, P2_1, and P2_21).
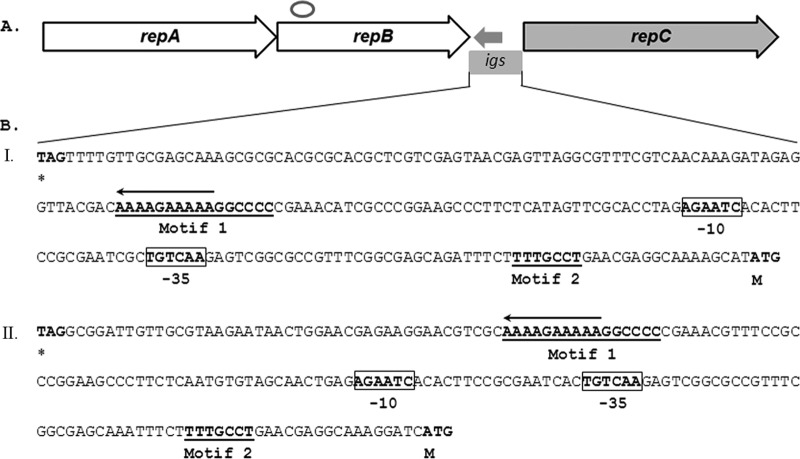
Fig 3.
Schematic representation of the repABC replication module in pBSC2-1 and pBSC2-2. (A) Open arrows (repA and repB) and the filled arrow (repC) indicate repABC, with the arrowheads showing their transcription directions. The small arrow in the opposite direction between repB and repC indicates the position of the putative antisense RNA (ctRNA). The position of the putative parS site is marked as an open circle. In both plasmids, a parS-like consensus sequence (GTTNNCNGCNGNNAAC) (10) is present within repB. The conserved intergenic region (igs) between repB and repC is marked by a box. (B) Nucleotide sequences of the intergenic region (igs) in pBSC2-1 (I) and pBSC2-2 (II). In both cases, sequences are shown from the stop codon of repB to the start codon of repC (both highlighted by boldface letters and indicated below by the respective codes, * and M). The −35 and −10 elements of the ctRNA promoter are indicated, boldface and boxed. The motifs 1 and 2, indicated in boldface and underlined, are conserved in the igs of repABC replicons. The T-tract, which probably constitutes the end of the transcript, is marked by an arrow, with an arrowhead showing the direction of transcription.
Eighty-eight ORFs (37%) in pBSC2-1 and 28 ORFs (18%) in pBSC2-2 showed maximum homology in BLASTX searches with proteins encoded by the genome of Methylocystis sp. strain Rockwell. When similar BLAST searches were done with remaining ORFs from both plasmids, proteins encoded by the genomes of Methylocystis sp. strain Rockwell and/or Ms. trichosporium OB3b appeared in the first five hits in most of them. This indicates that almost all plasmid-carried ORFs in strain SC2 are present in both strain Rockwell and strain OB3b. This, however, does not prove that the two latter strains contain similar plasmids, as the homologous ORFs were found scattered across different contigs of the released genome sequences and the regions of homology were never in reasonably large contigs. Interestingly, we could identify the alphaproteobacterial plasmid-specific replication module in both draft genomes. While Methylocystis sp. strain Rockwell was found to contain the genes in three distinct contigs (ctg177 [NZ_AEVM01000027], ctg148 [NZ_AEVM01000047], and ctg161 [NZ_AEVM01000028]), they were detected in two different contigs in Ms. trichosporium OB3b (ctg00105 [NZ_ADVE01000033] and ctg00122 [NZ_ADVE01000105]). The presence of these multiple repABC operon copies in the genome sequences of Methylocystis sp. strain Rockwell and Ms. trichosporium OB3b strongly supports the existence of plasmids, which is further corroborated by previous studies that reported the occurrence of plasmids in Methylocystis spp. and Ms. trichosporium OB3b (24). Thus, after whole-genome gap closure, such plasmids may be identified as separate entities in strains Rockwell and OB3b. However, we could not identify the repABC genes in the recently released plasmid sequence of Mm. alcaliphilum strain 20Z, which might propagate via some different method. The absence of the repABC genes appears logical, as Mm. alcaliphilum is a member of the Gammaproteobacteria.
Additional plasmid-related features of pBSC2-1 and pBSC2-2.
Apart from the repAB genes, which provide a partition function, additional loci that could ensure the faithful segregation of the two plasmids to daughter cells were identified. In pBSC2-2, a parAB locus (P2_23 to P2_24) was identified. These genes could encode a ParA-like ATPase and a ParB-like partition protein. pBSC2-1 was found to encode a RelBE family toxin-antitoxin system (P1_206 to P1_207), which is commonly present on large plasmids and chromosomes. The toxin component (RelE) represses translation, probably by binding to ribosomes, and the antidote protein (RelB, a member of the Arc/MetJ repressor family) stably binds and deactivates the toxin (26). A complete type IV secretion system locus (avhB1 [virB1] to avhB11 [virB11]) was identified in both plasmids (P1_220 to P1_230 and P2_39 to P2_50), followed by traG in pBSC2-1 (P1_219) and virD4 in pBSC2-2 (P2_38) (Fig. 1; see also Tables S2 and S3 in the supplemental material). In plasmids like RP4, Ti, and pBTK445, it has been shown by mutational analysis that their avhB (virB) loci are involved in the conjugative transfer function (14, 18). Thus, both pBSC2-1 and pBSC2-2 might also have a conjugative transfer function.
During PCR-based screening for the loss of plasmids, all colonies obtained after treatment with the DNA-intercalating agents (ethidium bromide and acridine orange) tested positive for both the plasmids and the genome. Thus, the plasmids were not cured by either of the DNA-intercalating agents. This may be accounted to the segregation mechanisms encoded by pBSC2-1 and pBSC2-2, which confer high plasmid stability. The incapability of plasmid curing by DNA-intercalating agents is quite often observed for similar large, low-copy-number plasmids (14, 37).
ORFs with significant similarity to metabolically important genes.
Mercury resistance (mer) genes were identified in both plasmids. While only merR and merT were identified in pBSC2-2 (P2_70 to P2_71), a mer operon, constituting the essential genes merR, merT, merP, and merA (P1_153 to P1_156), was identified in pBSC2-1. The characterized mer operons from different bacteria vary in structure and are generally composed of genes that encode the functional proteins for regulation (merR), transport (merT, merP, and/or merC), and reduction (merA) (3, 35). Several functional mer operons from Proteobacteria have been reported to contain the minimum number of genes. These include merRTPAD and merRTPCAD, carried by Tn501 and Tn21, respectively (3, 19). While the former operon does not contain a merC, the one present in the latter was shown by mutational analysis to have no effect on mercury resistance (19). Moreover, the merD gene, which is present in both operons, encodes an additional regulatory component. Its deletion does not hamper the mercury resistance phenotype (3). Based on these findings, we anticipated that the mer operon carried by pBSC2-1 might be functional and could provide resistance to only inorganic mercurial compounds (narrow spectrum), as it lacks merB, which together with merA confers resistance to organomercuric compounds (35). When grown on AMS plates supplemented with various concentrations of HgCl2, strain SC2 could resist only up to 0.1 μg ml−1 of HgCl2. This is at least 10- to 100-fold less than in other bacteria that harbor well-characterized mercury resistance plasmids (19). Some other heavy metal resistance genes were also identified in pBSC2-1, and these included three sets of genes encoding a heavy metal efflux pump belonging to CzcA family and an upstream efflux transporter belonging to the RND family (P1_88 to P1_90, P1_124 to P1_127, and P1_174 to P1_175). The presence of heavy metal resistance genes could be due to the fact that strain SC2 was isolated from a highly polluted aquifer in Germany (16). In addition, several copper transport-related genes were identified in pBSC2-1. These include multiple copies of genes encoding either the copper-binding protein CusF (P1_123, P1_130, and P1_167) or a CtpA-like copper-transporting P-type ATPase (P1_50, P1_55, P1_120, and P1_121). This transporter is known to supply copper to membrane-associated proteins that require this metal as a cofactor (20). The presence of copper transport genes in the plasmid is interesting, as several enzymes, including the primary metabolic enzyme (pMMO), are copper dependent (29).
Both plasmids also carry genes that generally encode proteins with housekeeping functions. In pBSC2-2, these include genes that encode a DNA-directed DNA polymerase (P2_29) and the sigma factor of RNA polymerase (P2_6 and P2_117). pBSC2-1 encodes subunits of the FoF1-ATP synthase complex (P1_97 to P1_105). It also contains genes encoding key enzymes of glycolysis, namely, phosphofructokinase (P1_63), phosphoenolpyruvate synthase (P1_64, P1_145, and P1_151), and glyceraldehyde-3-phosphate dehydrogenase (P1_66). Interestingly, all these ORFs are surrounded by transposase-like proteins, suggesting possible genomic rearrangement or duplication events, as all these genes are also present in the genome of strain SC2.
A nitric oxide reductase gene was identified in both plasmids (P1_45 and P2_130). In addition, a complete nos operon (nosRZDFYX) (P2_139 to P2_144) for nitrous oxide reductase was identified in pBSC2-2 (21, 28). Genes encoding a hybrid cluster protein (hydroxylamine reductase) (P2_147 and P2_148) was found adjacent to the nos operon, suggesting operation of a pathway for hydroxylamine detoxification by reduction to ammonium (32, 33). Methanotrophs are known to produce nitrite and nitrous oxide during growth on methane, apparently due to aerobic oxidation of ammonia and hydroxylamine (32). The genes for nitrification and denitrification are widespread in methanotrophs, and the encoded enzymes show functional redundancy. However, the genes for nitrous oxide reductase have not been detected in any of the genome-sequenced methanotrophs (32). Thus, the presence of a complete plasmid-borne nos operon in strain SC2 is unique among genome-sequenced methanotrophs and might enable SC2 cells to convert the greenhouse gas nitrous oxide to dinitrogen. A functional nos operon has been detected earlier on the megaplasmid pSymA of the symbiotic nitrogen-fixing soil bacterium Sinorhizobium meliloti (21).
A singleton pmoC is present in pBSC2-2.
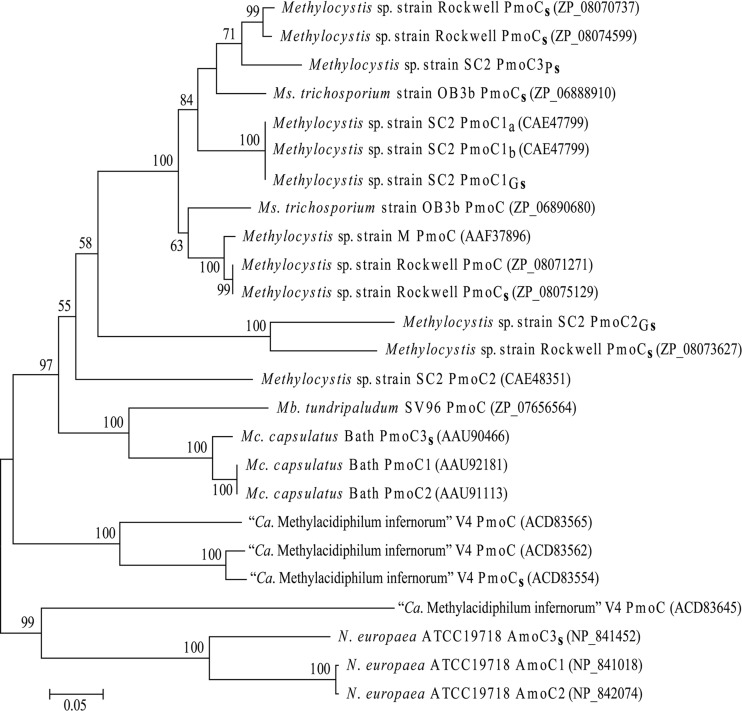
An interesting feature is the presence of a singleton pmoC (pmoC3Ps) in pBSC2-2 (P2_56), which could encode the PmoC subunit of pMMO. Five additional copies of pmoC were identified in the genome of strain SC2, including three as part of the pmoCAB gene clusters (pmoC1a, pmoC1b and pmoC2) encoding pMMO isozymes (2) and two singletons (pmoC1Gs and pmoC2Gs; unpublished data). The amino acid sequence deduced from the plasmid-borne pmoC (PmoC3Ps) showed greatest homology (88%) to the conventional PmoC1a and PmoC1b and contains the conserved Asp129, His133, and His146 residues. These three residues are present in all well-characterized PmoC proteins and are involved in the coordination of copper along with Glu200 of PmoA in the mononuclear copper-binding site of pMMO (30). Isolated copies of pmoC have been repeatedly found in type I and type II methanotrophs, and these include four copies in Methylocystis sp. strain Rockwell (31) and one copy each in Mc. capsulatus Bath (39) and Ms. trichosporium OB3b (33). One singleton pmoC was identified even in the distantly related “Ca. Methylacidiphilum infernorum” V4 genome (22). To determine the evolutionary origin of the plasmid-borne pmoC, a tree was constructed by using the amino acid sequences deduced from all the singleton pmoC genes reported above and representatives from the pmoCAB operons. PmoC3Ps showed a clear affinity to two pmoC singletons of Methylocystis sp. strain Rockwell and the lone copy from Ms. trichosporium OB3b (Fig. 4). A transcriptional regulator (P2_55) containing an N-terminal amidase domain and a C-terminal AraC-type DNA-binding helix-turn-helix (HTH) domain was identified upstream of pmoC3Ps and the two singletons in strain Rockwell. In Ms. trichosporium OB3b, the upstream transcriptional regulator belongs to the LysR family. Conclusive results on the functional role of the singleton pmoC copies are not yet available for any methanotrophic bacterium. Attempts to generate pmoC3 chromosomal insertion null mutants in Mc. capsulatus Bath were made but were unsuccessful. Therefore, Stolyar et al. suggested that pmoC3 may play an essential role in growth on methane (34). Our attempts to generate knockout mutants of singleton pmoC genes, whether present in the genome or present in the plasmid, also failed. Information on the methods used to achieve pmoC knockout mutants is given in the supplemental material (supplemental methods and Fig. S2). After the second recombination, no in-frame deletions were obtained and all tested clones were wild-type revertants. This may suggest that, like pmoC3 in Mc. capsulatus Bath, all three singleton pmoC genes present in strain SC2, including the plasmid-borne one, are essential for methanotrophic growth.
Fig 4.
Neighbor-joining tree showing the phylogenetic analysis of the derived amino acid sequences encoded by pmoC. The amino acid sequences of PmoC used in the tree construction were deduced from the singleton copies and pmoCAB operons present in the genomes of Mc. capsulatus Bath, Mb. tundripaludum SV96, Ms. trichosporium OB3b, Methylocystis sp. strain Rockwell, Methylocystis sp. strain SC2, Methylocystis sp. strain M, and “Ca. Methylacidiphilum infernorum” V4. The accession numbers of the respective PmoC proteins are given in parentheses. Singleton pmoC copies of all the methanotroph strains are marked with “s” in their designations. Singletons from strain SC2 are labeled with “G” or “P” to distinguish between the genome- and plasmid-carried copies. AmoC sequences from Nitrosomonas europaea ATCC 19718 (NC_004757) were also included. Bootstrap values above 50 are shown. The scale bar represents 0.05 substitutions per amino acid position.
In conclusion, strain SC2 was found to harbor two novel repABC-containing plasmids that are stably maintained due to the presence of dual partition systems and could be conjugative in nature. They also contain genes related to the methanotrophic mode of life. While one carries a singleton pmoC, the other has genes related to copper transport, which is indirectly related to proper functioning of pMMO. Further characterization of genes present in pBSC2-1 and pBSC2-2 will provide insights into the functional role these plasmids are playing in methanotrophs.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by the LOEWE Research Center for Synthetic Microbiology (SYNMIKRO) and the Max Planck Society. B.D. is grateful to the Alexander von Humboldt Foundation for his fellowship. S.D. is a postdoctoral fellow of the Max Planck Society.
We thank Klaas Schotanus for his help running PFGE.
Footnotes
Published ahead of print 13 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baani M, Liesack W. 2008. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc. Natl. Acad. Sci. U. S. A. 105:10203–10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barkay T, Miller SM, Summers AO. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355–384 [DOI] [PubMed] [Google Scholar]
- 4. Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 5. Boden R, et al. 2011. Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. J. Bacteriol. 193:7001–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carver T, et al. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. 2009. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25:119–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castillo-Ramirez S, Vazquez-Castellanos JF, Gonzalez V, Cevallos MA. 2009. Horizontal gene transfer and diverse functional constrains within a common replication-partitioning system in Alphaproteobacteria: the repABC operon. BMC Genomics 10:536 doi:10.1186/1471-2164-10-536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cervantes-Rivera R, Pedraza-Lopez F, Perez-Segura G, Cevallos MA. 2011. The replication origin of a repABC plasmid. BMC Microbiol. 11:158 doi:10.1186/1471-2180-11-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cevallos MA, Cervantes-Rivera R, Gutierrez-Rios RM. 2008. The repABC plasmid family. Plasmid 60:19–37 [DOI] [PubMed] [Google Scholar]
- 11. Chai Y, Winans SC. 2005. A small antisense RNA downregulates expression of an essential replicase protein of an Agrobacterium tumefaciens Ti plasmid. Mol. Microbiol. 56:1574–1585 [DOI] [PubMed] [Google Scholar]
- 12. Chen Y, et al. 2010. Complete genome sequence of the aerobic facultative methanotroph Methylocella silvestris BL2. J. Bacteriol. 192:3840–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information, p 45–56 Computer science and biology: Proceedings of the German Conference on Bioinformatics Research Centre for Biotechnology (GBF), Braunschweig, Germany [Google Scholar]
- 14. Dam B, Ghosh W, Das Gupta SK. 2009. Conjugative type 4 secretion system of a novel large plasmid from the chemoautotroph Tetrathiobacter kashmirensis and construction of shuttle vectors for Alcaligenaceae. Appl. Environ. Microbiol. 75:4362–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dunfield PF, et al. 2002. Isolation of a Methylocystis strain containing a novel pmoA-like gene. FEMS Microbiol. Ecol. 41:17–26 [DOI] [PubMed] [Google Scholar]
- 17. Gordon D. 2003. Viewing and editing assembled sequences using Consed, chapter 11, unit 11.2 In Current protocols in bioinformatics. John Wiley and Sons, Somerset, NJ: [DOI] [PubMed] [Google Scholar]
- 18. Hamilton CM, et al. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 182:1541–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamlett NV, Landale EC, Davis BH, Summers AO. 1992. Roles of the Tn21 merT, merP, and merC gene products in mercury resistance and mercury binding. J. Bacteriol. 174:6377–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hassani BK, Astier C, Nitschke W, Ouchane S. 2010. CtpA, a copper-translocating P-type ATPase involved in the biogenesis of multiple copper-requiring enzymes. J. Biol. Chem. 285:19330–19337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holloway P, McCormick W, Watson RJ, Chan YK. 1996. Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY, of Rhizobium meliloti. J. Bacteriol. 178:1505–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hou S, et al. 2008. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct 3:26 doi:10.1186/1745-6150-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lidstrom ME, Stirling DI. 1990. Methylotrophs: genetics and commercial applications. Annu. Rev. Microbiol. 44:27–58 [DOI] [PubMed] [Google Scholar]
- 24. Lidstrom ME, Wopat AE. 1984. Plasmids in methanotrophic bacteria: isolation, characterization and DNA hybridization analysis. Arch. Microbiol. 140:27–33 [DOI] [PubMed] [Google Scholar]
- 25. Mazur A, Majewska B, Stasiak G, Wielbo J, Skorupska A. 2011. repABC-based replication systems of Rhizobium leguminosarum bv. trifolii TA1 plasmids: incompatibility and evolutionary analyses. Plasmid 66:53–66 [DOI] [PubMed] [Google Scholar]
- 26. Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rasko DA, et al. 2007. Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J. Bacteriol. 189:52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richardson D, Felgate H, Watmough N, Thomson A, Baggs E. 2009. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle—could enzymic regulation hold the key? Trends Biotechnol. 27:388–397 [DOI] [PubMed] [Google Scholar]
- 29. Semrau JD, DiSpirito AA, Yoon S. 2010. Methanotrophs and copper. FEMS Microbiol. Rev. 34:496–531 [DOI] [PubMed] [Google Scholar]
- 30. Smith SM, et al. 2011. Crystal structure and characterization of particulate methane monooxygenase from Methylocystis species strain M. Biochemistry 50:10231–10240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stein LY, et al. 2011. Genome sequence of the methanotrophic alphaproteobacterium Methylocystis sp. strain Rockwell (ATCC 49242). J. Bacteriol. 193:2668–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stein LY, Klotz MG. 2011. Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem. Soc. Trans. 39:1826–1831 [DOI] [PubMed] [Google Scholar]
- 33. Stein LY, et al. 2010. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b. J. Bacteriol. 192:6497–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stolyar S, Costello AM, Peeples TL, Lidstrom ME. 1999. Role of multiple gene copies in particulate methane monooxygenase activity in the methane-oxidizing bacterium Methylococcus capsulatus Bath. Microbiology 145:1235–1244 [DOI] [PubMed] [Google Scholar]
- 35. Summers AO. 1986. Organization, expression, and evolution of genes for mercury resistance. Annu. Rev. Microbiol. 40:607–634 [DOI] [PubMed] [Google Scholar]
- 36. Svenning MM, et al. 2011. Genome sequence of the arctic methanotroph Methylobacter tundripaludum SV96. J. Bacteriol. 193:6418–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uraji M, Suzuki K, Yoshida K. 2002. A novel plasmid curing method using incompatibility of plant pathogenic Ti plasmids in Agrobacterium tumefaciens. Genes Genet. Syst. 77:1–9 [DOI] [PubMed] [Google Scholar]
- 38. Vuilleumier S, et al. 2012. Genome sequence of the haloalkaliphilic methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. J. Bacteriol. 194:551–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ward N, et al. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2:e303 doi:10.1371/journal.pbio.0020303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warner P, Higgins I, Drozd J. 1977. Examination of obligate and facultative methylotrophs for plasmid DNA. FEMS Microbiol. Lett. 1:339–342 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.