Abstract
Interaction of microbes with their environment depends on features of the dynamic microbial surface throughout cell growth and division. Surface modifications, whether used to acquire nutrients, defend against other microbes, or resist the pressures of a host immune system, facilitate adaptation to unique surroundings. The release of bioactive membrane vesicles (MVs) from the cell surface is conserved across microbial life, in bacteria, archaea, fungi, and parasites. MV production occurs not only in vitro but also in vivo during infection, underscoring the influence of these surface organelles in microbial physiology and pathogenesis through delivery of enzymes, toxins, communication signals, and antigens recognized by the innate and adaptive immune systems. Derived from a variety of organisms that span kingdoms of life and called by several names (membrane vesicles, outer membrane vesicles [OMVs], exosomes, shedding microvesicles, etc.), the conserved functions and mechanistic strategies of MV release are similar, including the use of ESCRT proteins and ESCRT protein homologues to facilitate these processes in archaea and eukaryotic microbes. Although forms of MV release by different organisms share similar visual, mechanistic, and functional features, there has been little comparison across microbial life. This underappreciated conservation of vesicle release, and the resulting functional impact throughout the tree of life, explored in this review, stresses the importance of vesicle-mediated processes throughout biology.
MICROBIAL MEMBRANE VESICLES
The production of spherical, membranous vesicles from microbial cell surfaces is conserved among organisms from all three branches of the tree of life, spanning both prokaryotes and eukaryotes: Gram-negative and Gram-positive bacteria (16, 47, 58, 73), archaea (17, 18), fungi (3, 65–67), and parasites (82, 83). For consistency in this review, we will refer to bacterial and archaeal structures as membrane vesicles (MVs) and fungal and parasitic vesicles as either exosomes or shedding microvesicles (two distinct populations referred to collectively as microvesicles [55]). The microscopic observation of microbial MVs spans more than 50 years, and numerous functions have been attributed to these extracellular vesicles by many investigators. The release of vesicles provides flexibility to respond to environmental cues, secrete components destined for the cell surface, virulence factors, and antigens, and interact with the host in the case of pathogens. Because MV release is conserved across many organisms, MV-mediated functions are likely to be critical to microbial life.
Both bacterial MVs and archaeal MVs are derived from the cell surface (Fig. 1A and 2A). Early observation of Gram-negative bacterial MVs revealed the release of an antigenic complex of lipopolysaccharide (LPS) and lipoprotein into the surrounding medium following amino acid deprivation of an Escherichia coli lysine auxotroph (40), which was initially proposed to be derived from the LPS-containing outer membrane (OM) of the bacteria (29). Since these early investigations, many groups have developed methodologies to isolate and analyze bacterial MVs. Although reconciling these differences in experimental design often makes it difficult to draw generalized conclusions, it is well accepted that Gram-negative bacterial MVs range from 10 to 300 nm in diameter and contain OM and periplasmic constituents, including proteins, lipoproteins, phospholipids, and LPS (43, 58). The contents of the inner membrane (IM) and cytoplasm were generally thought to be excluded from MVs, although recent analyses of the bacterial MV proteome suggest that some proteins typically annotated as having cytoplasmic localization consistently appear in MVs (15, 45, 92, 94). In addition to bacterial membrane proteins, toxins and signaling molecules can be incorporated into the membrane or lumen of the MV; MV release then serves as a secretion mechanism (42, 54, 95). Although Gram-negative bacterial MVs have been most rigorously studied, recent observation of Gram-positive MV release has demonstrated that this is a function more widely conserved across all bacteria. MVs derived from Gram-positive bacteria, such as Bacillus spp., are similarly sized (50 to 150 nm in diameter [47, 73]) and are rich in membrane lipids as well as toxins (including the anthrax toxin).
Fig 1.
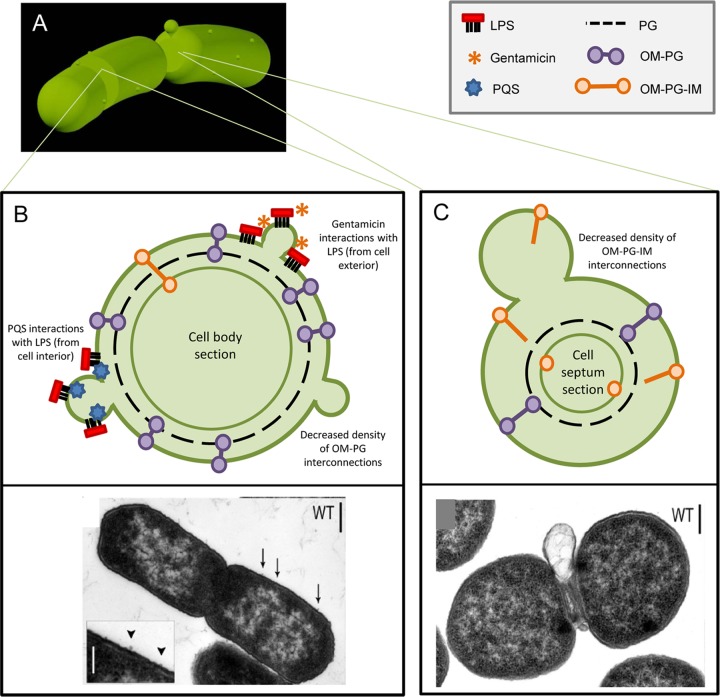
MV production by Bacteria. (A) Gram-negative bacteria release surface-derived membrane vesicles (MVs) at division septa and along the length of the cell body. (B) Upper panel: proteins harboring domains that link the outer membrane (OM) to the peptidoglycan layer (PG) minimize MV release along the cell body; temporary disruption of these OM-PG interactions results in MV release. Membrane-active antibiotics (such as gentamicin) and signaling molecules (such as pqs) interact with the membrane surface to induce MV release. Lower panel: transmission electron micrograph (TEM) of wild-type S. Typhimurium with inset showing small MV release along the cell body. (Reprinted from reference 15 with permission of the publisher, John Wiley and Sons.) (C) Upper panel: at the constricted division septum, temporary dissociation of OM-PG-IM protein complexes spanning the OM and inner membrane (IM) occurs, facilitating the release of a large MV before completing cell division. Lower panel: TEM of wild-type S. Typhimurium releasing a septal MV. (Reprinted from reference 15 with permission of the publisher, John Wiley and Sons.)
Fig 2.
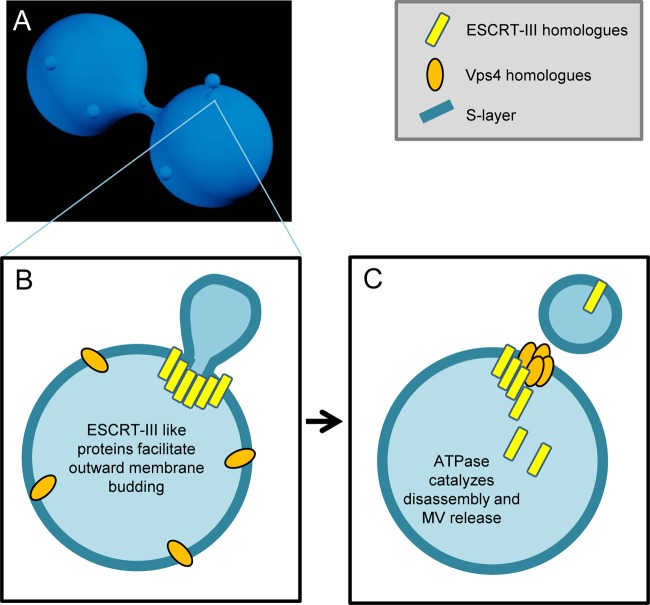
MV production by Archaea. (A) Archaea release MVs that are derived from the cell surface, similar to the process in Bacteria. (B) This process is facilitated through the coordinated action of ESCRT-III homologue proteins (yellow), conserved in archaeal and eukaryotic life. ESCRT-III homologues, known for membrane scission capabilities, are directed to surround the site of nascent MV formation and induce the outward protrusion of the membrane, including the archaeal S-layer. (C) Vps4 homologue ATPases then catalyze the disassembly of ESCRT-III homologues, and MV release occurs.
Archaeal MVs, such as those released by Sulfolobus species, range from 90 to 230 nm in diameter and contain membrane lipids and S-layer proteins also derived from the archaeal cell surface (17, 69). A common functional theme begins to emerge: these MVs can also transport toxic compounds into the surrounding milieu (69), although toxin production is not required for vesicle release, as non-toxin-producing strains and other archaea such as Ignicoccus naturally release MVs as well (71).
Eukaryotic microbial vesicles, derived from fungi and parasites, include at least two vesicle populations (Fig. 3) (25, 66). Exosomes (40 to 100 nm in diameter) are derived from multivesicular bodies (MVBs) within the cell and are typically homogenously shaped (Fig. 3B) (55). Shedding microvesicles (SMVs) (100 to 1,000 nm in diameter) bud directly from the cell surface, resulting in more heterogeneous vesicle morphology (Fig. 3C) (13). Vesicles derived from eukaryotic microbes contain characteristic lipids and proteins that reflect both surface constituents and secreted cellular components. While these two processes are visually similar when observed microscopically, it is likely that the cellular machineries participating in formation and the downstream functions of these MV populations are distinct. The presence of multiple active vesicle secretion mechanisms is supported by work in Saccharomyces cerevisiae, in which extracellular MVs were released even in the absence of known secretory pathways (67).
Fig 3.
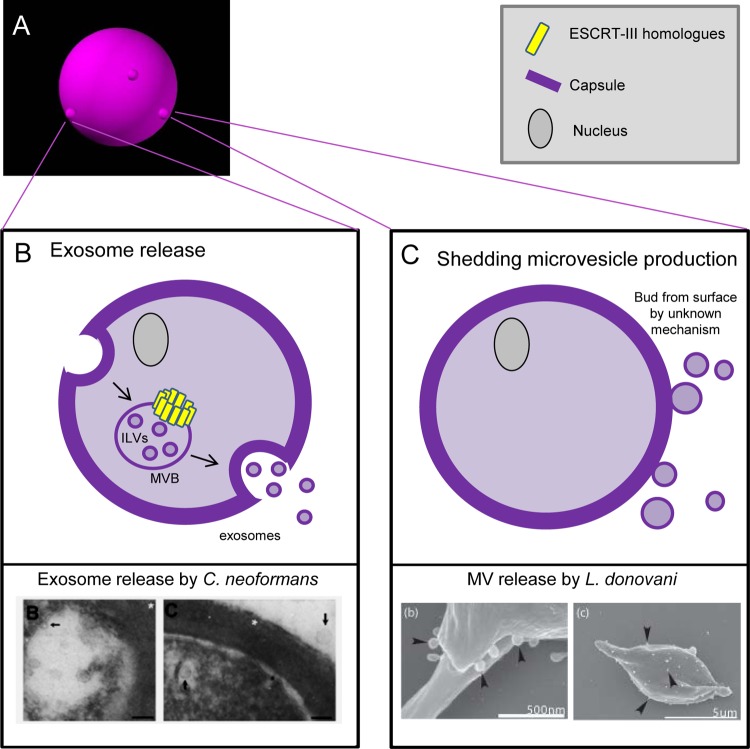
MV production by eukaryotic microbes. (A) Eukaryotic microbes, including fungi (shown here) and parasites, release MVs at the cell surface, although these MVs may be derived from multiple sources. (B) Upper panel: exosome release is a conserved process in eukaryotic microbes. An endosome is created in the cytosol, which traffics through the microbial cell. In transit, the ESCRT-III homologues (also conserved in Archaea and higher eukaryotes) induce formation of intraluminal vesicles (ILVs), creating multivesicular bodies (MVBs). MVBs fuse to the cell surface and release the vesicular content as exosomes. Lower panel: electron micrograph demonstrating release of exosomes by the fungus Cryptococcus neoformans. (Reprinted from reference 75 with permission.) (C) An additional pathway for MV release in eukaryotic microbes exists, appearing to produce surface-derived MVs reminiscent of those in Bacteria and Archaea. In this process, for which mechanistic details are as yet unknown, shedding microvesicles bud directly from the cell surface. Lower panel: electron micrograph demonstrating MV release by the parasite Leishmania donovani. (Reprinted from reference 82 with permission of the publisher, BioMed Central.)
The release of vesicles has been demonstrated for both pathogenic and nonpathogenic microbes under a range of growth conditions, including in liquid broth and on agar plates in the laboratory (15, 82, 90), in biofilms (6, 78), upon infection with bacteriophage (50), and by pathogenic organisms growing within an animal host (9, 20, 23, 52, 63, 88). In addition, modification of medium conditions, such as the presence of serum (62), limitation of essential amino acids (40), or treatment with subinhibitory concentrations of membrane active antibiotics (33), stimulates MV production, suggesting that vesicle release is both dynamic and manipulable, essential characteristics for microbes subjected to ever-changing environments. Conservation of this process during both in vitro and in vivo growth and the difficulty (or inability [57, 58, 67]) of genetic approaches to identify mutations that abrogate vesicle release reinforce the idea that this process is integral to microbial life.
MECHANISMS OF VESICLE BIOGENESIS
Bacteria.
As this process in Gram-positive bacteria is only beginning to be explored (16, 47, 56, 73), we will focus our discussion on the understanding of the process in Gram-negative bacteria. Gram-negative bacterial MV release, observed microscopically over several decades (7, 40), has been proposed to occur by many different mechanisms (5, 29, 42, 54, 58, 60, 96). Reconciliation of these mechanisms, however, is complicated by the variability under which these studies were completed (i.e., using mutant bacterial strains with altered LPS and/or nutrient requirements or using varied growth conditions, such as amino acid deprivation or the presence of antibiotics). Most models of MV release, studied primarily in Proteobacteria, propose an “either-or” involvement of proteins or LPS in this process. However, it is likely that MV formation results from the contribution of multiple dynamic surface components (Fig. 1), and there are several proposed mechanisms of MV release. As detailed reviews exist on this topic that are outside the scope of this review (43), we will limit our discussion here and include recent developments in the field.
LPS is integral to the structure and physiology of Gram-negative bacteria. Disruption of this molecule, therefore, impacts the stability and architecture of the microbial surface, including vesicle release. For example, in Pseudomonas aeruginosa, perturbation of the bacterial surface with subinhibitory concentrations of the antibiotic gentamicin stimulates increased MV release. Gentamicin is thought to interact uniquely with the two structurally distinguishable LPS species of P. aeruginosa, based on charge attraction (44, 74). Gentamicin treatment not only induces MV release but also results in enrichment of certain LPS species in MVs (64). A similar phenomenon is found in Porphyromonas gingivalis, which also expresses two differentially charged O antigens. Significant decrease in the abundance of one LPS species in the OM altered protein cargo incorporated into MVs (27). Furthermore, treatment of the E. coli cell surface with Mg2+ resulted in decreased MV production, supporting the idea that charge interactions on the cell surface (via the LPS O-antigen polysaccharide) influence MV release (89), and that substances acting on the cell surface in different environments may induce release of MVs with specific contents. Additionally, stimuli from within the bacterial cell can also influence MV release. The interaction of the lipid A portion of LPS with the P. aeruginosa-produced quorum-sensing molecule pqs (54) alters membrane curvature (53), thereby inducing MV production (Fig. 4B).
Fig 4.
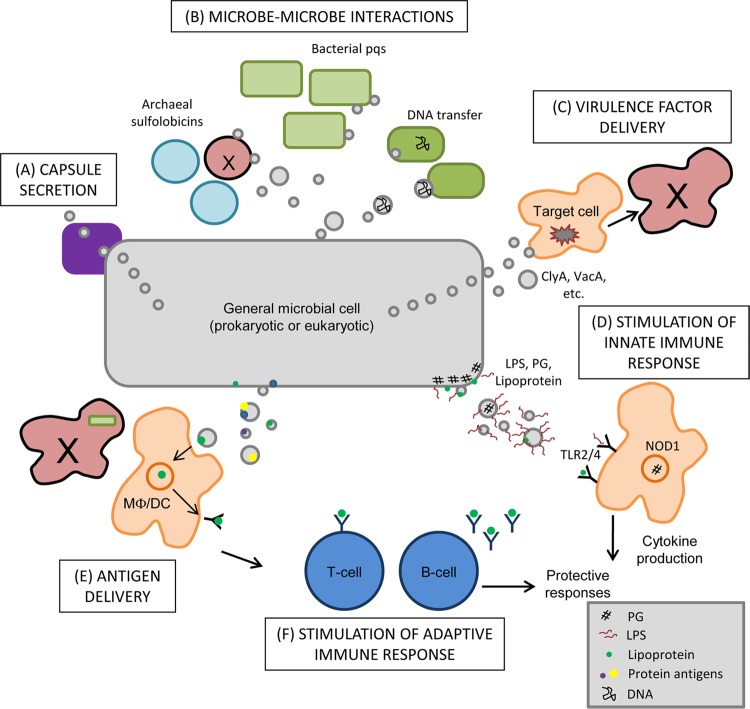
Biological impact of MV release. MVs, originating from bacteria, fungi, archaea, or parasites, possess many functions in microbial physiology and pathogenesis. MVs promote the secretion of capsular polysaccharide to the cell surface (A) and are utilized for cell-cell communication between Bacteria (release of quorum-sensing molecules and transfer of DNA) and Archaea (secretion of antimicrobials) (B). (C) For pathogenic microbes, the role of MVs in vivo is likely multifaceted, including the ability to directly deliver virulence factors, such as toxins, to target host cells. (D) Natural microbial structures present in MVs can act to stimulate the innate immune system through activation of TLRs (via LPS and/or lipoprotein sensing) and NLRs (via peptidoglycan detection). (E) These antigenic structures contained in MVs likely interact with antigen-presenting cells (APC) when released in vivo during infection, facilitating the presentation of MV antigens in cases where APC may be impaired by cytotoxic organisms. (F) Antigen presentation can lead to the stimulation of adaptive immune responses, triggering T-cell and B-cell responses that are directed toward MV antigens. The ways in which MVs are utilized by microbes during growth (and during infection in the case of pathogenic organisms) are complex and they underscore the importance of these structures for all microbial life.
The role that bacterial surface proteins play in MV release must also be considered, as they are integral to both the structure and function of the cell, and there are several nonmutually exclusive mechanisms discussed here in which proteins may be involved. The first model considers MV release in the absence of environmental stressors (like antibiotics) or mutations affecting nutrient requirements or LPS structure. A systematic and quantitative approach to analyze the MVs produced by Salmonella enterica serovar Typhimurium (S. Typhimurium) revealed that specific major envelope proteins modulate MV release by wild-type (WT) bacteria (15). WT MV production occurred along the cell body and at division septa (Fig. 1A). When specific envelope proteins were deleted, cells exhibited an enrichment of MV release at either cell body (Fig. 1B) or septa (Fig. 1C). This localization was due to the absence of important envelope interconnections normally present in envelope proteins of interest; when interacting domains as small as one amino acid were removed, the resulting phenotype mimicked a full deletion, underscoring the importance of these interactions. Specifically, proteins such as OmpA and LppAB are anchored in the OM but interact with the peptidoglycan (PG) layer (Fig. 1B, purple OM-PG interconnections), while Pal, TolB, and TolA form a protein complex that spans the OM-IM (Fig. 1C, orange OM-PG-IM interconnections). In WT cells, these tethers normally maintain MV production at minimal levels. OM-PG linked proteins dampen the release of small MVs from the cell body of S. Typhimurium, whereas formation of the cell division plane requires movement of OM-PG-IM protein complexes, facilitating septal MV release (15). Migration of these proteins along the dynamic cell surface promotes enrichment of a subset of constituents into septal or cell body-derived MVs, suggesting that Gram-negative bacteria may be able to control the distribution and abundance of envelope interconnections such that the site and content of released MVs are correspondingly regulated. This idea was confirmed by proteomic analysis of MV purified from septa and cell body, respectively. Based on these data, we proposed the following model: MV production occurs at envelope regions where the density of OM-PG and/or OM-PG-IM interconnections has been temporarily decreased (Fig. 1B and C) (15). While some organisms may have evolved distinct means of modulating LPS structure (61) resulting in modifications of MV formation, the high degree of conservation among the protein domains facilitating OM-PG and OM-PG-IM envelope interconnections across diverse Gram-negative bacteria (15) supports the idea that these connections have widespread importance in the process of bacterial MV release.
Other proteins may also contribute to MV formation. Accumulation of overexpressed periplasmic proteins promotes increased MV release, possibly via induction of outward budding of the membrane (59). However, these models assume that MVs are released into an aqueous milieu where they are able to freely travel. Although this may be true for pathogenic organisms that live in association with host cells and tissues, it is not the case in partially hydrated environments like microbe-rich soil. Recent studies have suggested a novel mechanism by which organisms may release MVs in water-restricted conditions. The construction of nanopods allows for MVs to travel from bacterial cells such as Delftia species (81). Nanopods are seemingly protective tubular structures made of surface layer protein through which MV-like structures containing LPS and OM proteins are released from cells. Interestingly, nanopod formation is conserved in other organisms, may be facilitated by LPS-surface layer protein interactions (19), and may represent a conserved mechanism for MV release in aqueously poor environments.
Fungi, parasites, and archaea.
Although taxonomically distant, many of the basic features of vesicle production by fungi, parasites, and archaea appear to be conserved, including surface release and the important protein homologues regulating the mechanisms of release. Archaeal vesicle release shares similar features with both prokaryotic and eukaryotic mechanisms of MV production, perhaps representing the most evolutionarily basic process upon which other microbes have adapted additional mechanisms (Fig. 2). Archaeal MVs are surface derived and are released by “pinching off” the cell surface, a phenomenon reminiscent of bacterial MVs and eukaryotic SMVs (Fig. 1B and 3C). While microbial SMV release may be controlled via mechanisms used in mammalian SMV release (via various enzymes, including calpain, flippase, floppase, scramblase, and gelsolin [68]), this process is currently uncharacterized. However, evidence supports a mechanism in which surface MV release by archaea is controlled by a regulated mechanism involving the conserved membrane scission machinery endosomal sorting complex required for transport (ESCRT-III) and vacuolar sorting protein (Vps4) homologues (Fig. 2B and C), as these proteins are found to be released in vesicles (17, 51). Interestingly, the absence of ESCRT-III-like homologues in some vesicle-producing archaea (such as the Thermococcales) suggests that multiple vesicle release mechanisms may exist (BLAST [86]). We focus our discussion on the currently proposed mechanism of archaeal MV release, thought to be modulated by ESCRT-III-like proteins.
To further underscore the utilization of similar mechanisms in phylogenetically distinct organisms, the involvement of ESCRT-III homologue proteins has been suggested for archaeal MV release, as well as the creation of vesicles destined to be released as eukaryotic microbial exosomes. Exosome release is a well-characterized process in multicellular eukaryotes. Released exosomes perform functions such as cell-cell communication and secretion and are characterized by specific protein markers, including HSP70, Alix, and clatherin (55). As mentioned previously, when an endocytic vesicle is created, the endosomal membrane is capable of protruding inward, resulting in the accumulation of intraluminal vesicles (ILVs). An MVB containing ILVs then travels to the cell surface, fuses, and releases exosomes into the surrounding environment (Fig. 3B) (37, 55).
Microbial ILV formation and surface MV formation in archaea are likely facilitated by (or likely involve) proteins which are homologous to mammalian ESCRT-III proteins. ESCRT-III machinery is well conserved between mammals, fungi, and archaea (48) (a full review of this machinery can be found elsewhere [26, 32, 98]). Simply stated, the function of these proteins is to cleave membranous “necks” in processes such as archaeal surface MV release, cell division, viral budding from mammalian cell surfaces, and ILV formation in eukaryotic microbes. These proteins have the ability to cleave membranes from the inside of a cell or vesicle instead of constricting the membrane from the outside. As in bacterial MV release, localization of protein participants in the membrane is a crucial factor in the successful directed release of vesicles. The coordinated actions of the ESCRT-III and Vps4 homologues ensure this critical localization. In eukaryotic systems, monomeric subunits of the ESCRT-III complex are present in the cytoplasm in an inactive form and, upon recruitment to the membrane, become active via conformational changes. ESCRT-III protein complex formation occurs at the membrane, allowing for constriction of the associated membrane (Fig. 2B and 3B). These proteins are only transiently membrane associated and are therefore returned to the cytosol via dissociation of oligomerized ESCRT machinery, a process catalyzed by VPS4 homolog ATPases (Fig. 2C). Although ESCRT-III-dependent ILVs have been shown to be targeted for degradation (not secretion) in a multicellular mammalian system (91), the involvement of these proteins in targeting eukaryotic microbial MVs has yet to be established. Not only are the processes of vesicle release visually similar in eukaryotes and archaea, but the protein participants are conserved and may share similar roles in the mechanistic regulation of release in both types of organisms, although the involvement of the archaeal ESCRT-III homologue proteins remain to be fully elucidated. Utilization of similar processes by such varied organisms for similar functional outcomes, like other essential biological processes, supports the importance of vesicle release for microbes.
FUNCTIONAL SIGNIFICANCE OF MEMBRANE VESICLES
Release of membrane vesicles during infection: in vitro and in vivo studies.
The outcome of any infection is determined by the complex interplay that balances pathogen replication and persistence with host defenses. Integral to this balance is microbial flexibility to adapt to different environments. The release of vesicles is a conserved surface feature that is poised to contribute to the adaptive capabilities of microbial cells. This is supported by the observation that MV production occurs in many pathogens both in vivo and during interaction with host cells in vitro. For example, Salmonella Typhimurium releases MVs during intracellular growth in epithelial cells and macrophages (24), and Helicobacter pylori, a causative agent of gastric ulcers, exhibits release of virulence factor-containing MVs upon interaction with gastric tissue (23, 63, 72, 87). Similarly, increased levels of PagC and OmpX, which are upregulated by S. Typhimurium under conditions that mimic the host intracellular environment, stimulate MV release (39). During infections caused by Neisseria meningitidis, in which the disease pathology is characterized by life-threatening sepsis, the production of endotoxic (LPS-containing) MVs in the bloodstream has been observed, and it likely contributes to pathogenesis of sepsis through Toll-like receptor (TLR)- and nucleotide-binding oligomerization domain-containing protein (NOD)-like receptor (NLR)-mediated inflammatory cytokine production (62, 88). Complementary to direct microscopic observation of MV release, complexes containing OM proteins and LPS have been isolated from blood during experimental sepsis (28), suggesting that this material may be vesicular in nature and could persist in vivo separately from parent bacteria. In addition, systemic effects of heart disease may be promoted by MVs from the oral pathogen Porphyromonas gingivalis, as MVs have been shown to aggregate platelets in vitro (80). Similarly, eukaryotic microbes have also been shown to release vesicular structures during interaction with host cells and tissues, such as the release of vesicles by Leishmania donovani during macrophage infection in vitro (83). Vesicle release is also likely the route by which Cryptococcus neoformans accumulates capsular polysaccharide on its surface during murine infection (Fig. 4A) (20). If microbes possessed the ability to modulate MV formation, potentially in response to environmental cues sensed within the host, it could provide a mechanism to shape the host-pathogen interaction. The presence of serum (9, 62) or antimicrobial peptides (10), temperature and pH variations (83), and as-yet-identified signals within host cells (20–22, 24, 100) and tissues (23, 52) stimulate microbial MV release. The ability to release MVs upon host cell interaction is, therefore, a seemingly widespread feature among microbial pathogens.
Membrane vesicle-mediated secretion of bioactive molecules.
Microbes use MV production to communicate with each other as well as with a host. Intermicrobial communication may promote either positive or negative interactions (Fig. 4B). P. aeruginosa releases the quorum-sensing signaling molecule pqs via MVs (54). The archaeon Sulfolobus solfataricus uses MV release to discourage nearby growth of other (competing) Sulfolobus species by secreting sulfolobicin toxins (69). Haemophilus influenzae releases DNA in MVs (termed transformasomes), and archaea within the order Thermococcales release DNA-associated MVs, likely to facilitate genetic exchange (34). Thus, intermicrobe cross talk, important for coordination of pathogenic mechanisms and destruction of competing species, is promoted by the conserved feature of MV release.
The use of MV-mediated secretion mechanisms to release virulence factors has also been increasingly demonstrated for many organisms (Fig. 4C) (31, 41, 49). These factors often do not have identifiable signal sequences (82, 95) or are secreted by strains lacking known secretion systems (67) and therefore are thought to be released from the microbe via MV production. For example, the cytotoxin ClyA of E. coli and Salmonella enterica serovar Typhi associates with the Gram-negative bacterial OM, promoting release in MVs; this affinity for lipid membranes shapes its interaction with the host cell membrane as well (95). The lytic activity of MV-derived ClyA against eukaryotic host cells was 8-fold more potent and occurred more quickly than that derived in soluble form from the periplasm, potentially due to direct toxin delivery via fusion of bacterial MVs with the host membrane. A targeted toxin delivery system is also observed for the toxin-containing Bacillus anthracis MVs (73) and the heat-labile enterotoxin-containing MVs of enterotoxigenic E. coli (ETEC). The association of ETEC toxin with both the MV exterior and lumen (30) facilitates the uptake of the MVs by susceptible target host cells via direct receptor binding and entry (38).
The vacuolating cytotoxin, VacA, of H. pylori, is a virulence factor expressed by 50 to 60% of clinical H. pylori isolates and can be released from the bacterium in soluble or MV-associated form (87). VacA-containing MVs associate with host cell surfaces and then enter and traverse host cells in vitro and in vivo (23, 87). Although the function and biological impact of soluble versus MV-associated VacA have not been fully elucidated, it is known that 25% of VacA is secreted in MV-associated form. In contrast to the ClyA toxin, MV-associated VacA exerts a less potent vacuolating activity on host cells (72), potentially modulating toxin potency or overall virulence during infection. Interestingly, MV-associated and soluble forms of the heat-labile ETEC toxin elicit similar cytokine responses from eukaryotic cells yet appear to achieve this via distinct cell signaling pathways (12). These data support the hypothesis that microbes may actively regulate the content of MVs (72) to manipulate the host-pathogen interplay.
The biological importance of vesicle release by eukaryotic microbes is highlighted by studies of the mechanism by which capsule molecules reach and accumulate at the Cryptococcus neoformans cell surface (Fig. 4A). Investigation of C. neoformans cells using advanced microscopy techniques revealed intracellular, spherical vesicles migrating through the cell, traversing the cell membrane layer, and being released from the cryptococcal surface in an intact, membrane-bound state (77). Release of these vesicles coincided with extracellular capsule accumulation, and immunoelectron microscopy revealed that capsular polysaccharide was MV associated (75, 76). MVs were also associated with the cell wall in acapsular cells, indicating that a vesicle transport mechanism is likely not restricted to capsule secretion but may be used for a broad range of large molecules and by other organisms that do not express capsular polysaccharide (76). In support of this hypothesis, acapsular fungi and Leishmania, Histoplasma, Candida, Sporothrix, and Saccharomyces produce microvesicles ranging from 10 to 350 nm in diameter. These vesicles have been observed intracellularly, in association with the cell wall and/or in the extracellular space, and contain microbial lipids, proteins, and carbohydrates thought to contribute to virulence (3, 75, 76, 82), including fungal lipids (such as glucosylceramide and ergosterol), superoxide dismutase, catalase, laccase, urease, and acid phosphatase (3, 75, 76). It is possible that MVs could be released during the close interaction between host cell and pathogen, facilitating direct delivery of virulence factors to the target host cell rather than the external milieu where diffusion could lessen the functional impact of the molecule.
In support of the importance of vesicle release for parasitic microbes, proteomic analysis of the secretome of the parasite Leishmania donovani revealed that 98% of secreted proteins lack signal sequences for well-established secretion pathways. This suggests that these proteins require nonclassical mechanisms of secretion, such as MV release, from the cell (82). Extracellular MVs were visualized being released from the cell body and flagellar pocket of the microbe and were found to also contain putative virulence factors that may contribute to pathogenesis through targeted delivery into or proximal to host cells. It is likely that this MV population includes both exosomes and SMVs, as only 10% of the proteins secreted by L. donovani were identified as typical exosome protein constituents (based on mammalian exosome studies) (82). There remain a high fraction of secreted proteins that have not been associated with exosome content, consistent with coincident release of SMVs.
Biological impact of MV release during infection.
As carriers of antigens and virulence factors, as well as facilitators of microbial surface modifications, the biological impact of MVs is likely to be considerable. We will focus here on MV-mediated effects on complex host-pathogen interactions during infection. Directed release of MVs may influence the innate immune response to infection, including Toll-like receptor (TLR) and nucleotide-binding oligomerization domain-containing protein (NOD) signaling (Fig. 4D). TLR ligands in bacterial MVs (including LPS and lipoproteins) stimulate maturation of and cytokine release by macrophages and dendritic cells (reference 2 and unpublished observations) and likely contribute to the pathogenesis of inflammatory infections such as neisserial sepsis (8, 9). Likewise, MVs carrying peptidoglycan, the ligand for the cytosolic innate immune receptor NOD1, are able to initiate NOD1 signaling and downstream NF-κB-dependent inflammatory responses (35, 36). Furthermore, infected antigen-presenting cells may no longer be capable of properly processing and presenting antigens, potentially inhibiting initiation of adaptive immune responses. MVs released by pathogenic organisms are nonviable and contain antigens recognized by the adaptive immune system (2, 4). These surface organelles may therefore represent an important source of antigen in vivo (Fig. 4E).
MVs and SMVs, owing to their derivation from the microbial surface, naturally contain antigens important for the generation of an immune response to bacterial and fungal infection, such as immunogenic surface proteins, LPS, and capsular polysaccharide (1, 4, 11, 70, 79, 85, 99). This is supported by proteomic analysis of MV content, which has identified the presence of known B and/or T cell antigens (Fig. 4F) (references 4, 11, 14, 45, 70, 84, 92, 94 and unpublished observations). In addition, recent evidence suggests that MVs may act as antigen “decoys” in vivo to redirect the antibody response, resulting in the production of antibodies ineffective for clearance of intact organisms (93). The observations that MVs are produced in vivo during infection and that sera from patients following bacterial and fungal infections display reactivity to antigens contained in MVs (1, 11, 75) suggest that proteins and carbohydrates present in MVs may act as additional and potentially significant sources of antigen during infection beyond that provided by the intact organism itself.
Host responses to MVs can benefit either the pathogen or the host. Signaling the presence of a foreign invader initiates a host defense (advantage for host) which might provide the microbe with an environmental advantage or opportunity to misdirect the immune system (advantage for microbe). Thus, host responses may be modulated by microbial organisms to accommodate their lifestyles while inside the host. For example, S. Typhimurium thrives in an inflamed intestine because inflammation provides a metabolic advantage over commensal organisms (97). In addition, the host-pathogen balance may be influenced by active microbial modulation of MV formation and/or contents in response to differing environmental conditions, including growth in host tissues. This phenomenon is evidenced by the ability of S. Typhimurium to dynamically direct specific cell components to certain parts of the cell, resulting in the enrichment of these contents in vesicle populations released at different cellular locations (15). In addition, other studies have shown preferential exclusion or inclusion of cell components in MVs compared to the cell surface (46, 47, 58). The ability to manipulate this conserved process likely has broad implications for altering host-pathogen interactions and the resulting immune response.
CONCLUSIONS
The release of membrane vesicles, a phenomenon shared by organisms across all three branches of life (Fig. 1, 2, and 3), plays an integral role in cell physiology and pathogenesis of infection. MV release is essential for promoting interactions between microbial cells and between eukaryotic host cells and microbes, including communication, release of antigens, and secretion of virulence factors (Fig. 4). For pathogenic microbes, MVs may facilitate modifications of the microbial surface to avoid immune detection, provide additional protective measures against host defense molecules, interact with antigen-presenting cells, shuttle capsule to the cell surface to avoid uptake by host cells, or provide the host immune system with microbial antigen targets. The ability of microbes to select the contents of MVs is currently underappreciated and understudied. The widespread functional conservation of MV release, in addition to homologous mechanistic components involved in this phenomenon, such as the ESCRT-III homologue proteins in archaea and eukaryotic microbes, underscores the importance of the process. Further investigation of MV contents and release in various environmental conditions (including that of the host) will surely reveal further examples of how these surface organelles, long thought to be mere artifacts, are essential to microbial life.
ACKNOWLEDGMENTS
We thank Matthew Kaiser for assistance with figure generation and the members of the Cookson laboratory for helpful manuscript revisions. We apologize to those whose work we could not reference as a result of size limitations.
Our work on host immune control of Salmonella infection is supported by National Institutes of Health grant U19 AI090882, and B.L.D. was additionally supported by National Institutes of Health grant T32 AI55396.
Footnotes
Published ahead of print 12 March 2012
REFERENCES
- 1. Abadi J, Pirofski L. 1999. Antibodies reactive with the cryptococcal capsular polysaccharide glucuronoxylomannan are present in sera from children with and without human immunodeficiency virus infection. J. Infect. Dis. 180:915–919 [DOI] [PubMed] [Google Scholar]
- 2. Alaniz RC, Deatherage BL, Lara JC, Cookson BT. 2007. Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo. J. Immunol. 179:7692–7701 [DOI] [PubMed] [Google Scholar]
- 3. Albuquerque PC, et al. 2008. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell. Microbiol. 10:1695–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bergman MA, et al. 2005. CD4+ T cells and toll-like receptors recognize Salmonella antigens expressed in bacterial surface organelles. Infect. Immun. 73:1350–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubes R. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180:4872–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beveridge TJ, Makin SA, Kadurugamuwa JL, Li Z. 1997. Interactions between biofilms and the environment. FEMS Microbiol. Rev. 20:291–303 [DOI] [PubMed] [Google Scholar]
- 7. Birdsell DC, Cota-Robles EH. 1967. Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli. J. Bacteriol. 93:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brandtzaeg P, et al. 2001. Neisseria meningitidis lipopolysaccharides in human pathology. J. Endotoxin Res. 7:401–420 [PubMed] [Google Scholar]
- 9. Brandtzaeg P, et al. 1992. Meningococcal endotoxin in lethal septic shock plasma studied by gas chromatography, mass-spectrometry, ultracentrifugation, and electron microscopy. J. Clin. Invest. 89:816–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brodsky IE, Ghori N, Falkow S, Monack D. 2005. Mig-14 is an inner membrane-associated protein that promotes Salmonella typhimurium resistance to CRAMP, survival within activated macrophages and persistent infection. Mol. Microbiol. 55:954–972 [DOI] [PubMed] [Google Scholar]
- 11. Brown A, Hormaeche CE. 1989. The antibody response to salmonellae in mice and humans studied by immunoblots and ELISA. Microb. Pathog. 6:445–454 [DOI] [PubMed] [Google Scholar]
- 12. Chutkan H, Kuehn MJ. 2011. Context-dependent activation kinetics elicited by soluble versus outer membrane vesicle-associated heat-labile enterotoxin. Infect. Immun. 79:3760–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cocucci E, Racchetti G, Meldolesi J. 2009. Shedding microvesicles: artefacts no more. Trends Cell Biol. 19:43–51 [DOI] [PubMed] [Google Scholar]
- 14. Cummings LA, Deatherage BL, Cookson BT. 17 September 2009, posting date Chapter 8.8.11, Adaptive immune responses during Salmonella infection. In Fang F, Kagnoff MF. (ed), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: http://www.ecosal.org [DOI] [PubMed] [Google Scholar]
- 15. Deatherage BL, et al. 2009. Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72:1395–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dorward DW, Garon CF. 1990. DNA is packaged within membrane-derived vesicles of gram-negative but not gram-positive bacteria. Appl. Environ. Microbiol. 56:1960–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellen AF, et al. 2009. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 13:67–79 [DOI] [PubMed] [Google Scholar]
- 18. Ellen AF, Zolghadr B, Driessen AM, Albers SV. 2010. Shaping the archaeal cell envelope. Archaea 2010:608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Engelhardt H, Gerbl-Rieger S, Krezmar D, Schneider-Voss S, Engel A, Baumeister W. 1990. Structural properties of the outer membrane and the regular surface protein of Comamonas acidovorans. J. Struct. Biol. 105:92–102 [Google Scholar]
- 20. Feldmesser M, Kress Y, Casadevall A. 2001. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 147:2355–2365 [DOI] [PubMed] [Google Scholar]
- 21. Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68:4225–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fernandez-Moreira E, Helbig JH, Swanson MS. 2006. Membrane vesicles shed by Legionella pneumophila inhibit fusion of phagosomes with lysosomes. Infect. Immun. 74:3285–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fiocca R, et al. 1999. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J. Pathol. 188:220–226 [DOI] [PubMed] [Google Scholar]
- 24. Garcia-del Portillo F, Stein MA, Finlay BB. 1997. Release of lipopolysaccharide from intracellular compartments containing Salmonella typhimurium to vesicles of the host epithelial cell. Infect. Immun. 65:24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gyorgy B, et al. 2011. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 68:2667–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanson PI, Shim S, Merrill SA. 2009. Cell biology of the ESCRT machinery. Curr. Opin. Cell Biol. 21:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haurat MF, et al. 2011. Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 286:1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hellman J, et al. 2000. Release of gram-negative outer-membrane proteins into human serum and septic rat blood and their interactions with immunoglobulin in antiserum to Escherichia coli J5. J. Infect. Dis. 181:1034–1043 [DOI] [PubMed] [Google Scholar]
- 29. Hoekstra D, van der Laan JW, de Leij L, Witholt B. 1976. Release of outer membrane fragments from normally growing Escherichia coli. Biochim. Biophys. Acta 455:889–899 [DOI] [PubMed] [Google Scholar]
- 30. Horstman AL, Kuehn MJ. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275:12489–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hozbor D, et al. 1999. Release of outer membrane vesicles from Bordetella pertussis. Curr. Microbiol. 38:273–278 [DOI] [PubMed] [Google Scholar]
- 32. Hurley JH, Hanson PI. 2010. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 11:556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kadurugamuwa JL, Beveridge TJ. 1997. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 40:615–621 [DOI] [PubMed] [Google Scholar]
- 34. Kahn ME, Barany F, Smith HO. 1983. Transformasomes: specialized membranous structures that protect DNA during Haemophilus transformation. Proc. Natl. Acad. Sci. U. S. A. 80:6927–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaparakis M, et al. 2010. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell. Microbiol. 12:372–385 [DOI] [PubMed] [Google Scholar]
- 36. Kawai T, Akira S. 2009. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 21:317–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keller S, Sanderson MP, Stoeck A, Altevogt P. 2006. Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 107:102–108 [DOI] [PubMed] [Google Scholar]
- 38. Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. 2004. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23:4538–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kitagawa R, et al. 2010. Biogenesis of Salmonella enterica serovar Typhimurium membrane vesicles provoked by induction of PagC. J. Bacteriol. 192:5645–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Knox KW, Vesk M, Work E. 1966. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 92:1206–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kouokam JC, et al. 2006. Active cytotoxic necrotizing factor 1 associated with outer membrane vesicles from uropathogenic Escherichia coli. Infect. Immun. 74:2022–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kuehn MJ, Kesty NC. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645–2655 [DOI] [PubMed] [Google Scholar]
- 43. Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64:163–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lam MY, et al. 1989. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J. Clin. Microbiol. 27:962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee EY, et al. 2007. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7:3143–3153 [DOI] [PubMed] [Google Scholar]
- 46. Lee EY, Choi DS, Kim KP, Gho YS. 2008. Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom. Rev. 27:535–555 [DOI] [PubMed] [Google Scholar]
- 47. Lee EY, et al. 2009. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9:5425–5436 [DOI] [PubMed] [Google Scholar]
- 48. Leung KF, Dacks JB, Field MC. 2008. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9:1698–1716 [DOI] [PubMed] [Google Scholar]
- 49. Lindmark B, et al. 2009. Outer membrane vesicle-mediated release of cytolethal distending toxin (CDT) from Campylobacter jejuni. BMC Microbiol. 9:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loeb MR. 1974. Bacteriophage T4-mediated release of envelope components from Escherichia coli. J. Virol. 13:631–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Makarova KS, Yutin N, Bell SD, Koonin EV. 2010. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat. Rev. Microbiol. 8:731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marsollier L, et al. 2007. Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog. 3:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mashburn-Warren L, et al. 2008. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol. Microbiol. 69:491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425 [DOI] [PubMed] [Google Scholar]
- 55. Mathivanan S, Ji H, Simpson RJ. 2010. Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 73:1907–1920 [DOI] [PubMed] [Google Scholar]
- 56. Mayer F, Gottschalk G. 2003. The bacterial cytoskeleton and its putative role in membrane vesicle formation observed in a Gram-positive bacterium producing starch-degrading enzymes. J. Mol. Microbiol. Biotechnol. 6:127–132 [DOI] [PubMed] [Google Scholar]
- 57. McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188:5385–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McBroom AJ, Kuehn MJ. 12 May 2005, posting date Chapter 2.2.4, Outer membrane vesicles. In Finlay BB. (ed), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: http://www.ecosal.org [Google Scholar]
- 59. McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63:545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mug-Opstelten D, Witholt B. 1978. Preferential release of new outer membrane fragments by exponentially growing Escherichia coli. Biochim. Biophys. Acta 508:287–295 [DOI] [PubMed] [Google Scholar]
- 61. Munford RS, Varley AW. 2006. Shield as signal: lipopolysaccharides and the evolution of immunity to gram-negative bacteria. PLoS Pathog. 2:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Namork E, Brandtzaeg P. 2002. Fatal meningococcal septicaemia with “blebbing” meningococcus. Lancet 360:1741. [DOI] [PubMed] [Google Scholar]
- 63. Necchi V, et al. 2007. Intracellular, intercellular, and stromal invasion of gastric mucosa, preneoplastic lesions, and cancer by Helicobacter pylori. Gastroenterology 132:1009–1023 [DOI] [PubMed] [Google Scholar]
- 64. Nguyen TT, Saxena A, Beveridge TJ. 2003. Effect of surface lipopolysaccharide on the nature of membrane vesicles liberated from the Gram-negative bacterium Pseudomonas aeruginosa. J. Electron Microsc. (Tokyo) 52:465–469 [DOI] [PubMed] [Google Scholar]
- 65. Nosanchuk JD, Nimrichter L, Casadevall A, Rodrigues ML. 2008. A role for vesicular transport of macromolecules across cell walls in fungal pathogenesis. Commun. Integr. Biol. 1:37–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Oliveira DL, et al. 2010. Biogenesis of extracellular vesicles in yeast: many questions with few answers. Commun. Integr. Biol. 3:533–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oliveira DL, et al. 2010. Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS One 5:e11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Piccin A, Murphy WG, Smith OP. 2007. Circulating microparticles: pathophysiology and clinical implications. Blood Rev. 21:157–171 [DOI] [PubMed] [Google Scholar]
- 69. Prangishvili D, et al. 2000. Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J. Bacteriol. 182:2985–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Puohiniemi R, et al. 1990. A strong antibody response to the periplasmic C-terminal domain of the OmpA protein of Escherichia coli is produced by immunization with purified OmpA or with whole E. coli or Salmonella typhimurium bacteria. Infect. Immun. 58:1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rachel R, Wyschkony I, Riehl S, Huber H. 2002. The ultrastructure of Ignicoccus: evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 1:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ricci V, et al. 2005. Free-soluble and outer membrane vesicle-associated VacA from Helicobacter pylori: two forms of release, a different activity. Biochem. Biophys. Res. Commun. 337:173–178 [DOI] [PubMed] [Google Scholar]
- 73. Rivera J, et al. 2010. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. U. S. A. 107:19002–19007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rivera M, Bryan LE, Hancock RE, McGroarty EJ. 1988. Heterogeneity of lipopolysaccharides from Pseudomonas aeruginosa: analysis of lipopolysaccharide chain length. J. Bacteriol. 170:512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rodrigues ML, et al. 2008. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 7:58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rodrigues ML, et al. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 6:48–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sakaguchi N, Baba T, Fukuzawa M, Ohno S. 1993. Ultrastructural study of Cryptococcus neoformans by quick-freezing and deep-etching method. Mycopathologia 121:133–141 [DOI] [PubMed] [Google Scholar]
- 78. Schooling SR, Beveridge TJ. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188:5945–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schop J. 2007. Protective immunity against cryptococcus neoformans infection. McGill J. Med. 10:35–43 [PMC free article] [PubMed] [Google Scholar]
- 80. Sharma A, et al. 2000. Porphyromonas gingivalis platelet aggregation activity: outer membrane vesicles are potent activators of murine platelets. Oral Microbiol. Immunol. 15:393–396 [DOI] [PubMed] [Google Scholar]
- 81. Shetty A, Chen S, Tocheva EI, Jensen GJ, Hickey WJ. 2011. Nanopods: a new bacterial structure and mechanism for deployment of outer membrane vesicles. PLoS One 6:e20725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Silverman JM, et al. 2008. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 9:R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Silverman JM, et al. 2010. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J. Cell Sci. 123:842–852 [DOI] [PubMed] [Google Scholar]
- 84. Singh SP, Williams YU, Benjamin WH, Klebba PE, Boyd D. 1996. Immunoprotection by monoclonal antibodies to the porins and lipopolysaccharide of Salmonella typhimurium. Microb. Pathog. 21:249–263 [DOI] [PubMed] [Google Scholar]
- 85. Singh SP, Williams YU, Klebba PE, Macchia P, Miller S. 2000. Immune recognition of porin and lipopolysaccharide epitopes of Salmonella typhimurium in mice. Microb. Pathog. 28:157–167 [DOI] [PubMed] [Google Scholar]
- 86. Soler N, Marguet E, Verbavatz JM, Forterre P. 2008. Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res. Microbiol. 159:390–399 [DOI] [PubMed] [Google Scholar]
- 87. Sommi P, et al. 1998. Persistence of Helicobacter pylori VacA toxin and vacuolating potential in cultured gastric epithelial cells. Am. J. Physiol. 275:G681–688 [DOI] [PubMed] [Google Scholar]
- 88. Stephens DS, Edwards KM, Morris F, McGee ZA. 1982. Pili and outer membrane appendages on Neisseria meningitidis in the cerebrospinal fluid of an infant. J. Infect. Dis. 146:568. [DOI] [PubMed] [Google Scholar]
- 89. Tashiro Y, Ichikawa S, Nakajima-Kambe T, Uchiyama H, Nomura N. 2010. Pseudomonas quinolone signal affects membrane vesicle production in not only gram-negative but also gram-positive bacteria. Microbes Environ. 25:120–125 [DOI] [PubMed] [Google Scholar]
- 90. Tetz VV, Rybalchenko OV, Savkova GA. 1990. Ultrastructural features of microbial colony organization. J. Basic Microbiol. 30:597–607 [DOI] [PubMed] [Google Scholar]
- 91. Trajkovic K, et al. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319:1244–1247 [DOI] [PubMed] [Google Scholar]
- 92. Vaughan TE, et al. 2006. Proteomic analysis of Neisseria lactamica and Neisseria meningitidis outer membrane vesicle vaccine antigens. Vaccine 24:5277–5293 [DOI] [PubMed] [Google Scholar]
- 93. Vidakovics ML, et al. 2010. B cell activation by outer membrane vesicles—a novel virulence mechanism. PLoS Pathog. 6:e1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Vipond C, et al. 2006. Proteomic analysis of a meningococcal outer membrane vesicle vaccine prepared from the group B strain NZ98/254. Proteomics 6:3400–3413 [DOI] [PubMed] [Google Scholar]
- 95. Wai SN, et al. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25–35 [DOI] [PubMed] [Google Scholar]
- 96. Wensink J, Witholt B. 1981. Outer-membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. Eur. J. Biochem. 116:331–335 [DOI] [PubMed] [Google Scholar]
- 97. Winter SE, et al. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wollert T, et al. 2009. The ESCRT machinery at a glance. J. Cell Sci. 122:2163–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Xu HR, Tan YY, Hsu HS, Moncure CW, Wang XM. 1993. Comparative antibody response to Salmonella antigens in genetically resistant and susceptible mice. Clin. Exp. Immunol. 91:73–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yoon H, Ansong C, Adkins JN, Heffron F. 2011. Discovery of Salmonella virulence factors translocated via outer membrane vesicles to murine macrophages. Infect. Immun. 79:2182–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]