Abstract
Helminth parasites ensure their survival by regulating host immunity through mechanisms that dampen inflammation. These properties have recently been exploited therapeutically to treat human diseases. The biocomplexity of the intestinal lumen suggests that interactions between the parasite and the intestinal microbiota would also influence inflammation. In this study, we characterized the microbiota in the porcine proximal colon in response to Trichuris suis (whipworm) infection using 16S rRNA gene-based and whole-genome shotgun (WGS) sequencing. A 21-day T. suis infection in four pigs induced a significant change in the composition of the proximal colon microbiota compared to that of three parasite-naive pigs. Among the 15 phyla identified, the abundances of Proteobacteria and Deferribacteres were changed in infected pigs. The abundances of approximately 13% of genera were significantly altered by infection. Changes in relative abundances of Succinivibrio and Mucispirillum, for example, may relate to alterations in carbohydrate metabolism and niche disruptions in mucosal interfaces induced by parasitic infection, respectively. Of note, infection by T. suis led to a significant shift in the metabolic potential of the proximal colon microbiota, where 26% of all metabolic pathways identified were affected. Besides carbohydrate metabolism, lysine biosynthesis was repressed as well. A metabolomic analysis of volatile organic compounds (VOCs) in the luminal contents showed a relative absence in infected pigs of cofactors for carbohydrate and lysine biosynthesis, as well as an accumulation of oleic acid, suggesting altered fatty acid absorption contributing to local inflammation. Our findings should facilitate development of strategies for parasitic control in pigs and humans.
INTRODUCTION
The whipworm Trichuris suis is a widespread helminth parasite of pigs. Common manifestations of T. suis infection include diarrhea, anorexia, and retarded growth. The high prevalence of intestinal nematodes, including T. suis, in pig production systems is one of the major factors constraining global food availability. This severely impacts small-scale farmers in developing countries. Coprological examinations suggest that the percentages of pigs infected with T. suis is 25% in Japan (29), 32% in Kenya (31), 38% in Netherlands (11), and 17% in Uganda (32). Upon an animal's ingestion of infective T. suis eggs, the larvae penetrate the cecal and colonic mucosa in the pig intestine, where all subsequent stages of larval development take place. After approximately 2 weeks of a histotrophic phase and ensuing vertical migration to areas immediately beneath the surface mucosal epithelium, the posterior ends of larvae protrude into the gut lumen. By week three postinfection (p.i.), the entire posterior body becomes exposed to the gut lumen (6), and mating and egg excretion begin around 41 days p.i. Adult worms begin to be expulsed from the intestine by week nine p.i. (21). Morphological observations indicate that infection results in local tissue disruption in the intestinal mucosal layer and profound histopathological changes, including eosinophil and mast cell infiltration, crypt hyperplasia, goblet cell hyperplasia, and mucosal hypertrophy (20, 28).
Infection has a systemic effect on several blood parameters, including a decrease in albumin and amylase, an increase in globulins (5), and an increase in basophils and eosinophils starting at 5 weeks p.i. (20). Infection also leads to infiltration of leukocytes in intestinal mucosa and subsequent increase of Th2-related cytokines, such as interleukin 4 (IL-4), IL-5, and IL-13 and related receptors (53). Serum T. suis-specific antibodies (IgG1, IgG2, and IgA) become elevated at 8 weeks p.i. during a trickle infection (30).
Trichuris suis has the potential to play an important role in human health as well. Structural and physiological similarities between the porcine and human gastrointestinal tracts and cross-reactivity between T. suis and Trichuris trichiura, an intestinal nematode that infects approximately 1 billion people worldwide, suggest that the T. suis-pig system can be developed as a model to understand T. trichiura infection in humans. Most importantly, helminths have evolved elegant mechanisms to modulate host immune responses and dampen inflammation. The immune modulation property of helminths has been exploited to test the “helminths and the IBD hygiene hypothesis” (49) and to treat several immune-mediated diseases, including Crohn's disease (39), ulcerative colitis (40), and allergic rhinitis (3). As a result, as many as 28 clinical trials using helminths to prevent or treat human diseases have been entered in the Human Helminth Co-infections Clinical Trials Database (www.niaid.nih.gov). While many trials have documented positive clinical outcomes, T. suis therapy has nevertheless drawn criticism over concerns of the invasiveness of worms for human physiology (19, 47).
It has long been recognized that the pathology of pig trichuriasis is not limited to its direct effect on the host but can also promote secondary infections resulting from tissue damage that leads to invasion by pathogenic microbes (6, 28). The gut microflora of the porcine large intestine acts synergistically with T. suis to produce severe clinical symptoms (36) that can be reversed by broad-spectrum antibiotic treatment of pigs (28). In addition, a case study documented an association between human infection with T. trichiura and the increased pathogenicity of Campylobacter jejuni (37). Clearly, the indirect impact of T. suis infection on host gut microbiota represents an essential part of host-parasite interactions and the expression of disease. In this study, we used metagenomic tools to examine changes in the porcine proximal colon microbiota in response to an experimental T. suis infection and associated changes in metabolic composition using a gas chromatography-mass spectrometry (GC-MS)-based global metabolomic approach.
MATERIALS AND METHODS
Animals and parasitology.
Seven female piglets (Crossbred: Landrace × Yorkshire × Poland China) were obtained from four different litters shortly after weaning at 4 weeks of age from the Beltsville Area Swine Facility. The pigs were maintained indoors on sealed concrete floors throughout the experiment and fed a balanced corn/soybean meal ration containing 16% protein and vitamins and minerals that exceeded National Research Council guidelines (44) and ad lib access to water; no antibiotics were used during the study. Pig management and handling procedures were based on a protocol approved by the Beltsville Area Animal Care and Use Committee (protocol 10-011). A single dose of infective T. suis eggs (2 × 104 eggs/pig) (14) was inoculated per os when the pigs were approximately 3 months of age (n = 4). Three other pigs were orally dosed with the supporting medium of phosphate-buffered saline (PBS) as a placebo and served as uninfected controls. All seven pigs were killed 21 days after inoculation. Luminal contents were collected from the proximal colon at ∼30 cm from the ileal/cecal junction. The pH of the contents was measured using a pH meter for semisolid materials. The collected samples were snap-frozen in liquid nitrogen prior to storage at −80°C until DNA was extracted. Colon pathology was examined by microscopy (28). Trichuris suis worms were counted after incubating the cecum and proximal colon in 10 mM EDTA for 2.5 h at 37°C, followed by manual removal and counting of the worms under a dissecting microscope. The pigs were free of inadvertent Ascaris suum infection based on the absence of worms from the small intestines and white spot lesions on the liver. No other helminth infections have been detected by routine screening in the herd.
Metagenomic DNA extraction, amplicon preparation, and sequencing.
The metagenomic DNA was extracted from the proximal colon contents. The detailed protocol was previously provided (26, 38) and can be founded in Tables S1 and S2 in the supplemental material. Both 16S sequences and whole-genome shotgun (WGS) sequences were deposited in the NCBI Sequence Read Archive (SRA accession no. SRA037229.2).
Sequence analysis, protein prediction, and annotation.
16S raw sequence reads were first decoded based on sample-specific eight-bp bar codes (2); their quality was checked. All sequence reads (mean counts per sample = 38,304 ± 6,349.6 and 40,639 ± 10,212.3 for three control and four infected pigs, respectively) were analyzed using RDP Classifier (48) at an 80% confidence threshold for taxonomic classification and taxonomic classification. The abundance data obtained from the RDP Classifier were input into the Vegan package in the R software program to generate Bray-Curtis (Steinhaus or Czekanowski) dissimilarity coefficients (function vegdistr in Vegan) and for cluster analysis and principal component analysis (PCA). Briefly, the relative abundance (%) was first transformed using the square root, and a dissimilarity index was first calculated. The dissimilarity was used as an input, and hierarchic clustering was performed using the standard R function hclust. Similarity was obtained by subtracting dissimilarity from 1.0.
The pipeline for WGS sequences was listed in Table S2 in the supplemental material. Briefly, quality control filters were first applied to WGS raw reads before analysis. Host contaminants (sequences of porcine origin) were removed. Possible artifacts (redundant reads) were identified and excluded using the software program CD-Hit (27). Quality WGS sequences were de novo assembled using the Newbler software program (v2.5.3). Open reading frames (ORFs) were predicted from all contigs and singletons of ≥300 bp in size using the FragGeneScan software program (v1.14), a recently developed program combining sequencing error models and codon usages in a hidden Markov model (HMM) to improve the prediction of protein-coding regions in short reads (35). Functional annotation was performed according to the COG (Clusters of Orthologous Groups), KEGG, and Pfam (v24.0) databases. Pfam 24.0 seed alignments were downloaded, and a database of core profile HMMs was compiled using the HMMSCAN software package (v3.0), which was used to annotate predicted proteins. Of note, the denominator (the number of total Pfam families or COGs) was very large, which resulted in a very small-number difference between control and infected groups. Predicted ORFS were further analyzed using the software program BLASTP (v2.2.21) with default parameters against the NCBI NR database (downloaded on 21 January 2011). The obtained hit table was analyzed using the software program MEGAN (v3.9) for taxonomic classification (13, 17). Statistical analysis was carried out using the program MetaStats (50), based on a modified t test.
Metabolomic analysis.
Samples of frozen luminal contents from the proximal colon were dispensed in 0.2-g aliquots into 4-ml screw-thread amber glass vials and sealed with black Top Hat polytetrafluoroethylene (PTFE)/silicone caps (J. G. Finneran, Vineland, NJ). All samples were stored at −80°C until analyzed. A headspace solid-phase microextraction (SPME) of the samples was analyzed essentially as described previously (10). The solid-phase microextraction fiber assembly was manually positioned in the headspace above the sample, and the fiber was exposed to the volatiles for 18 h (the sample vial temperature was held at 60°C for the duration of the exposure). The fiber assembly was then placed in the GC inlet for thermal desorption of the analytes. The following SPME fibers (Supelco, Bellefonte, PA) were used: polyacrylate (85 μm), carboxen-polydimethylsiloxane (75 μm), and carboxen-divinylbenzene-polydimethylsiloxane (50/30 μm), with GC inlet temperatures of 280, 300, and 270°C, respectively. All fibers were preconditioned before use, as per the manufacturer's instructions.
GC-MS instrument used for analysis was an Agilent 7890A GC and 5975C inert XL mass selective detector (MSD) with a triple-axis detector (Agilent, Palo Alto, CA). The GC-MS was equipped with a DB5-MS capillary column (Agilent), 30 m in length, with a 0.25-mm inside diameter (i.d.) and 0.25-μm film thickness, and a 0.75-mm-i.d. SPME injection port liner operated in splitless mode. Helium carrier gas was set to a 1.17-ml/min flow rate, and the GC oven was held at an initial temperature of 35°C for 1 min and ramped to 80°C at 3°C/min, to 120°C at 10°C/min, and finally to 260°C at 40°C/min. The final temperature of 260°C was held for 1.5 min. The total run time for the analysis method was 25.0 min. The MSD was scanned from 30 to 550 amu at a rate of 2.81 scans/s. Compounds were identified using the National Institute of Standards and Technology (NIST) (Washington, DC) Automated Mass Spectral Deconvolution and Identification System (AMDIS), (ver 2.69) software program and mass spectral library (NIST08). Only compounds with a spectral match score greater than or equal to 90% were considered.
RESULTS
Alterations in the porcine colon microbiota composition induced by infection.
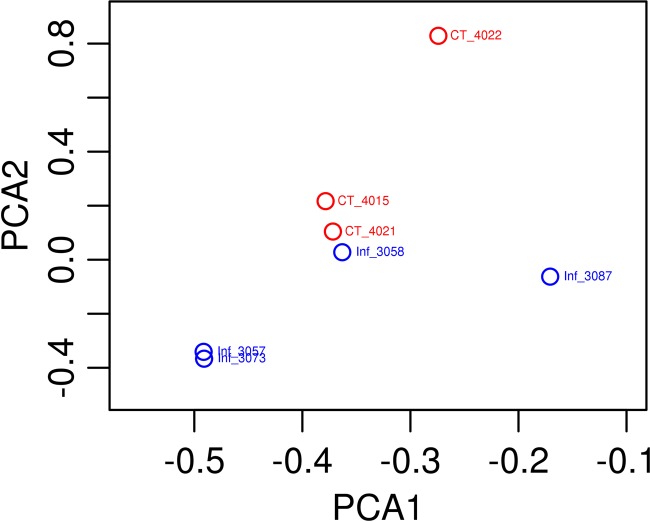
All three uninfected control pigs had a normal smooth and tan mucosal surface, while the four T. suis-infected pigs expressed a hemorrhagic surface with increased mucus production. The mean number of worms recovered from the intestines of infected pigs was 3,222 (±1,347.4, standard deviation [SD]), while no worms were detected from parasite-naive control pigs. The infection did not change the luminal colon pH. The infection level was subclinical, because no overt signs of trichuriasis, including diarrhea and anorexia, were observed. The taxonomic composition of the porcine proximal colon microbiota in T. suis-infected pigs was first analyzed based on the 16S sequences using RDP Classifier (48). Our quality control filtering indicated that artifacts (identical sequences) were negligible in this data set, and therefore, all raw reads were used for classification by RDP Classifier. Taxonomic classification using RDP classifier was able to assign 96.63%, 78.83%, and 54.33% of all 16S raw reads to a given phylum, family, and genus, respectively, at an 80% confidence threshold. This approach identified 15 phyla, where members of the Bacteroidetes, Firmicutes, Spirochaetes, and Proteobacteria were the most abundant, in this order, accounting for >99.4% of the assigned sequences. Sequences assigned to archaea were rare and were detected only in two of seven pigs studied. Collectively, RDP Classifier assigned the 16S rRNA sequences to 134 genera at an 80% confidence threshold. The mean number of genera per sample was 83.4 ± 10.0 (mean ± SD). The 10 most abundant genera accounted for ∼90% of all 16S sequences that were positively assigned to any genus in control pigs. Prevotella was the most abundant; this agrees with recently published reports (25, 34), accounting for 62.3% and 68.2% of 16S sequences assigned in control and infected pigs, respectively. The second-most-abundant genus was Oscillibacter (7.8% in control pigs), followed by Treponema (7.5%), Succinivibrio (3.6%), Anaerovibrio (2.2%), and Roseburia (1.9%). Forty-three genera were shared by all samples tested from control and infected pigs, possibly consisting of the core microbiota of the porcine proximal colon community. These genera (the core microbiota) remarkably represent 97.9% of the entire microbial community based on their relative abundance. Hierarchical clustering grouped all three control pig samples together based on relative abundances of the organisms making up the microbial community (Fig. 1). PCA confirmed the results from clustering analysis indicating that the three samples from control pigs were closely related; however, the separation between samples from all the infected pigs and control pigs was not distinct (Fig. 2).
Fig 1.
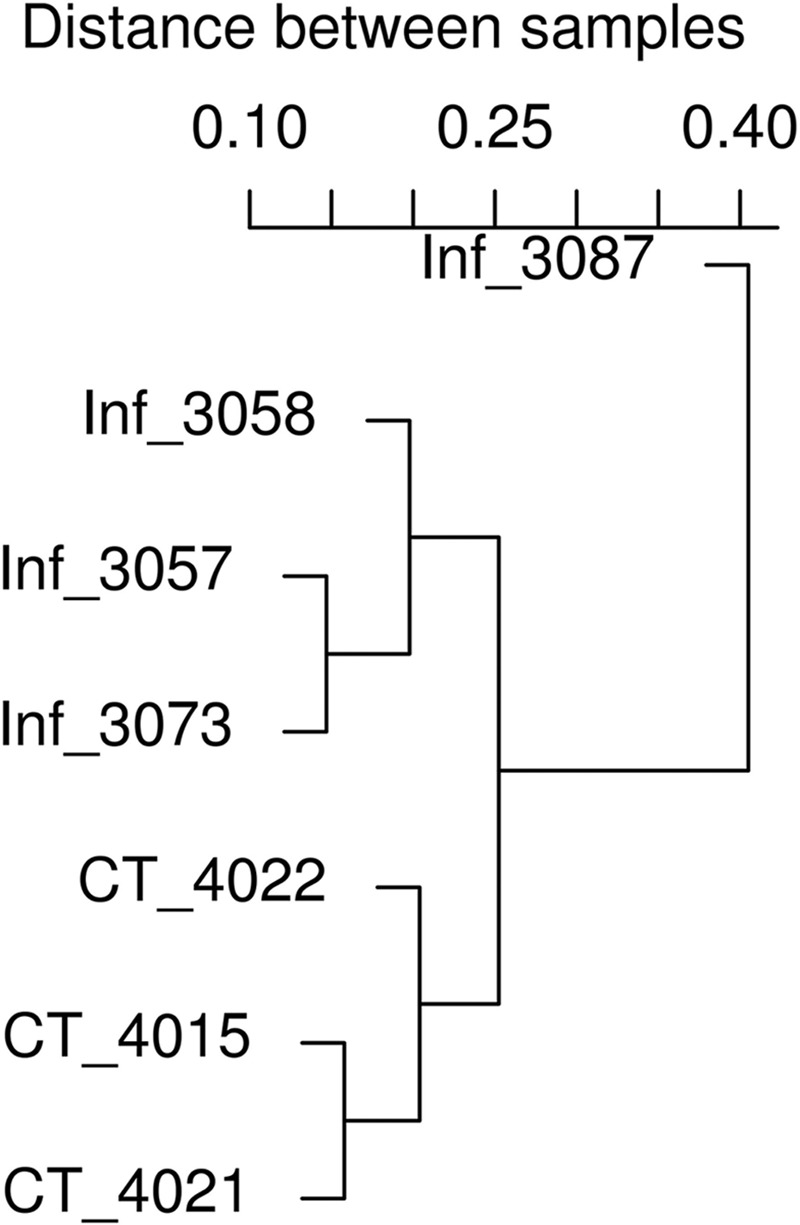
Clustering analysis based on Bray-Curtis similarity generated using the Vegan package in the R software program.
Fig 2.
Principal component analysis (PCA) based on Bray-Curtis similarity generated using the Vegan package in the R program. Of note, while control and infected samples were separate, the distinction was not clear due to the closeness of one of the infected samples to the control group.
Infection of pigs with T. suis had a significant impact on the proximal colon microbiota composition at both the phylum and genus levels (see Table S3 in the supplemental material). In particular, the relative abundances of 2 of the 15 phyla identified, Proteobacteria and Deferribacteres, were significantly altered by infection. Among 25 classes and 64 families positively assigned by RDP Classifier, the abundances of two classes and seven families were significantly impacted by infection (P < 0.05). As Table 1 shows, 17 genera (∼13% of all genera) identified were also significantly affected (P < 0.05). These genera included important taxa, such as Oscillibacter, Succinivibrio, Sporobacter, Spirochaeta, Paraprevotella, and Mitsuokella. The relative abundance of Oscillibacter, the second-most-abundant genus in the colon microbiota of control pigs, decreased from 7.8% (control) to 2.8% (infected). Similarly, the relative abundance of Succinivibrio decreased from 3.6% in control pigs to 0.4% in infected pigs. An 86-fold expansion in the relative abundance of Mucispirillum to 0.09% in infected pigs accounts for all observed changes in the phylum Deferribacteres. On the other hand, the relative abundance of the genus Paraprevotella increased from 0.5% in control to 3.0% as a result of infection. The relative abundance of Desulfovibrio followed a similar trend. While T. suis infection resulted in a significant alteration in the microbiota composition, it did not seem to change the biodiversity as judged by similar richness (ACE and Chao1) and diversity (Shannon-Weaver) indices at the genus level in samples from both control and infected pigs (data not shown). Community dissimilarity analysis based on the Bray-Curtis index suggested that control samples displayed a high degree of similarity of 0.92 and 0.81 at the phylum and genus levels, respectively (1.0 = identical), whereas within-group similarity for infected pigs was lower at 0.84 and 0.72 for the phylum and genus levels, respectively (Table 2). Of note, the control and infected microbiota shared the least similarity (0.70) at the genus level. Intuitively, the matrix revealed that the porcine colon microbial communities became altered as a result of infection.
Table 1.
Genera significantly impacted by Trichuris suis infection in porcine colon microbiotaa
| Genus | % abundance |
P value | |
|---|---|---|---|
| Control | Infected | ||
| Asteroleplasma | 0.279 ± 0.155 | 0.060 ± 0.042 | 0.0391 |
| Blautia | 0.425 ± 0.253 | 0.103 ± 0.032 | 0.0483 |
| Coprobacillus | 0.010 ± 0.006 | 0.000 ± 0.000 | 0.0171 |
| Desulfovibrio | 0.005 ± 0.009 | 0.315 ± 0.157 | 0.0207 |
| Dorea | 0.302 ± 0.178 | 0.027 ± 0.019 | 0.0250 |
| Eubacterium | 0.108 ± 0.036 | 0.031 ± 0.006 | 0.0075 |
| Mucispirillum | 0.001 ± 0.002 | 0.086 ± 0.036 | 0.0104 |
| Oribacterium | 0.298 ± 0.117 | 0.135 ± 0.037 | 0.0438 |
| Oscillibacter | 7.766 ± 2.370 | 2.778 ± 1.443 | 0.0174 |
| Paraprevotella | 0.473 ± 0.326 | 3.026 ± 1.379 | 0.0278 |
| Peptococcus | 0.015 ± 0.007 | 0.002 ± 0.003 | 0.0217 |
| Ruminococcus | 0.405 ± 0.058 | 0.176 ± 0.135 | 0.0423 |
| Schwartzia | 0.037 ± 0.014 | 0.010 ± 0.010 | 0.0299 |
| Selenomonas | 0.116 ± 0.051 | 0.026 ± 0.039 | 0.0448 |
| Spirochaeta | 0.535 ± 0.020 | 0.088 ± 0.079 | 0.0002 |
| Sporobacter | 0.664 ± 0.080 | 0.244 ± 0.187 | 0.0158 |
| Succinivibrio | 3.645 ± 1.534 | 0.358 ± 0.288 | 0.0076 |
Each set of numbers denotes the relative abundance in percentage (mean ± standard deviation; n = 4 for infected, and n = 3 for control). P values were calculated using a modified t test.
Table 2.
Bray-Curtis similarity matrix calculated using the Vegan package in the R program based on square-root-transformed abundance at a genus level
| Site | Similarity |
||||||
|---|---|---|---|---|---|---|---|
| CT_#4015 | CT_#4021 | CT_#4022 | Inf_#3057 | Inf_#3058 | Inf_#3073 | Inf_#3087 | |
| CT_#4015 | 1.000 | 0.842 | 0.785 | 0.771 | 0.745 | 0.724 | 0.550 |
| CT_#4021 | 0.842 | 1.000 | 0.807 | 0.776 | 0.751 | 0.719 | 0.550 |
| CT_#4022 | 0.785 | 0.807 | 1.000 | 0.753 | 0.778 | 0.711 | 0.575 |
| Inf_#3057 | 0.771 | 0.776 | 0.753 | 1.000 | 0.801 | 0.853 | 0.615 |
| Inf_#3058 | 0.745 | 0.751 | 0.778 | 0.801 | 1.000 | 0.804 | 0.621 |
| Inf_#3073 | 0.724 | 0.719 | 0.711 | 0.853 | 0.804 | 1.000 | 0.659 |
| Inf_#3087 | 0.550 | 0.550 | 0.575 | 0.615 | 0.621 | 0.659 | 1.000 |
The porcine proximal colon microbiota composition was also analyzed using WGS sequences. Shotgun sequences were evaluated using the MEGAN program (17), which facilitates the taxonomic binning of sequences of both prokaryotic and eukaryotic origins. This differs from the 16S rRNA gene approach, which assigns only sequences of bacterial and archaeal origins. While the order of the four most abundant phyla identified by MEGAN was the same as that identified by the 16S approach, some differences were notable. WGS sequences were assigned to a total of 45 phyla (including 29 prokaryotic and eukaryotic phyla) by MEGAN, while only 15 prokaryotic phyla were identified from the 16S sequences by RDP Classifier. At the genus level, the differences were even greater: MEGAN identified 723 genera, compared to the 134 identified by RDP. Among the 723 genera assigned by MEGAN, the relative abundances of 56 genera were significantly different between samples from the control and infected pigs (∼8%) and agreed with the 16S results. The 16S sequences assigned by RDP to archaea were virtually zero. However, 0.21% (control) and 0.34% (infected) of the WGS-generated sequences were positively assigned to the Archaea (the phylum Euryarchaeota) by MEGAN. The difference in the number of WGS sequences assigned to archaea between the two groups was significant, suggesting that the porcine proximal colon microbial community may harbor many unknown archaea that have yet to be documented in the existing 16S databases, such as RDP. The numbers of sequences assigned to Heliobacter by MEGAN were much higher in the infected pigs (P < 0.05), in agreement with the finding from the 16S sequences. Interestingly, MEGAN was able to assign WGS sequences to several genera that were unique to one of the two groups. For example, the sequences assigned to the genus Calditerrivibrio were observed in each of the three control pigs but were absent from infected pigs. Infection with T. suis resulted in the appearance of and possible colonization by more bacteria and viruses in infected pigs. In support of this argument, the sequences of five genera, Haloterrigena, Kineococcus, Nitrosomonas, Sagittula, and Phi29-like viruses, were observed in infected pigs only. Our finding suggest that both methods for taxonomic classification (RDP based on 16S sequences and MEGAN based on WGS sequences) may be complementary and that the true extent of microbial diversity in the porcine proximal colon microbiota may lie within the synthesis of markers identified by both methods.
Functional changes and shifts in metabolic potentials in the proximal colon microbiota induced by infection.
WGS sequence reads were first de novo assembled using the Newbler software program, and the open reading frames (ORF) were predicted using FragGeneScan from contigs and singletons greater than 300 bp in length. These ORFs were annotated against the COG, Pfam, and KEGG (Kyoto Encyclopedia of Genes and Genomes) databases in order to characterize functional changes and shifts in metabolic potentials of the proximal colon microbiota induced by T. suis infection. Approximately 3,989 COGs (roughly 77% of COGs in the database) were identified in the porcine proximal colon microbiota. These COGs were organized into 25 functional categories. Infection had a significant effect on the proximal colon microbial community function, since the number of ORFs assigned to 7 of the 25 COG functional categories was significantly different between samples from control and infected pigs (see Table S4 in the supplemental material). The affected categories included some of the most abundant assignments, such as general function predicted only (% assigned = 10.6%), replication, recombination and repair (8.6%), carbohydrate transport and metabolism (8.5%), and cell wall/membrane/envelope biogenesis (7.8%). In addition, approximately 5,085 Pfam protein families were identified from the WGS sequences obtained in the proximal colon microbial communities. ABC transporter (PF00005) was most abundant protein family in the porcine proximal colon, as in many other microbial communities (38), and the only protein family with >1% of ORF hits assigned. TonB-dependent receptor (PF00593), ATPase family associated with various cellular activities (PF00004), histidine kinase-, DNA gyrase B-, and HSP90-like ATPase (PF02518), response regulator receiver domain (PF00072), and glycosyl transferase family 2 (PF00535) were among the most abundant protein families in the proximal colon microbial community of control pigs. Approximately 272 Pfam protein families (roughly 5.35% of all Pfam families identified) were likely to be affected by infection, since the number of ORFs assigned to these Pfam families was significantly different between samples from control and infected pigs (P < 0.05). Gene ontology terms (GO) were identified in order to gain further insight into the general function of these Pfam families. Among 1,346 GO identified, infection resulted in significant changes in 112 GO (or ∼8.3% of all GO). Among the 112 GO that were significantly affected by infection, the 10 most abundant GO in terms of the numbers of ORFs assigned in the proximal colon microbiota are listed in Table 3. In addition to catalytic activity (GO:0003824), which accounted for 2.9% of ORFs assigned, carbohydrate metabolic process (GO:0005975), hydrolase activity, hydrolyzing O-glycosyl compounds (GO:0004553), and DNA repair (GO:0006281) were significantly altered by infection. Infection apparently repressed the carbohydrate metabolic potential of the proximal colon microbial community, since the number of ORF hits assigned to this metabolic process displayed a significant decrease in infected pigs, which is in agreement with the result of the COG functional analysis. KEGG pathway assignment provided an additional piece of evidence that infection may have a repressive impact on the metabolic potential of carbohydrates. Among the 291 KEGG pathways assigned, 75 displayed a significant difference in the number of hits assigned between samples from control and infected pigs. Carbohydrate and amino acid metabolisms were the two most important functional classes that were altered by infection. As shown in Table 4, starch and sucrose metabolism (KEGG 00500), fructose and mannose metabolism (00051), pyruvate metabolism (00620), and galactose metabolism (00052) were among the most repressed. In addition, lysine biosynthesis (00300) in the proximal colon microbiota was significantly decreased by infection (Table 4).
Table 3.
Gene ontology impacted by Trichuris suis infection in pigs
| Accession no. | Gene ontology description | P valuea | % hitsb |
|
|---|---|---|---|---|
| Control | Infected | |||
| GO:0003824 | Catalytic activity | 0.0030 | 2.87 ± 0.03 | 2.74 ± 0.01 |
| GO:0005975 | Carbohydrate metabolic process | 0.0088 | 2.31 ± 0.08 | 2.06 ± 0.03 |
| GO:0004553 | Hydrolase activity, hydrolyzing O-glycosyl compounds | 0.0299 | 1.09 ± 0.07 | 0.95 ± 0.02 |
| GO:0030170 | Pyridoxal phosphate binding | 0.0353 | 0.61 ± 0.01 | 0.59 ± 0.01 |
| GO:0006281 | DNA repair | 0.0093 | 0.50 ± 0.01 | 0.53 ± 0.00 |
| GO:0008170 | N-methyltransferase activity | 0.0233 | 0.19 ± 0.01 | 0.22 ± 0.01 |
| GO:0008415 | Acyltransferase activity | 0.0275 | 0.18 ± 0.00 | 0.16 ± 0.01 |
| GO:0000105 | Histidine biosynthetic process | 0.0215 | 0.17 ± 0.01 | 0.14 ± 0.00 |
| GO:0016868 | Intramolecular transferase; activity, phosphotransferase | 0.0241 | 0.16 ± 0.01 | 0.15 ± 0.00 |
| GO:0016874 | Ligase activity | 0.0204 | 0.12 ± 0.00 | 0.13 ± 0.01 |
P values were calculated based on a modified t test.
The number denotes the percentage of Pfam-annotated ORF (open reading frame) hits assigned to a given gene ontology (GO) category (mean ± SEM; n = 3).
Table 4.
Trichuris suis infection in pigs had profound impact on pathways related to carbohydrate and amino acid metabolisms in the porcine colon microbial community
| Accession no. | KEGG pathway description (function class)a | P valueb | q valueb | Ratio of ORF assignmentc |
|
|---|---|---|---|---|---|
| Control | Infected | ||||
| 00520 | Amino sugar and nucleotide sugar metabolism (C) | 0.0004 | 0.0349 | 0.0148 ± 8.99E−05 | 0.0136 ± 1.59E-04 |
| 00500 | Starch and sucrose metabolism (C) | 0.0071 | 0.0624 | 0.0125 ± 2.88E−04 | 0.0109 ± 2.29E-04 |
| 00330 | Arginine and proline metabolism (AA) | 0.0185 | 0.0624 | 0.0119 ± 2.03E−04 | 0.0109 ± 2.15E−04 |
| 00051 | Fructose and mannose metabolism (C) | 0.0085 | 0.0624 | 0.0109 ± 2.20E−04 | 0.0099 ± 1.25E−04 |
| 00620 | Pyruvate metabolism (C) | 0.0466 | 0.0624 | 0.0098 ± 1.87E−04 | 0.0091 ± 1.63E−04 |
| 00270 | Cysteine and methionine metabolism (AA) | 0.0353 | 0.0624 | 0.0095 ± 2.66E−04 | 0.0086 ± 1.52E−04 |
| 00260 | Glycine, serine, and threonine metabolism (AA) | 0.0077 | 0.0624 | 0.0073 ± 1.05E−04 | 0.0066 ± 1.14E−04 |
| 00052 | Galactose metabolism (C) | 0.0238 | 0.0624 | 0.0072 ± 2.78E−04 | 0.0062 ± 1.97E−04 |
| 00350 | Tyrosine metabolism (AA) | 0.0110 | 0.0624 | 0.0066 ± 1.11E−04 | 0.0062 ± 5.95E−05 |
| 00300 | Lysine biosynthesis (AA) | 0.0350 | 0.0624 | 0.0061 ± 1.43E−04 | 0.0056 ± 1.09E−04 |
| 00360 | Phenylalanine metabolism (AA) | 0.0023 | 0.0491 | 0.0055 ± 4.72E−05 | 0.0051 ± 4.96E−05 |
| 00040 | Pentose and glucuronate interconversions (C) | 0.0123 | 0.0624 | 0.0051 ± 1.73E−04 | 0.0045 ± 4.24E−05 |
| 00030 | Pentose phosphate pathway (C) | 0.0356 | 0.0624 | 0.0046 ± 1.46E−04 | 0.0042 ± 2.45E−05 |
| 00053 | Ascorbate and aldarate metabolism (C) | 0.0098 | 0.0624 | 0.0011 ± 2.78E−05 | 0.0010 ± 2.19E−05 |
C, carbohydrate metabolism; AA, amino acid metabolism.
P and q values were calculated based on a modified t test.
The number denotes the ratio of ORF hits assigned to a given KEGG pathway divided by the total number of ORFs assigned to any pathways (mean ± SE; n = 3).
A global volatile organic compound (VOC) metabolomic analysis of luminal contents from the proximal colon was performed with analytes matched to the NIST08 mass spectral library at a 90% confidence level. A total of 199 metabolites were identified for control pigs and 197 total metabolites were identified for infected pigs, and 85% (170) of these metabolites are common to the proximal colon contents of both groups. Twenty-nine metabolites were unique to the control cohort, and 27 metabolites were unique to the infected cohort (see Table S5 in the supplemental material). Most notably, the proximal colon contents from all four infected pigs contained oleic acid and 2-butenedioic acid, dibutyl esters which were not detected in the control pigs. Interestingly, oleic acid is well known to have antibacterial activity, which may contribute to the alteration of the microbiota composition. Oleic acid in the proximal colon may represent an alteration in fatty acid absorption due to infection. In contrast, all three control pigs contained 2-methyl-2-butenal, 2,6-dimethyl-4-heptanone, and 2-octene, which were not detected in the luminal contents obtained from any of the infected pigs. A KEGG search indicted that the first two of these molecules are reflective of carbohydrate metabolism and lysine and tryptophan metabolism, respectively.
DISCUSSION
Trichuris suis is a widespread intestinal parasitic nematode in pigs. However, studies of this helminth species have been motivated by its therapeutic potential in modulating human diseases (39). While several clinical trials have exploited the immune-suppressive and anti-inflammation properties of T. suis, its interactions with host and associated microbiota have received little attention. Understanding three-way interactions between the host, its microbiota, and the parasite will provide novel insights into physiological consequences of parasitic infection, which should facilitate the formulation of an optimal strategy for T. suis therapy, including reducing its possible negative impact on human pathophysiology (19) and the control of infection in swine and humans.
In this study, we characterized the porcine proximal colon microbiota in response to T. suis infection using both 16S rRNA gene-based and WGS sequencing. Due to physical and fiscal constraints, our sample size was small in the present study, and we increased the sample size in a follow-up study to examine other time points after inoculation with infective T. suis eggs (51). Nevertheless, our results demonstrated that T. suis infection in pigs resulted in a significant alteration in the microbiota composition of the proximal colon. The time of infection chosen for evaluation was 21 days after inoculation, when developing larvae emerge from the mucosal surface coincident with catarrhal enteritis and edema (4). The relative abundance of ∼27% of the phyla and 10% of the genera identified was significantly altered by infection. Infection substantially elevated the population of Mucispirillum bacteria, which colonize the intestinal mucus, in infected pigs relative to controls. We hypothesize that mucosal disruption induced by invading T. suis, which is purported to feed on mucopolysaccharides and mucoproteins (18), altered the composition of the proximal colon microbial ecosystem in favor of those organisms, such as Mucispirillum, which have a predilection for mucosal surfaces. Notably, the genera harboring carbohydrate- and/or fiber-utilizing bacteria were significantly affected by T. suis infection. Ruminococcus, arguably the most predominant cellulolytic bacterium isolated from the gastrointestinal tract of animals, including the rumen, was significantly reduced by infection at both days 21 and 53 postinfection (51). Planned experiments will examine local physiological changes in barrier function, secretion, and absorption associated with infection in the proximal colon and if noninfectious purgative agents can mimic parasite-induced modulation of the microbiome.
Fermentability and physical properties of dietary carbohydrates have a significant effect on the establishment, intestinal distribution, and fecundity of intestinal worms in pigs (33). Readily fermentable carbohydrates in the diet do not seem to affect the establishment of T. suis in naive pigs but result in reduced (fecundity) fecal egg counts, early expulsion, and reduced growth of the worms in the intestine (42). Both infection and dietary carbohydrates significantly influence intestinal morphology and mucin biosynthesis in the porcine large intestine (43). Metabolism of these dietary carbohydrates is controlled by intestinal microorganisms and takes place mainly in the hindgut of monogastric animals, such as pigs and humans. The end product of the fermentation is short-chain fatty acids (SCFA), such as butyrate and acetate, which are absorbed by the intestinal epithelium, enter into circulation as energy sources, and improve local epithelial cell function. Total SCFA concentrations are significantly greater in the large intestine of pigs fed fermentable carbohydrates, while the luminal pH value is reduced (42), which in turn affects the intestinal microbiota composition. Our study found that the number of ORFs identified for the carbohydrate metabolic process (GO:0005975; Table 3) was significantly reduced by T. suis infection from that for control pigs (2.06% versus 2.31%). As many as eight KEGG pathways related to carbohydrate metabolism were significantly repressed by infection (Table 4), including starch and sucrose metabolism and fructose and mannose metabolism. The 16S rRNA gene-based taxonomic classification indicated that several bacterial genera whose major activities related to carbohydrate metabolism were reduced by infection. Together, these results suggested that T. suis infection may have impaired the ability of the proximal colon microbiota to utilize carbohydrates. Dietary supplements with readily fermentable carbohydrates could help restore or even enhance the function of the colon microbiota in T. suis-infected pigs, especially in metabolizing carbohydrates to generate energy sources. Furthermore, the restored function of the microbiota may lead to the repair of colonic epithelium damaged by infection. This in turn could enhance protective immunity and reduce inflammation through restored capacity of mucin biosynthesis (45) and subsequent worm expulsion through epithelial cell turnover (52). These findings support the use of diets rich in highly fermentable carbohydrates as a nematode control strategy in pigs (33). Moreover, it is reasonable to consider concurrent administration of T. suis eggs with selected probiotic bacterial populations that are reduced in the colon microbiota by parasite infection to ameliorate any negative influence of the worm on human and porcine physiology (7). In another study, we demonstrated that feeding pigs probiotic Bifidobacterium lactis (Bb12) during an infection with Ascaris suum prevented the reduced glucose absorption in the small intestine that normally facilitates worm expulsion (G. Solano-Aguilar, T. Shea-Donohue, K. Madden, H. Dawson, Y. Jones, M. Restrepo, N. Schoene, and J. Urban, personal communication).
Our findings indicate that the biosynthesis capacity of certain essential amino acids of the proximal colon microbiota may be reduced by T. suis infection, since the number of ORFs assigned to the lysine biosynthesis pathway (KEGG no. 00300) was significantly decreased in infected pigs (Table 4), as was the number of ORFs assigned to the cysteine and methionine metabolism pathway (KEGG 00270). The percentage of ORFs predicted from the WGS sequences assigned to this pathway was 0.86% in the proximal colon microbiota of infected pigs, compared to 0.95% in control pigs. The difference was small but nevertheless significant. A published observation suggests that intestinal infestation with parasites increased the dietary requirement for lysine, especially in chronically undernourished men in India (24). One of the hallmarks of host immune responses to intestinal parasitic infection in animals, including ruminants, is increased hepatic synthesis and gastrointestinal losses of protein high in sulfur-containing amino acids, such as cysteine and methionine (46). Animals demand extra amounts of sulfur-containing amino acids to meet their particular nutrient partition and mount an effective cell-mediated immune response during periods of pathological and physiologic stress, such as parasite infestation. A compromised biosynthesis capacity for essential amino acids in the gut microbiota of infected pigs, as well as an increased demand for such amino acids, suggested that nutritional status should be prominently considered in strategies to control the modulating activity of parasites.
Changes in fecal VOCs have been associated with gastrointestinal diseases (10, 12). A GCMS-based VOC analysis of feces obtained from the proximal colon of the control and infected pig cohorts identified (using a strict >90% identity to the NIST mass spectral library) 170 core metabolites common to both cohorts, while 29 and 27 metabolites were uniquely detected in the control and infected cohorts, respectively (see Table S5 in the supplemental material). Most notably, 2-methyl-2-butenal and 2,6-dimethyl-4-heptanone were uniquely detected in the luminal contents of all control uninfected pigs. KEGG links 2-methyl-2-butenal as a derivative of crotonaldehyde coming from crotonoyl-coenzyme A (CoA), part of the butanoate metabolism of carbohydrates and involved in the production of the amino acids lysine and tryptophan. The absence of 2-methyl-2-butenal from T. suis-infected pigs could reflect reduced carbohydrate metabolism locally. In addition, 2,6-dimethyl-4-heptanone is a product of butyric acid, generated by bacterial anaerobic fermentation in control pigs, but was absent in all T. suis-infected pigs. Interestingly, 2-octene was detected in the proximal colon contents of all control pigs. The physiological significance is not clear; however, 2-octene has also been detected in a longitudinal study of feces from human subjects, although it is largely absent from patients with ulcerative colitis and others infected with Campylobacter jejuni (12). The detection of 2-butenedioic acid, dibutyl ester, commonly found in the manufacture of plastics, was detected in all of the T. suis-infected pigs, although the relevance is not known.
The detection of oleic acid exclusively in the proximal luminal contents of all T. suis-infected pigs but not the control pigs was also notable because normally its absorption is stable throughout the intestine of pigs (41). A relative increase in oleic acid in the proximal colon of infected pigs could contribute to localized mucosal inflammation (22, 23) and change the microbiome due to its antibacterial activity (9, 16). A further consideration would be to determine if T. suis-induced mucosal damage in the proximal colon facilitated systemic exposure to local lipopolysaccaride, which has been shown recently to alter the oxidation of oleic acid in piglets and contribute to malabsorption by enterocytes (15).
Parasitic infection in pigs was accompanied by a concomitant change in the production of a class of molecules involved in bacterial invasion of host epithelial cells (1), including invasins and internalins. Internalins are bacterial leucine-rich repeat-containing surface proteins involved in mediating recognition and invasion of epithelial cells or acting as a growth factor (invasin) in infection processes (8). In addition, infection may indirectly modulate host immune responses via the alteration of the colon microbiota. Pathways such as T-cell receptor signaling and Toll-like receptor signaling, as well as the intestinal immune network for IgA production (KEGG no. 04672), and localized host expression for Th2-related genes that are worm burden dependent (51) were significantly affected in infected pigs.
One limit on interpretation of the data is the relatively small sample size of parasite-naive and T. suis-infected pigs that was evaluated. However, the depth of the analysis points to a remarkable similarity in genera representing a core microbiota between control and infected pigs, distinguished by differences in some key genera, such as Mucispirillum, Succinivibrio, and Ruminococcus and supported by metabolic and functional data. The inherent response of the host to maintain homeostasis in the face of a dose of T. suis eggs inducing a largely local and subclinical infection would predictably exacerbate and more profoundly alter the microbiome and related metabolic network activated by an infection expressing overt disease. While detailed mechanisms require further definition, our findings suggested that a holistic understanding of three-way interactions among the host, its microbiota, and the parasite will undoubtedly optimize strategies for helminth control in pigs and humans and for more effective therapeutic use of helminths in humans to modulate inflammation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dante Zarlenga for his constructive comments.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
Footnotes
Published ahead of print 9 April 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Abner SR, et al. 2002. Response of intestinal epithelial cells to Trichuris suis excretory-secretory products and the influence on Campylobacter jejuni invasion under in vitro conditions. J. Parasitol. 88:738–745 [DOI] [PubMed] [Google Scholar]
- 2. Andersson AF, et al. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 3:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bager P, et al. 2010. Trichuris suis ova therapy for allergic rhinitis: a randomized, double-blind, placebo-controlled clinical trial. J. Allergy Clin. Immunol. 125:123–130 [DOI] [PubMed] [Google Scholar]
- 4. Batte EG, Moncol DJ. 1972. Whipworms and dysentery in feeder pigs. J. Am. Vet. Med. Assoc. 161:1226–1228 [PubMed] [Google Scholar]
- 5. Batte EG, McLamb RD, Muse KE, Tally SD, Vestal TJ. 1977. Pathophysiology of swine trichuriasis. Am. J. Vet. Res. 38:1075–1079 [PubMed] [Google Scholar]
- 6. Beer RJ. 1973. Studies on the biology of the life-cycle of Trichuris suis Schrank, 1788. Parasitology 67:253–262 [DOI] [PubMed] [Google Scholar]
- 7. Bercik P, et al. 2010. Chronic gastrointestinal inflammation induces anxiety-like behavior and alters central nervous system biochemistry in mice. Gastroenterology 139:2102–2112 [DOI] [PubMed] [Google Scholar]
- 8. Bierne H, Sabet C, Personnic N, Cossart P. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes. Infect. 9:1156–1166 [DOI] [PubMed] [Google Scholar]
- 9. Chen CH, et al. 2011. An innate bactericidal oleic acid effective against skin infection of methicillin-resistant Staphylococcus aureus: a therapy concordant with evolutionary medicine. J. Microbiol. Biotechnol. 21:391–399 [PubMed] [Google Scholar]
- 10. Dixon E, et al. 2011. Solid-phase microextraction and the human fecal VOC metabolome. PLoS One 6:e18471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eijck IA, Borgsteede FH. 2005. A survey of gastrointestinal pig parasites on free-range, organic and conventional pig farms in The Netherlands. Vet. Res. Commun. 29:407–414 [DOI] [PubMed] [Google Scholar]
- 12. Garner CE, et al. 2007. Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. FASEB J. 21:1675–1688 [DOI] [PubMed] [Google Scholar]
- 13. Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME. J. 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hill DE, Gamble HR, Rhoads ML, Fetterer RH, Urban JF., Jr 1993. Trichuris suis: a zinc metalloprotease from culture fluids of adult parasites. Exp. Parasitol. 77:170–178 [DOI] [PubMed] [Google Scholar]
- 15. Hou Y, et al. 2011. Effects of α-ketoglutarate on energy status in the intestinal mucosa of weaned piglets chronically challenged with lipopolysaccharide. Br. J. Nutr. 106:357–363 [DOI] [PubMed] [Google Scholar]
- 16. Huang CB, George B, Ebersole JL. 2010. Antimicrobial activity of n-6, n-7 and n-9 fatty acids and their esters for oral microorganisms. Arch. Oral Biol. 55:555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res. 17:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jenkins T. 1970. A morphological and histochemical study of Trichuris suis (Schrank, 1788) with special reference to the host-parasite relationship. Parasitology 61:357–374 [DOI] [PubMed] [Google Scholar]
- 19. Kradin RL, Badizadegan K, Auluck P, Korzenik J, Lauwers GY. 2006. Iatrogenic Trichuris suis infection in a patient with Crohn disease. Arch. Pathol. Lab Med. 130:718–720 [DOI] [PubMed] [Google Scholar]
- 20. Kringel H, Iburg T, Dawson H, Aasted B, Roepstorff A. 2006. A time course study of immunological responses in Trichuris suis infected pigs demonstrates induction of a local type 2 response associated with worm burden. Int. J. Parasitol. 36:915–924 [DOI] [PubMed] [Google Scholar]
- 21. Kringel H, Roepstorff A. 2006. Trichuris suis population dynamics following a primary experimental infection. Vet. Parasitol. 139:132–139 [DOI] [PubMed] [Google Scholar]
- 22. Knoch B, et al. 2009. Genome-wide analysis of dietary eicosapentaenoic acid- and oleic acid-induced modulation of colon inflammation in interleukin-10 gene-deficient mice. J. Nutrigenet. Nutrigenomics 2:9–28 [DOI] [PubMed] [Google Scholar]
- 23. Knoch B, et al. 2010. Dietary arachidonic acid-mediated effects on colon inflammation using transcriptome analysis. Mol. Nutr. Food Res. 54(Suppl 1):S62–S74 [DOI] [PubMed] [Google Scholar]
- 24. Kurpad AV, et al. 2003. Intestinal parasites increase the dietary lysine requirement in chronically undernourished Indian men. Am. J. Clin. Nutr. 78:1145–1151 [DOI] [PubMed] [Google Scholar]
- 25. Lamendella R, Santo Domingo JW, Ghosh S, Martinson J, Oerther DB. 2011. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 11:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li RW, Connor EE, Li C, Baldwin RL, Spark ME. 2012. Characterization of the rumen microbiota in pre-ruminant calves using metagenomic tools. Environ. Microbiol. 14:129–139 [DOI] [PubMed] [Google Scholar]
- 27. Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659 [DOI] [PubMed] [Google Scholar]
- 28. Mansfield LS, Urban JF., Jr 1996. The pathogenesis of necrotic proliferative colitis in swine is linked to whipworm induced suppression of mucosal immunity to resident bacteria. Vet. Immunol. Immunopathol. 50:1–17 [DOI] [PubMed] [Google Scholar]
- 29. Matsubayashi M, et al. 2009. Coprological survey of parasitic infections in pigs and cattle in slaughterhouse in Osaka, Japan. J. Vet. Med. Sci. 71:1079–1083 [DOI] [PubMed] [Google Scholar]
- 30. Nejsum P, et al. 2009. Population dynamics of Trichuris suis in trickle-infected pigs. Parasitology 136:691–697 [DOI] [PubMed] [Google Scholar]
- 31. Nganga CJ, Karanja DN, Mutune MN. 2008. The prevalence of gastrointestinal helminth infections in pigs in Kenya. Trop. Anim. Health Prod. 40:331–334 [DOI] [PubMed] [Google Scholar]
- 32. Nissen S, et al. 2011. Prevalence of gastrointestinal nematodes in growing pigs in Kabale District in Uganda. Trop. Anim. Health Prod. 43:567–572 [DOI] [PubMed] [Google Scholar]
- 33. Petkevicius S, et al. 2007. The effect of inulin on new and on patent infections of Trichuris suis in growing pigs. Parasitology 134:121–127 [DOI] [PubMed] [Google Scholar]
- 34. Poroyko V, et al. 2010. Gut microbial gene expression in mother-fed and formula-fed piglets. PLoS One 5:e12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rho M, Tang H, Ye Y. 2010. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 38:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rutter JM, Beer RJ. 1975. Synergism between Trichuris suis and the microbial flora of the large intestine causing dysentery in pigs. Infect. Immun. 11:395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shin JL, Gardiner GW, Deitel W, Kandel G. 2004. Does whipworm increase the pathogenicity of Campylobacter jejuni? A clinical correlate of an experimental observation. Can. J. Gastroenterol. 18:175–177 [DOI] [PubMed] [Google Scholar]
- 38. Sparks ME, et al. 2011. Detection of functional shifts in the rumen microbiota in response to propionate intake in cattle, p 165–183 In Li RW. (ed), Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, New York, NY [Google Scholar]
- 39. Summers RW, et al. 2003. Trichuris suis seems to be safe and possibly effective in the treatment of inflammatory bowel disease. Am. J. Gastroenterol. 98:2034–2041 [DOI] [PubMed] [Google Scholar]
- 40. Summers RW, Elliott DE, Urban JF, Jr, Thompson RA, Weinstock JV. 2005. Trichuris suis therapy for active ulcerative colitis: a randomized controlled trial. Gastroenterology 128:825–832 [DOI] [PubMed] [Google Scholar]
- 41. Swiss LD, Bayley HS. 1976. Influence of the degree of hydrolysis of beef tallow on its absorption in the young pig. Can. J. Physiol. Pharmacol. 54:719–727 [DOI] [PubMed] [Google Scholar]
- 42. Thomsen LE, Petkevicius S, Bach Knudsen KE, Roepstorff A. 2005. The influence of dietary carbohydrates on experimental infection with Trichuris suis in pigs. Parasitology 131:857–865 [DOI] [PubMed] [Google Scholar]
- 43. Thomsen LE, Knudsen KE, Hedemann MS, Roepstorff A. 2006. The effect of dietary carbohydrates and Trichuris suis infection on pig large intestine tissue structure, epithelial cell proliferation and mucin characteristics. Vet. Parasitol. 142:112–122 [DOI] [PubMed] [Google Scholar]
- 44. Urban JF, Jr, Romanowski RD, Steele NC. 1989. Influence of helminth parasite exposure and strategic application of anthelmintics on the development of immunity and growth of swine. J. Anim. Sci. 67:1668–1677 [DOI] [PubMed] [Google Scholar]
- 45. Van der Sluis M, et al. 2006. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131:117–129 [DOI] [PubMed] [Google Scholar]
- 46. Van Houtert MFJ, Sykes AR. 1996. Implications of nutrition for the ability of ruminants to withstand gastrointestinal nematode infections. Int. J. Parasitol. 26:1151–1167 [DOI] [PubMed] [Google Scholar]
- 47. Van Kruiningen HJ, West AB. 2005. Potential danger in the medical use of Trichuris suis for the treatment of inflammatory bowel disease. Inflamm. Bowel. Dis. 11:515. [DOI] [PubMed] [Google Scholar]
- 48. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weinstock JV, Elliott DE. 2009. Helminths and the IBD hygiene hypothesis. Inflamm. Bowel. Dis. 15:128–133 [DOI] [PubMed] [Google Scholar]
- 50. White JR, Nagarajan N, Pop M. 2009. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 5:e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu S, et al. Worm burden-dependent disruption of the porcine colon microbiota by Trichuris suis infection. Plos One, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zaiss DM, et al. 2006. Amphiregulin, a TH2 cytokine enhancing resistance to nematodes. Science 314:1746. [DOI] [PubMed] [Google Scholar]
- 53. Zarlenga D, Dawson H, Kringel H, Solano-Aguilar G, Urban JF. 2004. Molecular cloning of the swine IL-4 receptor alpha and IL-13 receptor 1 chains: effects of experimental Toxoplasma gondii, Ascaris suum and Trichuris suis infection on tissue mRNA levels. Vet. Immunol. Immunopathol. 101:223–234 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.