Abstract
Invasive amebiasis due to Entamoeba histolytica infection is an important cause of morbidity in developing countries. The E. histolytica genome contains two homologues to the metalloprotease leishmanolysin gene, Entamoeba histolytica MSP-1 (EhMSP-1) and EhMSP-2, while the commensal ameba Entamoeba dispar has lost EhMSP-1. In this study, we sought to characterize E. histolytica metallosurface protease 1 (EhMSP-1). Using immunoprecipitation and a model substrate, we found that EhMSP-1 was a functional metalloprotease. Confocal microscopy and flow cytometry revealed that EhMSP-1 localized to the cell surface and revealed the existence of distinct, nonclonal trophozoite populations with high and low EhMSP-1 surface abundance that became synchronized following serum starvation. Phenotypic assays were performed after silencing EhMSP-1. Adherence of EhMSP-1-deficient trophozoites to tissue culture cell monolayers was more than five times greater than that of control amebas, but surface staining of several antigens, including the galactose adherence lectin, was unchanged. EhMSP-1 silencing similarly increased adherence to both viable and apoptotic Jurkat lymphocytes. Tissue culture cell monolayer destruction was reduced by EhMSP-1 silencing, although it was blocked almost completely by inhibiting cysteine proteases. Consistent with a primary defect in regulation of amebic adherence, EhMSP-1 silencing also resulted in reduced mobility on tissue culture cell monolayers and in increased phagocytosis. In conclusion, EhMSP-1 was shown to be a surface metalloprotease involved in regulation of amebic adherence, with additional effects on cell motility, cell monolayer destruction, and phagocytosis.
INTRODUCTION
Amebiasis, which is caused by invasive Entamoeba histolytica infection, remains an important cause of morbidity and mortality globally (19). After excystation, invasive amebic infection begins with adherence of trophozoites to intestinal mucus and epithelial cells (13, 14, 26, 42). Trophozoites then degrade the mucus and epithelial layers by secreting proteases and contact-dependent cell killing, which is followed by amebic phagocytosis of killed cells and migration into the tissue (26–28, 39, 44).
Numerous proteases are encoded by the Entamoeba histolytica genome. Of these, to date, research has focused primarily on a large family of secreted and cell surface cysteine proteases, which have been firmly linked to amebic virulence (1, 10, 38, 43). As is the case for many proteases, the amebic cysteine proteases are promising drug targets, and specific cysteine protease inhibitors have been identified that prevent E. histolytica-induced tissue destruction in animal models (24). In contrast to the case with cysteine proteases, very little is known about the other types of E. histolytica proteases.
Leishmanolysin (also called gp63), the founding member of the M8 family of metalloendopeptidases, is essential for virulence of Leishmania major and is a leading candidate for vaccine development (55). M8 metalloproteases are defined by presence of a zinc-binding HEXXH catalytic site motif, a third zinc-binding His residue located further toward the C terminus, and a highly conserved Met residue C terminal to the third His (45). Leishmanolysin is a glycosylphosphatidylinositol (GPI)-anchored surface metalloprotease that is expressed abundantly on Leishmania promastigotes (55). It degrades extracellular matrix proteins during tissue invasion and prevents complement-mediated lysis of promastigotes by inactivating C3b deposited on the cell membrane (i.e., converting C3b to iC3b) and degrading C5 (9, 40). Orthologous surface metallopeptidases in both Trypanosoma brucei and Trypanosoma cruzi are also required for virulence, although their function differs from that of leishmanolysin (18, 23, 33). For example, Trypansoma brucei leishmanolysin orthologues function during antigenic variation by removing the variant surface glycoprotein that is being replaced from the surface of bloodstream trypanosomes (33).
Gene sequences encoding orthologous M8 metalloproteases are also present in nonpathogenic species, such as Crithidia fasciculata, suggesting that the evolutionary pressure to maintain these proteins derives from functions that are not specific for virulence (29). Surface metalloproteases are critical for migration of cancer cells through the extracellular matrix, although whether they contribute to this process by degrading extracellular matrix proteins, by cleaving adhesins on migrating cells, or both remains unclear (15, 48). Deletion of the Drosophila melanogaster M8 metalloprotease invadolysin gene results in defects in cell migration during embryogenesis (37). Invadolysin localizes to the leading edge of migrating cells, suggesting that it plays an active role in cell migration, but this remains to be directly tested.
Two leishmanolysin homologues are encoded in the E. histolytica genome, but only one copy of the gene is present in the closely related commensal organism Entamoeba dispar. We propose the name E. histolytica metallosurface protease 1 (EhMSP-1) for the E. histolytica-specific protein and EhMSP-2 for the common homologue (NCBI GeneID numbers 3409717 and 3406949, respectively). EhMSP-2 was one of 87 genes with significant differential expression (≥2-fold) in a recent microarray analysis comparing gene expression in virulent and avirulent E. histolytica trophozoite strains derived from the same genetic background (mRNA levels were more than 20-fold higher in the avirulent strain) (5). On the basis of these data and the fundamental contributions of leishmanolysin and its orthologues to Leishmania and Trypanosoma virulence, we decided to characterize the E. histolytica surface metalloproteases, beginning with the pathogen-specific family member EhMSP-1. Here, we show that EhMSP-1 is an active metalloprotease whose presence on the cell surface is regulated during cell culture. Trophozoites made deficient in EhMSP-1 by gene expression knockdown were characterized in vitro. Our findings show that EhMSP-1 functions in regulation of adherence and also affects motility, tissue culture monolayer destruction, and phagocytosis. Surprisingly, it plays no role in in vitro resistance to complement.
MATERIALS AND METHODS
Antibodies.
The sources of commercially available antibodies are indicated where each use is described. Non commercially available antibodies were as follows. The anti-Gal-GalNAc lectin mouse monoclonal antibody (MAb) (clone 3D12) was a gift from William A. Petri (University of Virginia, Charlottesville), the antiproteophosphoglycan mouse monoclonal antibody (clone EH5) was a gift from Michael Duchene (University of Vienna, Vienna, Austria), and the anti-inhibitor of cysteine protease 1 (anti-ICP-1) rabbit polyclonal IgG antibody was a gift from Tomoyoshi Nozaki (National Institute of Infectious Diseases, Tokyo, Japan). The anti-serine-rich E. histolytica protein (anti-SREHP) (clone 10D11) and anti-E. histolytica surface antigen (clone 5E12) mouse monoclonal antibodies were previously described by us (51). The anti-EhMSP-1 rabbit polyclonal IgG was raised against purified recombinant EhMSP-1 protein by a commercial vendor (Antibodies Incorporated, Davis, CA) and affinity purified, and the anti-EhMSP-1 monoclonal single-chain Fv (scFv) recombinant antibody was selected by phage display using the Tomlinson I and J libraries (provided by Greg Winter, MRC Centre for Protein Engineering, Cambridge, United Kingdom) by panning against recombinant EhMSP-1 and was purified using protein A-agarose (25).
Cell cultures.
Entamoeba histolytica HM-1:IMSS or G3 strain trophozoites were cultivated axenically in TYI-S-33 medium (17). Entamoeba dispar strain SAW760 was grown in TYI-S-33 medium in xenic culture. Where indicated, E. histolytica HM-1:IMSS trophozoite clones were selected by dilution and growth in 0.55% agar with TYI-S-33, followed by recovery and amplification in broth medium as described previously (41). Also where indicated, amebas were grown in serum-free TYI-S-33 medium for 21 h prior to replacement of complete culture medium for the specified time and use in experiments. Trophozoites were harvested for all experiments during log-phase growth by chilling on ice for 20 min, centrifugation (400 × g, 10 min, 4°C), and suspension in phosphate-buffered saline (PBS) or M199s (M199 medium supplemented with 25 mM HEPES [pH 6.8], 5.7 mM cysteine, and 0.5% bovine serum albumin [BSA]).
Jurkat T lymphocytes (clone E-6) were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS), 200 U/ml penicillin G, and 200 μg/ml streptomycin sulfate. Chinese hamster ovary (CHO) cells were grown to confluence on 24-well plates in F-12K medium containing l-glutamine with 10% FBS, 200 U/ml penicillin G, and 200 μg/ml streptomycin sulfate.
Gene sequence analysis and phylogenetics.
Genes encoding EhMSP-1 homologues in E. dispar and Entamoeba invadens were identified in the EuPath database using pBLAST (2, 31). Amino acid sequences of 11 additional M8 family metalloendopeptidases from diverse species were selected from the MEROPS peptidase database (46). A human M10 matrix metalloprotease was included as an outgroup. The predicted amino acid sequences were aligned using ClustalW (52) and the alignment was checked manually for accuracy, as reflected by correct alignment of conserved motifs. Phylogenetic analysis was performed using PHYLIP, version 3.67, with the maximum parsimony method (PROTPARS) (21). A total of 1,000 bootstrap resampling iterations were performed. Bootstrap proportions greater than 60% are shown on the consensus tree, which was displayed using Treeview (version 1.6.6).
EhMSP-1 silencing.
EhMSP-1 was silenced in G3 strain trophozoites using the method described by Bracha et al. (7). For this, 500 bp of EhMSP-1 was amplified from cDNA using reverse transcriptase PCR (RT-PCR) and the primers 5′-ACTGCAAGGCCTATGGTAAGTACTCCACCT-3′ (forward) and 5′-GTCATCGAGCTCTTATCTTCCTTTAGAAAT-3′ (reverse) and cloned into the neo′ vector psAP2-Gunma at the 5′ StuI and 3′ SacI restriction digestion sites downstream of the 5′ upstream regulatory element of the amoebapore A (EhAP-A) gene. Entamoeba histolytica G3 strain trophozoites were then transfected with the psAP2-Gunma empty vector (G3 empty vector) or psAP2-EhMSP-1 silencing vector using 20 μg plasmid DNA and the transfection reagent Attractene (Qiagen) as previously described (11). Transfected parasites were selected with 1.5 μg/ml G418 sulfate beginning 24 h after transfection, the G418 concentration was increased to 24 μg/ml over a period of 4 weeks, and EhMSP-1 silencing was confirmed by quantitative RT-PCR (qRT-PCR) and Western blotting. G418 was then removed, ongoing gene silencing was confirmed, and phenotypic assays were performed using parasites grown without antibiotic selection.
qRT-PCR.
Total RNA isolation from G3 transfectants was performed using an RNeasy kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized from 500 ng amebic total RNA using oligo(dT) primers and following the instructions for the Improm-II reverse transcription system kit (Promega). Quantitative PCR was performed to measure ribosomal protein L10, EhMSP-1, and EhMSP-2 mRNA relative abundance using an ABI PRISM 7900HT sequence detection system, SYBR green, and the following primers: for EhMSP-1, 5′-GACAGTACCTGTTGTTACCAGTGAAAA-3′ (forward) and 5′-GCCAATGTGAAATTAGAAATGACTGA-3′ (reverse); for EhMSP-2, 5′-TGATTTTTTTGTTAGTCATTACAGCACTT-3′ (forward) and 5′-CCGTGTATTTGCACCATTGC-3′ (reverse); and for L10 (NCBI accession number XM_647426.2), 5′-CTACTGGGATCCAGGAAGATGTTATAGACTTG-3′ (forward) and 5′-CTACTGGAATTCTTAATTGAATACGTGCTGC-3′ (reverse). The primers were validated using standard curves, and relative gene expression was calculated using the comparative threshold cycle (CT) method (34).
Immunoblotting.
Amebas were harvested from subconfluent cultures and washed twice in PBS, and the pelleted cells were lysed in cold lysis buffer (50 mM Tris-Cl [pH 7.4], 300 mM NaCl, 1.0% Triton X-100) containing protease inhibitors (400 μM AEBSF, 200 μM EDTA, 60 nM aprotinin, 200 μM leupeptin, 2.8 μM E64, and 26 μM bestatin). Twenty-five micrograms of each sample was separated by SDS-PAGE using reducing or nonreducing conditions and a 10% gel and transferred to a polyvinylidene difluoride (PVDF) membrane that was blocked with 5% milk in TBS (100 mM Tris-Cl [pH 7.5], 0.1% Tween 20) and incubated with 1.5 μg/ml affinity-purified anti-EhMSP-1 polyclonal rabbit IgG or anti-ICP-1 polyclonal rabbit IgG diluted in blocking buffer. Bound antibody was detected with a goat anti-rabbit IgG–horseradish peroxidase (HRP) conjugate antibody (1:10,000 dilution in blocking buffer) (Jackson Laboratory) and enhanced chemiluminescence (Pierce Biotechnology).
Complement assays.
To assay complement-dependent cell lysis, amebas were washed twice with PBS and resuspended in M199 medium, and a 50-μl suspension of 7.0 × 104 parasites was incubated with either 36% normal human serum (NHS) or heat-inactivated NHS for 30 min at 37°C. Cells were washed once with 1.0 ml PBS and then stained in 100 μl of 0.2% trypan blue. The total number of parasites, including those which had excluded the dye, was counted, and complement resistance (cell viability) was calculated as the percentage of viable cells in the samples relative to the negative control.
To assay cell surface accumulation of iC3b and C7, 650-μl suspensions of 1.5 × 105 parasites were incubated with 36% C8-depleted human serum (Quidel Corp.) or heat-inactivated NHS for 10 min at 37°C and then washed twice with 1.0 ml cold PBS, fixed with 4% paraformaldehyde, blocked with 10% heat-inactivated goat serum–5% milk–2.5% BSA, and stained for flow cytometry. iC3b was stained with an anti-human iC3b mouse monoclonal antibody (10 μg/ml) (Quidel Corp.) and an anti-mouse IgG–PE Cy5.5 conjugate (1:150 dilution) (Caltag Laboratories). C7 was stained with sheep anti-human C7 IgG (400 μg/ml) (The Binding Site) and a donkey anti-sheep IgG–Alexa 488 conjugate (1:500) (Invitrogen). Cells were analyzed using a Beckman Coulter EPICS XL-MCL flow cytometer.
Immunofluorescent staining for flow cytometry and confocal microscopy.
The relative abundance of E. histolytica surface antigens was determined using a Beckman Coulter EPICS XL-MCL flow cytometer. Cells were immunostained as described above for staining of iC3b and C7, with or without membrane permeabilization with 0.2% Triton X-100 as indicated. The primary and secondary antibodies and concentrations used were the following: 10 μg/ml anti-EhMSP-1; 100 μg/ml anti-Gal/GalNAc lectin, anti-SREHP, and anti-E. histolytica surface antigen 5E12; a 1:25 dilution of antiproteophosphoglycan; and 4 μg/ml anti-rabbit or anti-mouse IgG–Alexa 488 conjugate (Invitrogen).
To prepare amebas for confocal microscopy, cells were distributed onto Cell-Tak (BD Biosciences)-coated glass coverslips and allowed to adhere for 15 min at 37°C. Adherent amebas were then fixed with 4% paraformaldehyde, and when indicated, cells were permeabilized with 0.2% Triton X-100. After blocking with 10% heat-inactivated goat serum–5% milk–2.5% BSA, amebas were stained with affinity-purified anti-EhMSP-1 polyclonal IgG (10 μg/ml) or protein A-purified preimmune rabbit IgG (10 μg/ml; negative control), anti-SREHP MAb IgG (50 μg/ml) diluted in blocking buffer, and an anti-rabbit IgG–Alexa 568 conjugate (2 μg/ml) (Invitrogen) and an anti-mouse IgG–Alexa 488 conjugate (2 μg/ml) (Invitrogen). Coverslips were inverted onto Fluorogel (Electron Microscopy Sciences), and slides were examined using a Zeiss 510 Meta confocal microscope. Emission from Alexa 488 and Alexa 568 was pseudocolored red and green, respectively.
Adherence assays.
CHO cells grown to confluence in 24-well plates were washed three times with PBS and fixed with 4% paraformaldehyde. Cells were washed three more times with PBS, followed by quenching with 250 mM glycine solution for 30 min at room temperature. Following three more washes, 1.0 ml of trophozoites (2.0 × 105/well) suspended in M199s medium was added on top of the monolayers and incubated for 30 min at 37°C. Nonadhered trophozoites were aspirated from the wells, and the cell monolayers were washed twice with cold PBS. The wells were then filled with 1.0 ml cold PBS, and the plate was placed on ice for 15 min. Again, nonadhered parasites were aspirated from the wells, and the CHO cells were washed twice with cold PBS. The remaining adherent trophozoites were fixed with 4% paraformaldehyde and stained for immunofluorescent microscopy with either an anti-GalNAc or anti-SREHP antibody and an anti-mouse IgG–Alexa 488 conjugate as described above. To quantify the number of adherent trophozoites remaining, randomly located images (magnification, ×200) within the central 60% of each well were acquired using a Nikon eTi-2000 microscope equipped with a programmable, motorized stage, and the number of adherent trophozoites in each low-power image was then counted.
Adherence to healthy and apoptotic Jurkat lymphocytes was assayed using a rosetting assay as previously described (27). Amebas and healthy or apoptotic Jurkat lymphocytes were mixed (1 × 104 amebas and 1 × 105 Jurkat cells), centrifuged at 200 × g for 5 min, and incubated for 60 min on ice. The ameba-host cell pellets were then vortexed for a duration empirically determined to leave 10 to 20% of control amebas adherent to Jurkat cells (to enable detection of increased adherence), and amebas were scored by microscopy for adherence. Greater than 100 trophozoites were scored for each condition, and amebas with ≥3 adherent Jurkat cells were defined as adherent.
Cytotoxicity assay.
Cytolysis of Jurkat lymphocytes by E. histolytica was assayed by measuring release of the fluorescent dye 2′,7′-bis(carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester (BCECF-AM; Invitrogen) (4). Jurkat lymphocytes were labeled with 10 μM BCECF in PBS at 37°C for 20 min. Amebas (3 × 104) and Jurkat cells (3 × 105) in PBS were centrifuged at 200 × g and 4°C and incubated at 37°C for 30 min. Cells were then centrifuged at 3,000 × g for 10 min, the supernatants were removed, and fluorescence of the supernatant was measured using a BioTek microtiter plate fluorimeter and excitation and emission wavelengths of 485 nm and 528 nm, respectively. Nonspecific release of BCECF was corrected for by subtraction of fluorescence released in the absence of trophozoites, and specific cytolysis was calculated as a percentage of fluorescence released during 100% lysis by Triton X-100. Data were normalized to percent lysis during incubation with G3 empty vector control trophozoites.
Cell monolayer destruction assay.
CHO cells were cultured in 24-well plates to confluence. A suspension of amebas in M199s medium (1.0 × 105/well) was distributed into the wells and incubated at 37°C. The amebas were then removed from the monolayer by placing the plate on ice for 15 min and then washing gently with cold PBS. The remaining CHO cell monolayers were fixed with 4% paraformaldehyde and stained with 650 μl/well of 0.1 mg/ml methylene blue in 100 mM sodium borate buffer (pH 8.0) for 5 min at room temperature. Cells were washed twice with 1.0 ml sodium borate buffer, and 1.0 N HCl (1.0 ml/well) was added to the monolayers and incubated for 30 min at 37°C to extract the dye. The extracted dye was transferred into enzyme-linked immunosorbent assay (ELISA) plate wells (200 μl/well), and the optical density of the samples was measured at 600 nm on a microplate ELISA reader.
Phagocytosis assay.
The flow cytometry-based phagocytosis assay, which measures amebic uptake of fluorescently labeled lymphocytes, has been described previously (27). Jurkat lymphocytes were stained with carboxyfluorescein succinimidyl ester (CFSE) (30 μM, 37°C, 15 min). Cells were then washed and residual CFSE was quenched by addition of an equal volume of FBS and incubation for an additional 15 min. A suspension of 4.0 × 105 labeled lymphocytes was then added to 2.0 × 105 trophozoites in a final volume of 200 μl PBS, centrifuged at 400 × g for 5 min, and incubated at 37°C for 20 min. Cells were washed twice in cold PBS with 20 mg/ml d-galactose, fixed in 300 μl 4% paraformaldehyde, and washed, and amebic fluorescence was analyzed by flow cytometry with a Beckman Coulter EPICS XL-MCL flow cytometer. Amebas were distinguished from lymphocytes based on forward- and side-scatter characteristics. Phagocytic amebas were considered to be those with fluorescence above background, and data were expressed as a phagocytic index, defined as the percentage of amebas that are phagocytic multiplied by the mean fluorescence of phagocytic amebas.
Transwell migration assay.
A 100-μl/well suspension of 1.0 × 104 parasites in PBS was distributed on top of 6.5-mm transwell inserts (upper chamber) with polycarbonate membranes containing 8-μm pores (Costar). As a stimulus for trophozoite migration, 600 μl TYI-S-33 medium was placed in the lower chamber. The parasites were then incubated at 37°C, after which the plate was placed on ice for 15 min, and cells that migrated to the bottom chamber were harvested and counted with a hemacytometer.
Motility assay.
A novel assay was developed to quantify nondirected, 2-dimensional motility of E. histolytica trophozoites on fixed CHO cell monolayers. CHO cells grown to confluence in 24-well plates were washed and fixed as described above (adherence assays). Trophozoites were fluorescently labeled with CFSE (56 μM in PBS, 10 min on ice), nonreacted CFSE was quenched by addition of an equal volume of adult bovine serum, and amebas were then resuspended at 5 × 104 cells per ml in M199s, 20 μl of which was applied to each well along with a glass coverslip. Image acquisition began exactly 5 min after plating, and images were taken every 5 s for a total of 10 min. Video images were acquired using a Nikon eTi-2000 microscope utilizing the NIS Elements AR 3.1 software package.
Image stacks were imported into the NIH ImageJ program, and an unsharp mask filter (Σ = 2.5, mask weight = 0.8) followed by a rolling-ball background subtraction (radius = 12) was applied to all images. Images were then manually thresholded and binarized. A mask was created with a lower size limit of 150 to 400 μm2 (varied with each image stack) and excluded edges. The lookup table (LUT) was inverted. Image stacks were then analyzed using the Imaris 7.3.1 software package (Bitplane Scientific Software, South Windsor, CT) (54). Motion tracks were created using an autoregressive motion tracking algorithm with the region growing option enabled. The automated analysis was manually examined for incorrect centroid assignment (resulting from cell proximity, poor thresholding, or portions of cells migrating out of the acquisition field). Images of cells with incorrect centroid placement were removed from the analysis, and the mean speed measurement for each correctly tracked trophozoite was included in the statistical analysis.
Protease assay.
Protease activity of immunoprecipitated EhMSP-1 was measured using a commercial kit, Quanti Cleave (Thermo Scientific), by following the manufacturer's instructions. Briefly, 2.0 ×107 E. histolytica HM-1:IMSS trophozoites were lysed in 1.0 ml cold nondenaturing lysis buffer (50 mM Tris-Cl [pH 7.4], 300 mM NaCl, 1.0% Triton X-100) containing 400 μM AEBSF, 200 μM EDTA, 60 nM aprotinin, 200 μM leupeptin, 2.8 μM E64, and 26 μM bestatin. The cell lysate was split into two 500-μl samples and transferred to microcentrifuge tubes, and 15 μg of either scFv anti-EhMSP-1 MAb or an irrelevant scFv MAb was added to each sample. After incubation for 1 h at 4°C, 160 μl of a 25% suspension of protein A-agarose beads (Santa Cruz Biotechnology) was added and each sample further incubated overnight. Samples were washed four times in 1.0 ml cold PBS and the pelleted beads resuspended in 260 μl cold PBS. Fifty microliters of each resuspended sample was used in the protease assay (duplicates in three independent assays) with or without inclusion of 15 mM 1,10-phenanthroline monohydrate (Sigma) as indicated. Trypsin (66.7 μg/ml) was used as a control.
Preparation of figures and statistics.
Quantitative data were expressed as the mean and standard error (SE). Statistical analyses and graphs were prepared using GraphPad Prism 5 software. In most cases, significance (P ≤ 0.05) was determined using an unpaired Student t test with or without Welch's correction for nonparametric data sets as appropriate. For experiments with multiple groups for comparison, a one-way analysis of variance (ANOVA) and Bonferroni's correction were used. Flow cytometry data were analyzed using WinMDI software (version 2.9). Microscopy images were prepared for publication using either Zeiss LSM software (confocal microscopy), NIH ImageJ (adherence assay), or Imaris 7.3.1 software (motility assay). Final figure preparation was done using Adobe Illustrator CS5.
RESULTS
EhMSP-1 is an M8 family metalloendopeptidase that E. dispar has lost.
The predicted amino acid sequence of EhMSP-1 contains a fully conserved M8-type metalloprotease active site, including an HEXXH motif, a third His residue C terminal to this, and the conserved Met residue (see Fig. S1 in the supplemental material). Unlike leishmanolysin and its trypanosomal orthologues, EhMSP-1 and EhMSP-2 are predicted to be type I integral membrane proteins, since each has a putative N-terminal signal peptide and transmembrane domain (Fig. S1). The amino acid sequences of EhMSP-1 and EhMSP-2 are 34% identical and 54% similar to each other.
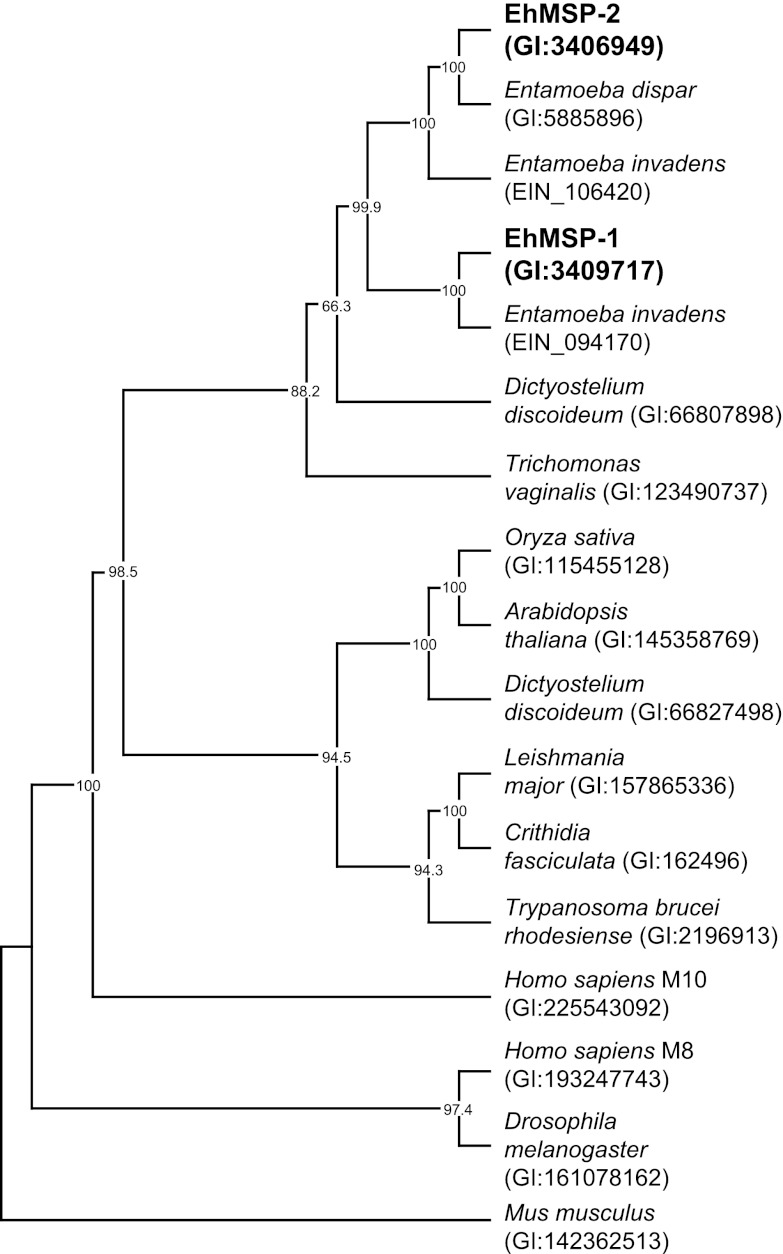
To explore the evolutionary origin of EhMSP-1, we made a phylogenetic tree using aligned amino acid sequences of 17 M8 family proteases from diverse species and a maximum parsimony algorithm (Fig. 1). Leishmanolysin and one of its T. brucei orthologues segregated with an orthologue from the nonpathogenic protozoan Crithidia fasciculata and within a larger subgroup that included plant orthologues. Not surprisingly, both EhMSP-1 and EhMSP-2 segregated with M8 family members from other Entamoeba species within a subgroup that also contained uncharacterized orthologues from Trichomonas vaginalis and the amoeboid slime mold Dictyostelium discoideum. Entamoeba dispar has only one gene copy that was most closely related to EhMSP-2. In contrast, the reptile pathogen Entamoeba invadens has two gene copies, which grouped with high confidence separately with each of the E. histolytica sequences, implying that EhMSP-1 and EhMSP-2 likely arose as a result of gene duplication prior to divergence of E. histolytica and E. invadens. Since E. histolytica is more closely related to E. dispar than to E. invadens, it follows that E. dispar must have once had both gene copies and lost MSP-1 after divergence from E. histolytica.
Fig 1.
Phylogenetic analysis of EhMSP-1 and EhMSP-2 suggests that E. dispar has lost the gene encoding EhMSP-1. The predicted amino acid sequences for EhMSP-1 and EhMSP-2 were used to identify genes encoding homologous proteins by pBLAST in E. dispar and E. invadens. The predicted amino acid sequences of these proteins were aligned with those of representative M8 metalloendopeptidases from diverse species and analyzed using maximum parsimony with the program PHYLIP. A human M10 family matrix metalloendopeptidase was included as an outgroup. The consensus tree from 1,000 bootstrap iterations and bootstrap proportions >60% are shown.
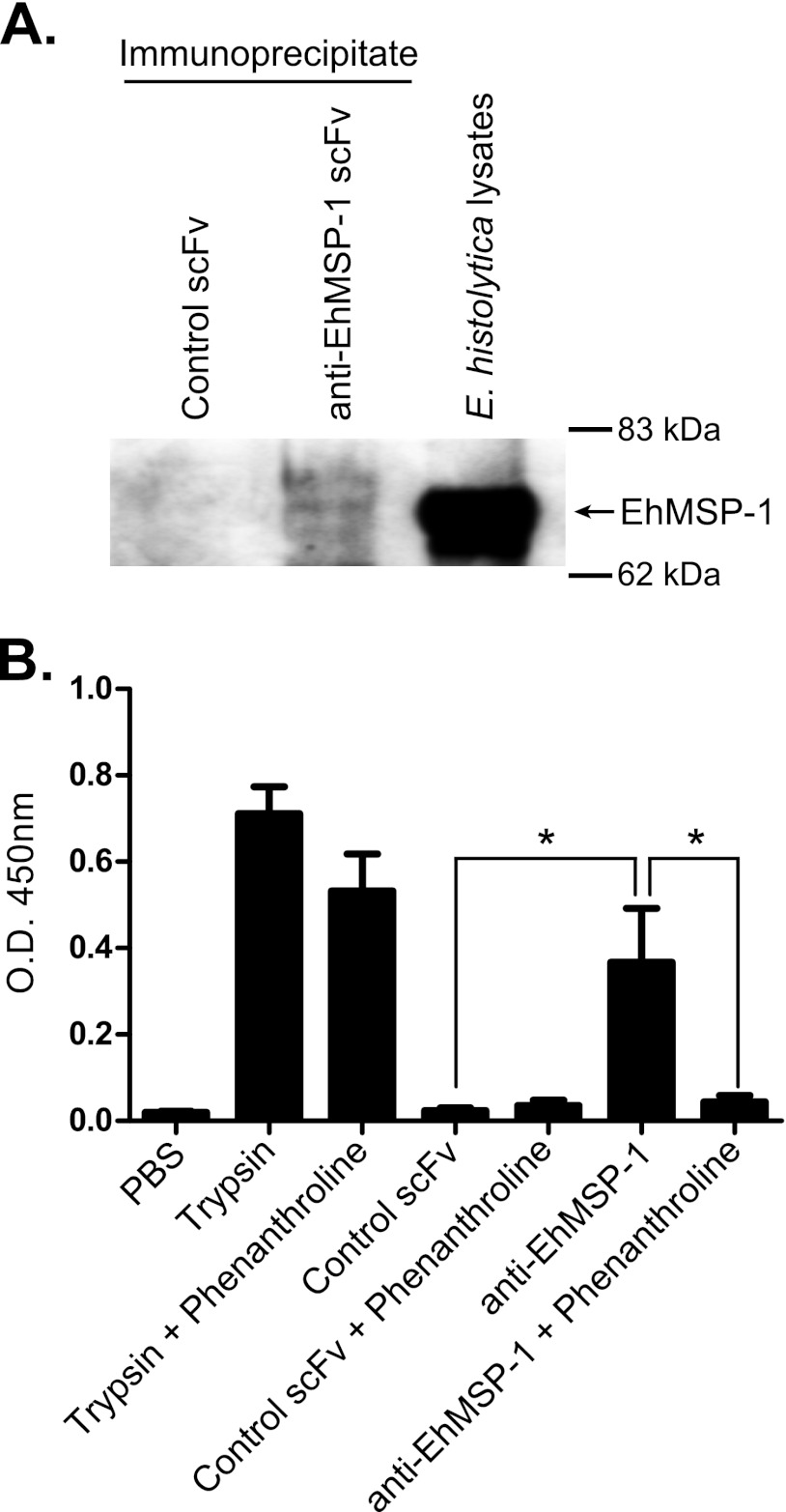
We immunoprecipitated EhMSP-1 and used an in vitro protease assay to determine whether it is a functional metalloprotease. A specific single-chain Fv (scFv) monoclonal antibody that was selected by phage display and validated by ELISA (see Fig. S2 in the supplemental material) was used to immunoprecipitate EhMSP-1. Precipitation of EhMSP-1 by the scFv monoclonal antibody but not a control scFv antibody was confirmed by Western blotting (Fig. 2A). Using succinylated casein as a model substrate, significant protease activity was detected following immunoprecipitation with the anti-EhMSP-1 scFv, while no detectable protease activity was precipitated using the irrelevant control scFv (Fig. 2B). Specificity of the detected protease activity was further implied by complete inhibition with the metalloprotease inhibitor phenanthroline (15 mM), which had no effect on activity of the serine protease trypsin. Because the results of both the bioinformatics analysis and protease assay were consistent, we concluded that EhMSP-1 is a bona fide M8 metalloprotease.
Fig 2.
EhMSP-1 has protease activity that is inhibited by the Zn2+ metalloprotease inhibitor phenanthroline. An anti-EhMSP-1 scFv recombinant monoclonal antibody or control scFv antibody was used for immunoprecipitation from HM-1:IMSS strain E. histolytica lysates. (A) Immunoblot demonstrating specific immunoprecipitation of EhMSP-1. Immunoprecipitated proteins were separated by SDS-PAGE, transferred to a PVDF membrane, and probed with affinity-purified anti-EhMSP-1 rabbit polyclonal IgG. Whole E. histolytica lysates were run in an adjacent lane as a positive control. (B) Detection of metalloprotease activity. Protease activity of immunoprecipitated protein was measured in the presence or absence of 15 mM phenanthroline. Trypsin (66.7 μg/ml) was used as a positive control for the assay and a negative control for phenanthroline inhibition. The graph shows mean optical density (O.D.) at 450 nm and SE measured for three independent immunoprecipitation experiments. *, P = 0.03 in each case.
Silencing EhMSP-1 expression reveals an important role in regulation of adherence.
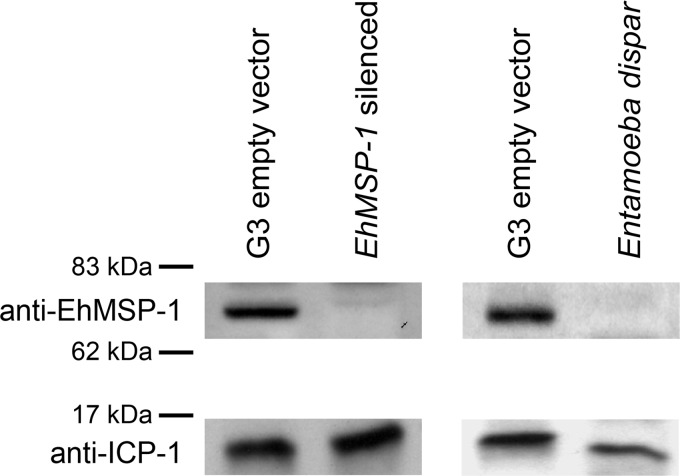
To begin assessing the function of EhMSP-1, EhMSP-1-deficient trophozoites were made using the gene silencing method recently described for G3 strain E. histolytica trophozoites (7). qRT-PCR demonstrated greater than 2,000-fold reduction in EhMSP-1 mRNA relative to G3 parasites transfected with the control empty plasmid, which persisted for greater than 1 year in the absence of G418 (Table 1). Furthermore, EhMSP-2 gene expression was not affected. Western blots using an affinity-purified anti-EhMSP-1 polyclonal antibody showed a single band in the control cell lysate that was not detected in lysates from the EhMSP-1-silenced trophozoites or Entamoeba dispar (Fig. 3). The same blot was probed with an antibody to the inhibitor of cysteine protease 1 (ICP-1) protein as a loading control, which bound a protein band of approximately 14 kDa in lysates from both E. histolytica and E. dispar.
Table 1.
qRT-PCR data demonstrating specific EhMSP-1 silencing in G3 strain E. histolytica trophozoitesa
| Time after G418 removal | Gene | Sample | CT (mean ± SD) | ΔCT | ΔΔCT | Fold difference |
|---|---|---|---|---|---|---|
| 1 mo | EhMSP-1 | G3 control | 19.4 ± 0.1 | 0.5 | ||
| G3 EhMSP-1 (−) | 31.3 ± 0.3 | 12.6 | 12.1 | 4,271.2 | ||
| EhMSP-2 | G3 control | 32.1 ± 1.0 | 13.2 | |||
| G3 EhMSP-1 (−) | 31.8 ± 0.7 | 13.0 | −0.2 | 0.9 | ||
| L10 | G3 control | 18.9 ± 0.3 | ||||
| G3 EhMSP-1 (−) | 18.8 ± 0.6 | |||||
| 1 yr | EhMSP-1 | G3 control | 19.7 ± 0.1 | 1.3 | ||
| G3 EhMSP-1 (−) | 31.3 ± 0.5 | 12.7 | 11.4 | 2,814.4 | ||
| EhMSP-2 | G3 control | 31.3 ± 0.6 | 12.9 | |||
| G3 EhMSP-1 (−) | 31.3 ± 0.5 | 12.8 | −0.1 | 0.9 | ||
| L10 | G3 control | 18.4 ± 0.2 | ||||
| G3 EhMSP-1 (−) | 18.6 ± 0.3 |
Trophozoite mRNA was isolated from the EhMSP-1-silenced (−) and empty vector control G3 strain trophozoites 1 month and 1 year after removal of G418 selection. cDNA was synthesized using oligo(dT) primers, and gene transcripts for EhMSP-1, EhMSP-2, and L10 (ribosomal protein) were quantified by qRT-PCR using SYBR green. Shown are the mean and SD for threshold cycle numbers (CT). The fold change in gene expression for EhMSP-1 and EhMSP-2 was calculated using the comparative CT method and L10 as the control gene (34).
Fig 3.
Immunoblot demonstrating reduced EhMSP-1 protein in E. histolytica G3 strain trophozoites. (Left) Lysates from EhMSP-1-silenced and G3 trophozoites transfected with empty plasmid (G3 empty vector) were separated under nonreducing conditions by SDS-PAGE, transferred to a PVDF membrane, and probed with affinity-purified anti-EhMSP-1 rabbit polyclonal IgG and an anti-rabbit IgG–HRP conjugate secondary antibody. As a loading control, the same blot was reprobed with a rabbit polyclonal anti-ICP-1 IgG antibody. (Right) Immunoblot of G3 empty vector control E. histolytica and Entamoeba dispar strain SAW760 lysates.
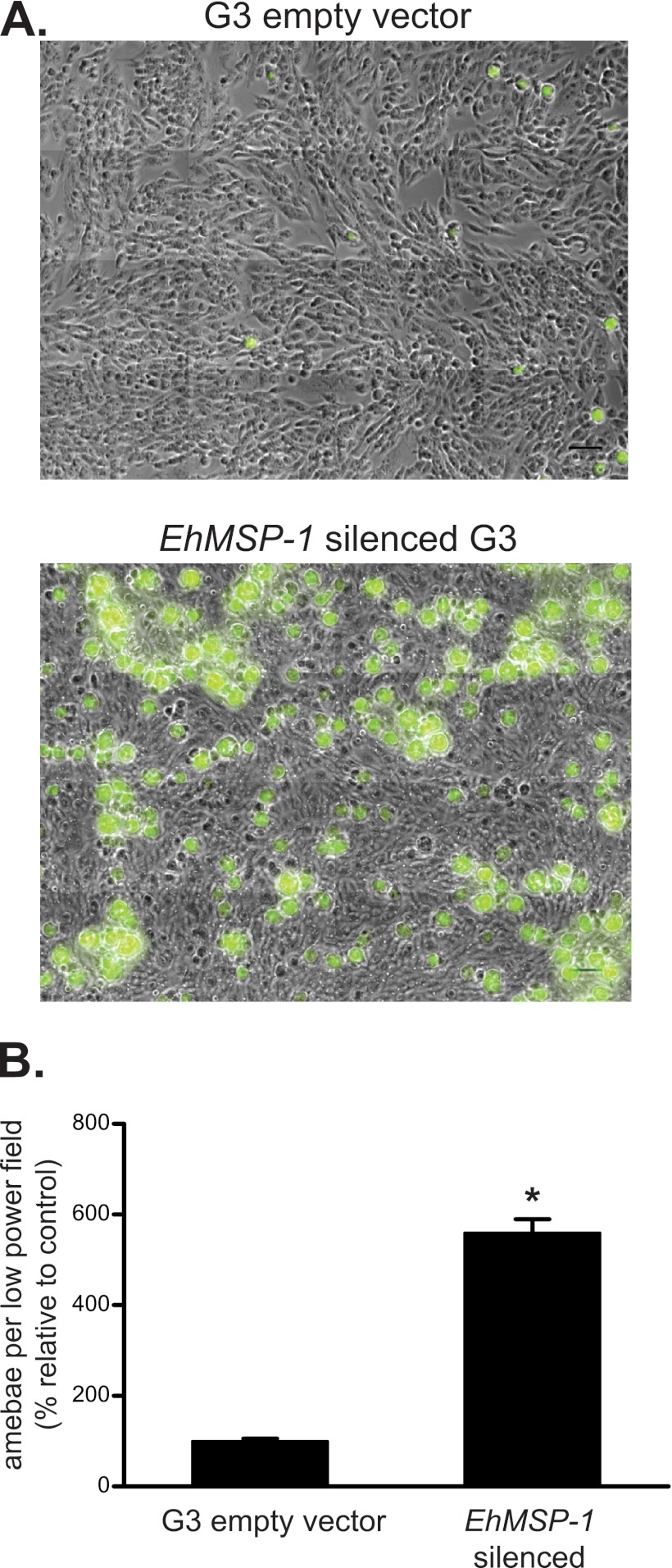
It was immediately evident that the EhMSP-1-deficient trophozoites were difficult to harvest from culture tubes for use in phenotypic assays, because they remained stuck to the glass walls of the tubes. Thus, we began characterizing the EhMSP-1-silenced trophozoites by measuring adherence to cell monolayers (Fig. 4). After allowing trophozoites to adhere to fixed Chinese hamster ovary (CHO) cell monolayers and washing rigorously, trophozoites that remained adherent to the cell monolayers were fixed, labeled, and visualized by immunofluorescence using an automated microscope. A dramatic increase in adherence of the EhMSP-1-deficient trophozoites was seen readily at low magnification. To provide this perspective, Fig. 4A shows a representative image with a four-by-four array of adjacent low-magnification images tiled together to create a single large image for each condition. Adherent trophozoites were visualized by immunofluorescent staining. The increase in adherence of the EhMSP-1-deficient trophozoites was quantified in three independent experiments (Fig. 4B). For this, random low-magnification images were acquired and the number of adherent trophozoites per low-power field was counted. The mean number of adherent EhMSP-1-silenced trophozoites per low-magnification image was more than five times the mean number of adherent control cells. We also measured adherence to healthy and apoptotic Jurkat lymphocytes using a rosetting assay. Similar to the adherence result obtained using fixed CHO cell monolayers, the EhMSP-1-silenced trophozoites were approximately three to five times more adherent to apoptotic and healthy Jurkat cells (see Fig. S3 in the supplemental material).
Fig 4.
EhMSP-1 silencing dramatically increases E. histolytica adherence to host cell monolayers. G3 empty vector control and EhMSP-1-silenced trophozoites were allowed to adhere to fixed CHO cell monolayers (2.0 × 105/well, 30 min, 37°C) prior to washing, immunofluorescent staining, and image acquisition. (A) Tiled low-magnification epifluorescent images showing increased adherence of EhMSP-1-silenced trophozoites. Cells were stained with an anti-GalNAc-lectin monoclonal antibody and anti-mouse IgG Alexa 488 (green)-conjugated secondary antibody. A four-by-four tiled array of images is shown (original magnification, ×200; scale bars are 100 μm). (B) Graph comparing the relative number of adherent trophozoites per low-power image for EhMSP-1-silenced and G3 empty vector control trophozoites. Adherent trophozoites were immunostained, randomly located low-magnification images (magnification, ×200) were acquired with an automated microscope (n = 416 images for EhMSP-1-silenced trophozoites and n = 440 for G3 empty vector control), and adherent trophozoites were counted. Combined results from three independent experiments are shown (mean and SE percent adherent trophozoites relative to G3 empty vector control). *, P < 0.0001 by unpaired t test with Welch's correction for nonnormal distributions.
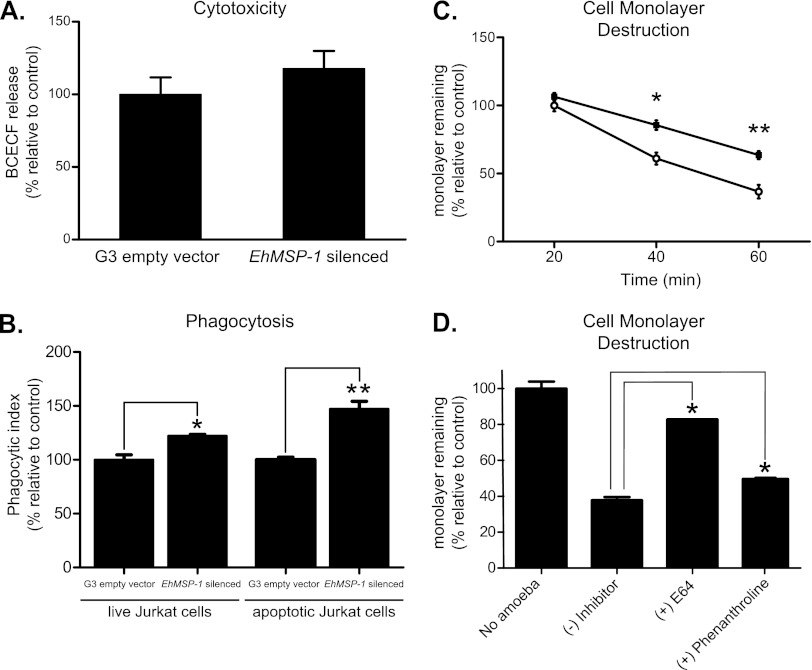
EhMSP-1 silencing did not significantly affect E. histolytica killing of Jurkat lymphocytes, as determined by measuring release of fluorescent dye from Jurkat cells incubated with trophozoites (Fig. 5A). However, the EhMSP-1-deficient cells were significantly more phagocytic than the G3 empty vector control cells (Fig. 5B). Although phagocytosis of both viable and apoptotic lymphocytes was significantly enhanced by EhMSP-1 silencing, the magnitude of the phenotype was larger for phagocytosis of apoptotic cells. This finding and the cytotoxicity data were consistent with previously published data suggesting that cell killing is the rate-limiting step in phagocytosis (27).
Fig 5.
EhMSP-1 silencing reduces E. histolytica cytopathic effect and enhances phagocytic ability. Cytotoxic ability is unaffected. (A) Effect of EhMSP-1 silencing on amebic lysis of lymphocytes, as reflected by release of the fluorescent dye BCECF (ameba/Jurkat cell ratio = 1:10, 30 min, 37°C) (mean and SE relative to G3 empty vector control, combined data from two experiments, n = 6). (B) Effect of EhMSP-1 silencing on amebic phagocytosis of live and apoptotic Jurkat lymphocytes. Phagocytosis of fluorescently labeled healthy or apoptotic Jurkat lymphocytes by EhMSP-1-silenced and control trophozoites was assayed by flow cytometry and expressed as the phagocytic index (ameba/Jurkat cell ratio = 1:2, 20 min, 37°C) (mean and SE relative to G3 empty vector controls, representative data from three experiments, n = 3). *, P = 0.01; **, P = 0.003. (C) Effect of EhMSP-1 silencing on amebic destruction of host cell monolayers. EhMSP-1-silenced (■) or G3 empty vector control (○) trophozoites (1 × 105/well) were added to confluent CHO cell monolayers and incubated at 37°C for the indicated times, after which the cell monolayers were washed, fixed, and stained with methylene blue. After washing, the remaining dye was extracted and quantified by measuring absorbance (600 nm) with a spectrophotometer. Data were expressed as the percentage of monolayer remaining relative to monolayers incubated with G3 empty vector control cells for 20 min (mean and SE, combined data from three independent experiments, n = 9). *, P = 0.005; **, P = 0.0003. (D) Effects of phenanthroline (100 μM) and E64 (100 μM) on tissue culture monolayer destruction by wild-type E. histolytica trophozoites. HM-1:IMSS strain E. histolytica trophozoites (1 × 105/well) were added to confluent CHO cell monolayers and incubated at 37°C for 40 min, after which cell monolayer destruction was assayed as for panel C. Data were expressed as the percentage of readings from monolayers to which no trophozoites were added (mean and SE, combined data from two independent experiments, n = 6). *, P = 0.05.
Interestingly, despite increased ability to adhere to CHO cell monolayers, the EhMSP-1-silenced trophozoites were significantly less efficient at destroying them (Fig. 5C). Phenanthroline at 100 μM partially inhibited cell monolayer destruction by HM-1:IMSS trophozoites (Fig. 5D), but to a lesser extent than silencing EhMSP-1 gene expression. Phenanthroline alone was toxic to the monolayer (data not shown), which prevented use of higher concentrations and may account for the inability to recapitulate the phenotype of the EhMSP-1-deficient cells completely. As has been shown previously, the cysteine protease inhibitor E64 (100 μM) almost completely blocked cell monolayer destruction. The fact that nearly all monolayer destruction can be blocked with E64 implies that EhMSP-1 does not participate directly in monolayer damage, which is due primarily to the cysteine proteases (32, 47). Rather, EhMSP-1 likely plays an indirect role in monolayer destruction.
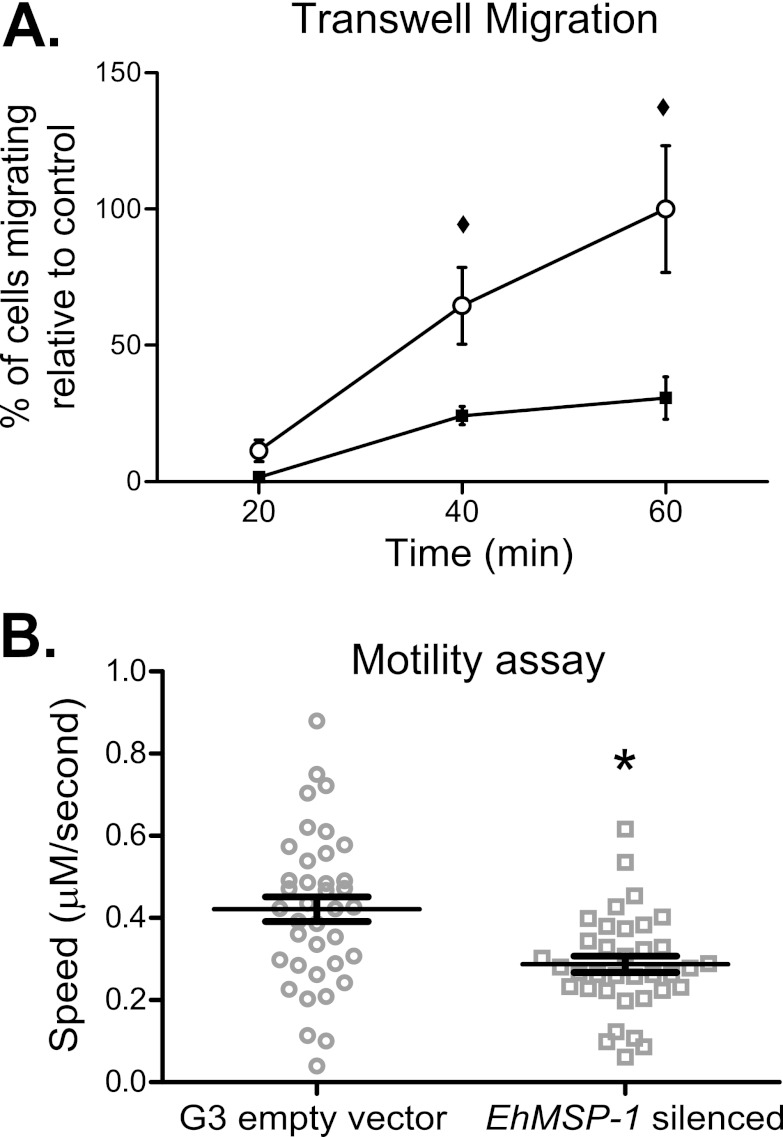
We reasoned that the EhMSP-1-deficient trophozoites might be less able to move due to increased adherence, which could account for the reduction in their ability to destroy cell monolayers. As a simple initial experiment, we examined transwell migration of the EhMSP-1 and G3 empty vector control cells. Indeed, the EhMSP-1-silenced cells were substantially less efficient than control parasites at transwell migration (Fig. 6A). Transwell migration reflects a complex phenotype which involves both chemotaxis and motility and might be affected by aggregation of trophozoites. Thus, we devised a novel assay to quantify nondirected, 2-dimensional motility of individual trophozoites during interaction with cell monolayers by using time-lapse video microscopy of fluorescently labeled EhMSP-1-silenced and G3 empty vector control parasites. To quantify amebic movement, the fluorescent trophozoites were identified by converting the images to binary images and thresholding, and motion tracks were created using Imaris motion tracking software (54). Representative green fluorescent videos (i.e., nonbinarized) for the control and EhMSP-1-silenced trophozoites are shown in Movies S1 and S2, respectively, in the supplemental material, and Movies S3 and S4 in the supplemental material show the corresponding binarized, tracked videos. As shown in Fig. 6B, EhMSP-1 silencing significantly reduced parasite motility.
Fig 6.
EhMSP-1 silencing reduces E. histolytica motility. (A) Effect of EhMSP-1 silencing on amebic transwell migration. Movement of EhMSP-1-silenced (■) or G3 empty vector control (○) trophozoites through 8-μm pores toward TYI-S-33 growth medium was assayed following incubation at 37°C for the indicated times. In each case, 1.0 × 104 cells were added to the upper well chamber. The number of migrating cells was expressed as the percentage of control cells found in the lower well after 60 min (mean and SE, combined data from two independent experiments, n = 5). ♦, P = 0.03. (B) Effect of EhMSP-1 silencing on nondirected amebic motility during incubation on tissue culture monolayers. CFSE-labeled EhMSP-1-silenced and G3 empty vector control trophozoites were observed interacting with fixed CHO cell monolayers by video microscopy. Fluorescent trophozoites were identified in binarized images by thresholding in the green channel, and motility tracks were generated using Imaris motility tracking software. Data points indicate the mean speed for individual trophozoites, and the overall mean and SE for each trophozoite population are shown (n = 38 for G3 empty vector control and n = 36 for EhMSP-1-silenced cells). *, P = 0.0005.
The adherence phenotype could not be explained simply by increased steady-state abundance of the d-galactose-specific adherence lectin, which might be expected if EhMSP-1 cleaves the lectin protein as has recently been shown for the rhomboid protease EhROM1 (3). As assessed by flow cytometry, there was no difference in steady-state surface abundance of the lectin between EhMSP-1-silenced and control cells (see Fig. S4A in the supplemental material). Similarly, no difference was detected for three other surface antigens: the serine-rich E. histolytica protein (SREHP), an uncharacterized surface antigen (5E12 antigen), and the lipoproteophosphoglycan (LPPG) (Fig. S4B to D). It should be noted that these steady-state data do not eliminate the possibility that any or all of these proteins are cleaved by EhMSP-1 but rapidly replaced on the cell surface during, for example, cell migration.
Finally, because leishmanolysin inactivates serum complement, we sought to determine whether EhMSP-1 participates in serum complement resistance. The EhMSP-1-silenced trophozoites and control trophozoites were equally susceptible to lysis following activation of the classical complement cascade (see Fig. S5A in the supplemental material). Inactivation of C3b by conversion to iC3b and accumulation of the terminal complement component C7 were also examined by flow cytometry, following activation of the classical complement pathway in the presence of C8-depleted human serum. The EhMSP-1-silenced and control cells converted C3b to iC3b with equal efficiencies (Fig. S5B), and no difference in accumulation of C7 was detected (Fig. S5C). Although we cannot exclude an in vivo role and/or a confounding effect due to ongoing EhMSP-2 expression, EhMSP-1 appears to play no significant role in complement resistance in vitro.
EhMSP-1 cell surface expression is regulated, with populations of high and low surface expression that vary during cell culture.
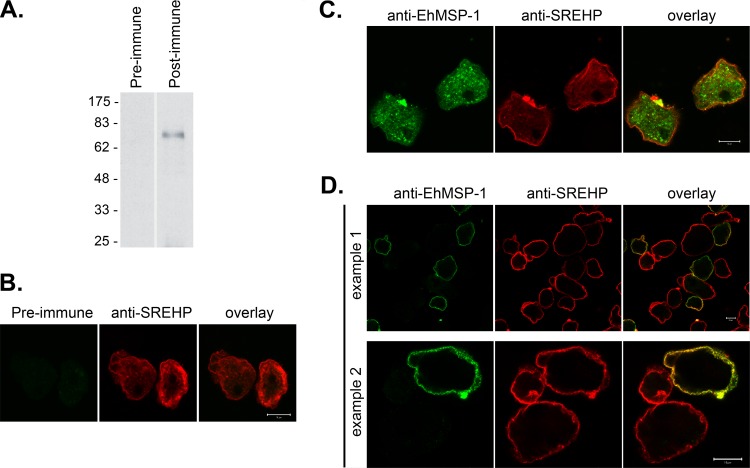
Immunofluorescent staining using an affinity-purified rabbit polyclonal anti-EhMSP-1 antibody and confocal microscopy were used to localize EhMSP-1. This antibody labeled a single band of approximately 70 kDa by Western blotting of E. histolytica lysates (Fig. 7A), and no immunofluorescent signal was detected by confocal microscopy after immunostaining with preimmune IgG (Fig. 7B). The specificity of the antibody for EhMSP-1 was further implied by its failure to detect any protein by Western blotting after EhMSP-1 silencing (which had no effect on EhMSP-2 mRNA abundance) or any protein in Entamoeba dispar lysates (Fig. 3 and Table 1). Costaining for SREHP was used to mark the cell membrane. EhMSP-1 had a vacuolar staining pattern that was readily detected in all detergent-permeabilized trophozoites, while less prominent staining could be seen localized to the cell membrane of some trophozoites (Fig. 7C). Staining without membrane permeabilization more clearly demonstrated the presence of EhMSP-1 on the surface of only a subset of cells (Fig. 7D). We used immunofluorescent staining and flow cytometry to assess EhMSP-1 whole-cell abundance (Fig. 8A, detergent permeabilized) and exposure on the cell surface (Fig. 8B, nonpermeabilized) for trophozoites in mid-log-phase growth. Consistent with the microscopy data, EhMSP-1 staining had a normal distribution for detergent-treated cells, but populations with low and high surface exposure were present.
Fig 7.
Immunolocalization of EhMSP-1 in E. histolytica trophozoites by confocal microscopy. (A) Western blot showing a single band in E. histolytica lysates detected using affinity-purified, rabbit polyclonal anti-EhMSP-1 IgG and an anti-rabbit IgG–HRP conjugate. (B) Immunofluorescent confocal microscopy showing control staining performed on detergent-permeabilized trophozoites using preimmune rabbit IgG. Trophozoites were revealed using an anti-SREHP monoclonal mouse IgG and an anti-mouse IgG–Alexa 488 conjugate (pseudocolored red). (C) Immunofluorescent confocal microscopy of permeabilized cells demonstrating the presence of EhMSP-1 in vesicles of all trophozoites. (D) Immunofluorescent confocal microscopy of nonpermeabilized trophozoites demonstrating the presence of EhMSP-1 on the surface of only a subset of amebas. EhMSP-1 was stained using affinity-purified anti-EhMSP-1 rabbit polyclonal IgG and an anti-rabbit IgG–Alexa 568 conjugate, and pseudocolored green. The SREHP was stained as for panel B. All scale bars shown are 10 μm.
Fig 8.
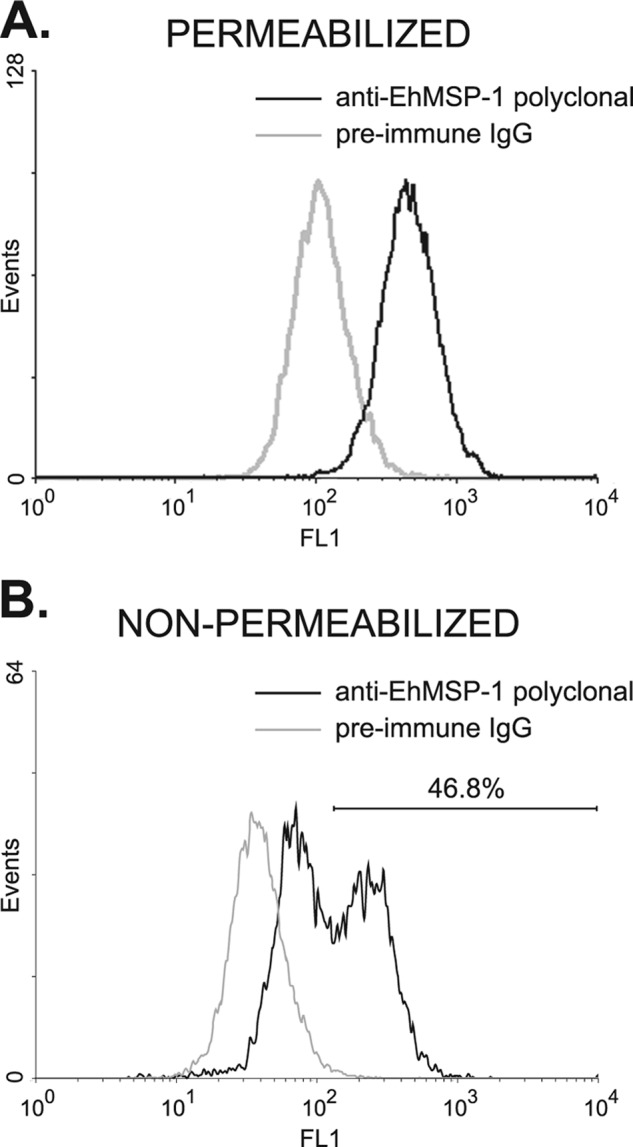
Whole-cell EhMSP-1 abundance versus surface exposure as assessed by flow cytometry. E. histolytica trophozoites harvested after 24 h of culture were fixed, permeabilized (A) or not permeabilized (B) with 0.2% Triton X-100, and stained with either anti-EhMSP-1 rabbit polyclonal IgG or preimmune rabbit IgG followed by an anti-rabbit IgG–Alexa 488 conjugate. EhMSP-1 staining was assessed by flow cytometry. Representative fluorescence-activated cell sorting histograms are shown. (A) Permeabilized trophozoites showing whole-cell staining of EhMSP-1. EhMSP-1 abundance had a normal distribution. (B) Nonpermeabilized trophozoites showing staining of cell surface EhMSP-1. Distinct populations with high and low surface EhMSP-1 exposure were evident.
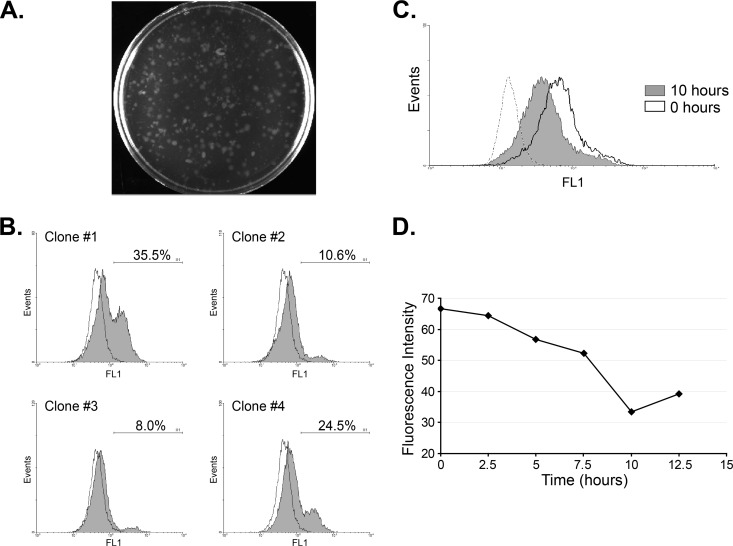
To distinguish between regulated exposure of EhMSP-1 on the amebic cell surface and the possibility that clonal populations with high and low surface exposure existed within our cultures, we isolated E. histolytica clones by growth in agar. We then selected and expanded four clones by cultivation in liquid media and assessed surface EhMSP-1 exposure (Fig. 9A and B). Populations with high and low surface EhMSP-1 levels were present for each of the clones examined, effectively eliminating the possibility of genetic heterogeneity affecting EhMSP-1 surface exposure in the original culture. Moreover, the relative sizes of the high and low EhMSP-1 populations varied from clone to clone, further implying that surface exposure of EhMSP-1 is regulated during cell culture.
Fig 9.
High versus low surface exposure of EhMSP-1 is not clonal and can be synchronized by serum starvation. (A and B) Clones were selected from cell culture in agar, and surface EhMSP-1 exposure was measured by flow cytometry. (A) Plate showing E. histolytica clones grown in agar suspension. (B) Surface EhMSP-1 staining for selected clones. Clones isolated by growth in agar were recovered in broth culture, and EhMSP-1 on the surface of nonpermeabilized cells was stained and analyzed by flow cytometry. Representative fluorescence-activated cell sorting histograms for four clones and background staining (preimmune serum; dashed line) are shown. The percentage of cells with high surface EhMSP-1 levels and the gate used is shown on each histogram. A mixed population of cells with high and low EhMSP-1 surface exposure was recovered in each case. (C) Serum starvation results in synchronized EhMSP-1 surface exposure. E. histolytica trophozoites were serum starved for 21 h, followed by readdition of serum. Representative fluorescence-activated cell sorting histograms are shown, displaying relative EhMSP-1 surface staining immediately following (0 h) and 10 h after readdition of serum. The dashed line shows staining with preimmune serum. Following serum starvation, a single peak which persisted for more than 10 h was observed at every time point. (D) EhMSP-1 surface exposure varies with time. EhMSP-1 surface exposure was synchronized by serum starvation followed by refeeding as for panel C. EhMSP-1 surface exposure was measured by immunostaining and flow cytometry every 2.5 h. The graph shows mean fluorescence intensity versus time.
Serum starvation has been shown to induce translocation of an E. histolytica Rab protein (EhRab11) to the cell periphery, to affect expression of several genes, and to partially synchronize the cell cycle of E. histolytica trophozoites in axenic culture (22, 30, 36, 49). We therefore examined whether serum starvation influenced surface exposure of EhMSP-1. By following surface staining with flow cytometry, we found that EhMSP-1 surface exposure was synchronized by serum starvation, as indicated by presence of a nearly normal distribution of EhMSP-1 surface expression levels after serum starvation and refeeding (Fig. 9C). Nearly all trophozoites had high EhMSP-1 surface levels after overnight serum starvation (0 h), followed by a progressive decline in surface abundance that occurred over a 10-h period after refeeding (Fig. 9D). The cells no longer had synchronized levels of surface EhMSP-1 exposure after 12.5 h, since high and low populations were again present (data not shown). As has been previously reported (22), serum starvation resulted in a more homogeneous distribution of amebic DNA content (by propidium iodide staining), but cell division as assessed by CFSE staining was not directly linked to DNA content (data not shown). Given this, it was not possible to determine whether EhMSP-1 surface abundance is linked to cell cycle or whether synchronization following serum starvation was simply a stress response. In either case, these data provided additional evidence that the surface abundance of EhMSP-1 is regulated.
DISCUSSION
This work demonstrates that EhMSP-1 is a novel M8 family surface metalloprotease that functions in regulation of E. histolytica adherence, with lesser effects on cell motility, cell monolayer destruction, and phagocytosis. That EhMSP-1 is an M8 family metalloendopeptidase was shown both bioinformatically and using protein immunoprecipitated from E. histolytica trophozoites. Silencing EhMSP-1 expression dramatically increases E. histolytica adherence to fixed Chinese hamster ovary cell monolayers and live or apoptotic Jurkat lymphocytes, while also substantially decreasing cell monolayer destruction. The ability of the cysteine protease inhibitor E64 to block cell monolayer destruction almost completely in this study and in others suggests that EhMSP-1 silencing affects monolayer destruction indirectly and that EhMSP-1 itself is not likely to directly degrade cell monolayers (32, 47). Silenced trophozoites were also less motile than control amebas when interacting with cell monolayers, less efficient at transwell migration, and more efficient at phagocytosis of Jurkat lymphocytes. These are all complex phenotypes, involving adherence in addition to other abilities. Thus, these differences may simply reflect the altered adherence phenotype (e.g., increased adherence results in less ability to move, which, in turn, causes them to be less able to destroy cell monolayers) or may be more complicated. The ultimate mechanism by which EhMSP-1 affects adherence remains unknown, but EhMSP-1 silencing does not affect steady-state surface abundance of several surface antigens, including the GalNAc-specific lectin and the serine-rich E. histolytica protein (SREHP), which have been previously implicated in adherence (42, 50, 51). Finally, the abundance of EhMSP-1 on the cell surface appears to be regulated, since nonclonal trophozoite populations with high and low EhMSP-1 surface abundance exist and EhMSP-1 surface abundance is synchronized by serum starvation.
Persistent EhMSP-1 silencing was achieved in this study using a method that requires the use of a trophozoite strain (G3) that is already silenced for the secreted pore-forming peptide amoebapore (7). Amoebapore silencing is known to reduce cytolytic ability and liver abscess formation (8, 56). Thus, although this gene silencing system enabled comparison of EhMSP-1-silenced and G3 control (transfected with empty vector) trophozoites for in vitro phenotypic characterization, alternative gene silencing methods or a specific inhibitor will be needed to study the contribution of EhMSP-1 to virulence in vivo. Furthermore, since the G3 parasite strain is already defective in cytotoxic ability, studies using G3 parasites to examine cell killing by E. histolytica are inherently limited. Although silencing EhMSP-1 in the G3 strain had no effect on amebic lysis of Jurkat lymphocytes, we cannot exclude the possibility that EhMSP-1 contributes to cytotoxic ability in wild-type E. histolytica. Attempts to silence EhMSP-1 in wild-type trophozoites using alternative methods to enable in vivo infection studies have been unsuccessful thus far (data not shown).
Matrix metalloproteinases for which the active form is localized to specialized adherence structures called invadosomes, which contain integrins and matrix metallopeptidases surrounding a dense actin bundle, have been implicated in regulation of cancer cell adherence and invasion (reviewed in references 6 and 16). In this context, matrix metalloproteinases are believed to affect intregrin signaling by altering the local density of extracellular matrix ligands required for adherence, and metalloproteinase activity reduces cancer cell adherence while facilitating cell motility and invasion. Our working hypothesis is that EhMSP-1 may function in E. histolytica adherence and motility during interaction with the host through an analogous mechanism. It also remains possible that EhMSP-1 cleaves E. histolytica surface adhesins, since although EhMSP-1 silencing did not alter steady-state levels of several E. histolytica surface antigens assessed in this study, the possibility that EhMSP-1 cleaves an untested amebic antigen or that one of the tested antigens is cleaved but rapidly replaced cannot be excluded. Although EhMSP-1 silencing did not alter E. histolytica sensitivity to complement, it also remains possible that EhMSP-1 participates in immune evasion such as during concanavalin A- or antibody-induced cap formation and shedding, as has recently been suggested for the rhomboid protease EhROM (3, 12, 20).
Ultimately, it will be essential to determine the substrate specificity of EhMSP-1 in order to determine the mechanism underlying the adherence defect observed in this study and the true function of EhMSP-1 in E. histolytica biology. It will also be necessary to determine whether the protease activity of EhMSP-1 is regulated and, if so, the precise location of EhMSP-1 activity within the cell. Along these lines, leishmanolysin and other M8-type proteases are synthesized as proenzymes that are activated by a “cysteine switch” mechanism (35, 53). Many of these questions could be addressed using functional recombinant EhMSP-1, which we are currently trying to produce. Additional work to characterize EhMSP-2 and to understand the mechanism and biological significance of the variation in EhMSP-1 surface expression that we observed during parasite growth will also be fruitful to determine the precise role these proteases play in Entamoeba biology and the pathogenesis of invasive amebiasis. Given the accepted roles of adherence, motility, and phagocytosis in amebic invasiveness, it seems likely that regulation of these functions by EhMSP-1 may profoundly affect amebic virulence. If future in vivo studies confirm this, it would establish EhMSP-1 as a new target for approaches to prevent or treat amebiasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of our laboratory for their helpful discussions and critique of the manuscript.
This work was supported by NIAID grant R01 AI072021 and NIH grant P20 RR021905 to C.D.H.
Footnotes
Published ahead of print 26 March 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Ankri S, Stolarsky T, Bracha R, Padilla-Vaca F, Mirelman D. 1999. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect. Immun. 67:421–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aurrecoechea C, et al. 2010. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 38:D415–D419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baxt LA, Baker RP, Singh U, Urban S. 2008. An Entamoeba histolytica rhomboid protease with atypical specificity cleaves a surface lectin involved in phagocytosis and immune evasion. Genes Dev. 22:1636–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berninghausen O, Leippe M. 1997. Necrosis versus apoptosis as the mechanism of target cell death induced by Entamoeba histolytica. Infect. Immun. 65:3615–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biller L, et al. 2010. Differences in the transcriptome signatures of two genetically related Entamoeba histolytica cell lines derived from the same isolate with different pathogenic properties. BMC Genomics 11:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bourboulia D, Stetler-Stevenson WG. 2010. Matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPS): positive and negative regulators of tumor cell adhesion. Semin. Cancer Biol. 20:161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bracha R, Nuchamowitz Y, Anbar M, Mirelman D. 2006. Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog. 2:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bracha R, Nuchamowitz Y, Leippe M, Mirelman D. 1999. Antisense inhibition of amoebapore expression in Entamoeba histolytica causes a decrease in amoebic virulence. Mol. Microbiol. 34:463–472 [DOI] [PubMed] [Google Scholar]
- 9. Brittingham A, et al. 1995. Role of the Leishmania surface protease gp63 in complement fixation, cell adhesion, and resistance to complement-mediated lysis. J. Immunol. 155:3102–3111 [PubMed] [Google Scholar]
- 10. Bruchhaus I, Loftus BJ, Hall N, Tannich E. 2003. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell 2:501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buss SN, et al. 2010. Members of the Entamoeba histolytica transmembrane kinase family play non-redundant roles in growth and phagocytosis. Int. J. Parasitol. 40:833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calderón J, De Lourdes-Munoz M, Acosta HM. 1980. Surface redistribution and release of antibody-induced caps in Entamoebae. J. Exp. Med. 151:184–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chadee K, Meerovitch E. 1985. Entamoeba histolytica: early progressive pathology in the cecum of the gerbil (Meriones unguiculatus). Am. J. Trop. Med. Hyg. 34:283–291 [DOI] [PubMed] [Google Scholar]
- 14. Chadee K, Petri WA, Jr, Innes DJ, Ravdin JI. 1987. Rat and human colonic mucins bind to and inhibit adherence lectin of Entamoeba histolytica. J. Clin. Invest. 80:1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deryugina EI, Quigley JP. 2006. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 25:9–34 [DOI] [PubMed] [Google Scholar]
- 16. Destaing O, Block MR, Planus E, Albiges-Rizo C. 2011. Invadosome regulation by adhesion signaling. Curr. Opin. Cell Biol. 23:597–606 [DOI] [PubMed] [Google Scholar]
- 17. Diamond LS, Harlow DR, Cunnick C. 1978. A new medium for axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431–432 [DOI] [PubMed] [Google Scholar]
- 18. El-Sayed NM, Donelson JE. 1997. African trypanosomes have differentially expressed genes encoding homologues of the Leishmania GP63 surface protease. J. Biol. Chem. 272:26742–26748 [DOI] [PubMed] [Google Scholar]
- 19. Epidemiological Bulletin 1997. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico, 28–29 January, 1997. Epidemiol. Bull. 18:13–14 [PubMed] [Google Scholar]
- 20. Espinosa-Cantellano M, Martinez-Palomo A. 1994. Entamoeba histolytica: mechanism of surface receptor capping. Exp. Parasitol. 79:424–435 [DOI] [PubMed] [Google Scholar]
- 21. Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 22. Gangopadhyay SS, Ray SS, Kennady K, Pande G, Lohia A. 1997. Heterogeneity of DNA content and expression of cell cycle genes in axenically growing Entamoeba histolytica HM1:IMSS clone A. Mol. Biochem. Parasitol. 90:9–20 [DOI] [PubMed] [Google Scholar]
- 23. Grandgenett PM, Coughlin BC, Kirchhoff LV, Donelson JE. 2000. Differential expression of GP63 genes in Trypanosoma cruzi. Mol. Biochem. Parasitol. 110:409–415 [DOI] [PubMed] [Google Scholar]
- 24. He C, et al. 2010. A novel Entamoeba histolytica cysteine proteinase, EhCP4, is key for invasive amebiasis and a therapeutic target. J. Biol. Chem. 285:18516–18527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoogenboom HR. 2005. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 23:1105–1116 [DOI] [PubMed] [Google Scholar]
- 26. Huston CD. 2004. Parasite and host contributions to the pathogenesis of amebic colitis. Trends Parasitol. 20:23–26 [DOI] [PubMed] [Google Scholar]
- 27. Huston CD, Boettner DR, Miller-Sims V, Petri WA. 2003. Apoptotic killing and phagocytosis of host cells by the parasite Entamoeba histolytica. Infect. Immun. 71:964–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huston CD, Houpt ER, Mann BJ, Hahn CS, Petri WA. 2000. Caspase 3-dependent killing of host cells by the parasite Entamoeba histolytica. Cell. Microbiol. 2:617–625 [DOI] [PubMed] [Google Scholar]
- 29. Inverso JA, Medina-Acosta E, O'Connor J, Russell DG, Cross GA. 1993. Crithidia fasciculata contains a transcribed leishmanial surface proteinase (gp63) gene homologue. Mol. Biochem. Parasitol. 57:47–54 [DOI] [PubMed] [Google Scholar]
- 30. Jain R, Shrimal S, Bhattacharya S, Bhattacharya A. 2010. Identification and partial characterization of a dynamin-like protein, EhDLP1, from the protist parasite Entamoeba histolytica. Eukaryot. Cell 9:215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson M, et al. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36:W5–W9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keene WE, Hidalgo ME, Orozco E, McKerrow JH. 1990. Entamoeba histolytica: correlation of the cytopathic effect of virulent trophozoites with secretion of a cysteine proteinase. Exp. Parasitol. 71:199–206 [DOI] [PubMed] [Google Scholar]
- 33. LaCount DJ, Gruszynski AE, Grandgenett PM, Bangs JD, Donelson JE. 2003. Expression and function of the Trypanosoma brucei major surface protease (GP63) genes. J. Biol. Chem. 278:24658–24664 [DOI] [PubMed] [Google Scholar]
- 34. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 35. Macdonald MH, Morrison CJ, McMaster WR. 1995. Analysis of active site and activation mechanism of the Leishmania surface metalloproteinase GP63. Biochim. Biophys. Acta 1253:199–207 [DOI] [PubMed] [Google Scholar]
- 36. McGugan GC, Temesvari LA. 2003. Characterization of a Rab11-like GTPase, EhRab11, of Entamoeba histolytica. Mol. Biochem. Parasitol. 129:137–146 [DOI] [PubMed] [Google Scholar]
- 37. McHugh B, et al. 2004. Invadolysin: a novel, conserved metalloprotease links mitotic structural rearrangements with cell migration. J. Cell Biol. 167:673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moncada D, Keller K, Ankri S, Mirelman D, Chadee K. 2006. Antisense inhibition of Entamoeba histolytica cysteine proteases inhibits colonic mucus degradation. Gastroenterology 130:721–730 [DOI] [PubMed] [Google Scholar]
- 39. Moncada D, Keller K, Chadee K. 2003. Entamoeba histolytica cysteine proteinases disrupt the polymeric structure of colonic mucin and alter its protective function. Infect. Immun. 71:838–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mosser DM, Brittingham A. 1997. Leishmania, macrophages and complement: a tale of subversion and exploitation. Parasitology 115:S9–S23 [DOI] [PubMed] [Google Scholar]
- 41. Mueller DE, Petri WA., Jr 1995. Clonal growth in Petri dishes of Entamoeba histolytica. Trans. R. Soc. Trop. Med. Hyg. 89:123. [DOI] [PubMed] [Google Scholar]
- 42. Petri WA, Jr, Smith RD, Schlesinger PH, Murphy CF, Ravdin JI. 1987. Isolation of the galactose-binding lectin that mediates the in vitro adherence of Entamoeba histolytica. J. Clin. Invest. 80:1238–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Que X, Reed SL. 2000. Cysteine proteinases and the pathogenesis of amebiasis. Clin. Microbiol. Rev. 13:196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ravdin JI, Guerrant RL. 1981. Role of adherence in cytopathic mechanisms of Entamoeba histolytica. Study with mammalian tissue culture cells and human erythrocytes. J. Clin. Invest. 68:1305–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rawlings ND, Barrett AJ. 1993. Evolutionary families of peptidases. Biochem. J. 15:205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rawlings ND, Barrett AJ, Bateman A. 2010. MEROPS: the peptidase database. Nucleic Acids Res. 38:D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reed SL, Keene WE, McKerrow JH. 1989. Thiol proteinase expression correlates with pathogenicity of Entamoeba histolytica. J. Clin. Microbiol. 27:2772–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sabeh F, Shimizu-Hirota R, Weiss SJ. 2009. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 185:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shrimal S, Bhattacharya S, Bhattacharya A. 2010. Serum-dependent selective expression of EhTMKB1-9, a member of Entamoeba histolytica B1 family of transmembrane kinases. PLoS Pathog. 6:e1000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stanley SL, Jr, Tian K, Koester JP, Li E. 1995. The serine-rich Entamoeba histolytica protein is a phosphorylated membrane protein containing O-linked terminal N-acetylglucosamine residues. J. Biol. Chem. 270:4121–4126 [DOI] [PubMed] [Google Scholar]
- 51. Teixeira JE, Huston CD. 2008. Participation of the serine-rich Entamoeba histolytica protein in amebic phagocytosis of apoptotic host cells. Infect. Immun. 76:959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: improving sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Van Wart HE, Birkedal-Hansen H. 1990. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. U. S. A. 87:5578–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Worbs T, Mempel TR, Bolter J, von Andrian UH, Forster R. 2007. CCR7 ligands stimulate the intranodal motility of T lymphocytes in vivo. J. Exp. Med. 204:489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yao C, Donelson JE, Wilson ME. 2003. The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol. Biochem. Parasitol. 132:1–16 [DOI] [PubMed] [Google Scholar]
- 56. Zhang X, et al. 2004. Expression of amoebapores is required for full expression of Entamoeba histolytica virulence in amebic liver abscess but is not necessary for the induction of inflammation or tissue damage in amebic colitis. Infect. Immun. 72:678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.