Abstract
Phagocytosis of host cells is characteristic of tissue invasion by the intestinal ameba Entamoeba histolytica, which causes amebic dysentery and liver abscesses. Entamoeba histolytica induces host cell apoptosis and uses ligands, including C1q, on apoptotic cells to engulf them. Two mass spectrometry analyses identified calreticulin in amebic phagosome preparations, and, in addition to its function as an endoplasmic reticulum chaperone, calreticulin is believed to be the macrophage receptor for C1q. The purpose of this study was to determine if calreticulin functions as an E. histolytica C1q receptor during phagocytosis of host cells. Calreticulin was localized to the surface of E. histolytica during interaction with both Jurkat lymphocytes and erythrocytes and was present in over 75% of phagocytic cups during amebic erythrophagocytosis. Presence of calreticulin on the cell surface was further demonstrated using a method that selectively biotinylated cell surface proteins and by flow cytometry using trophozoites overexpressing epitope-tagged calreticulin. Regulated overexpression of calreticulin increased E. histolytica's ability to phagocytose apoptotic lymphocytes and calcium ionophore-treated erythrocytes but had no effect on amebic adherence to or destruction of cell monolayers or surface expression of the GalNAc lectin and serine-rich E. histolytica protein (SREHP) receptors. Finally, E. histolytica calreticulin bound specifically to apoptotic lymphocytes and to human C1q. Collectively, these data implicate cell surface calreticulin as a receptor for C1q during E. histolytica phagocytosis of host cells.
INTRODUCTION
Entamoeba histolytica is the protozoan parasite that causes amebiasis, a disease characterized by dysentery and, more rarely, amebic liver abscess. It is estimated to cause about 100,000 deaths annually (55). Dysentery occurs following invasion of the colonic epithelium by E. histolytica trophozoites. Prominent pathological features of invasive disease include tissue destruction, acute inflammation, and amebic phagocytosis of human erythrocytes and immune cells (14, 26, 31, 46).
Amebic trophozoites phagocytose apoptotic cells more efficiently than live or necrotic cells, and a sequential model of cell killing and phagocytosis has been proposed (29). Trophozoites first adhere to host target cells, a step that is predominantly mediated by a d-galactose/N-acetyl-d-galactosamine (d-Gal/GalNAc)-specific surface lectin (42). Adherence is followed by contact-dependent cell killing (44). The mechanism of cell killing remains unclear, but secretion of amebic pore-forming peptides, an influx of host cytoplasmic Ca2+, production of reactive oxygen species, and activation of host cell phosphatases and caspase-3 all appear to contribute (30, 32, 47, 52). Finally, E. histolytica phagocytoses the dying cell (29). Phosphatidylserine is exposed on the plasma membrane outer leaflet early during apoptotic cell death and following Ca2+ ionophore treatment of erythrocytes, and phosphatidylserine is a ligand that stimulates E. histolytica phagocytosis of apoptotic lymphocytes and damaged erythrocytes (6, 10, 18, 29). Human C1q and the related collectin mannose binding lectin (MBL), both of which bind to apoptotic cells and trigger macrophage phagocytosis, are additional ligands that stimulate E. histolytica phagocytosis (39, 49). A number of E. histolytica cell surface proteins have been implicated in adherence and phagocytosis, including the GalNAc-specific adherence lectin (42), the serine-rich E. histolytica protein (SREHP) (51), an as yet unidentified mannose binding lectin (11), a phagosome-associated transmembrane kinase (TMK96 or PATMK) (9), and the EhCPAdh complex, which contains a cysteine protease (CP) and an adhesin (Adh) (3). However, the amebic C1q/collectin receptor remains unidentified.
In an effort to identify additional phagocytosis receptors, several groups have isolated E. histolytica phagosomes after incubation with either latex or paramagnetic beads and used mass spectrometry to identify proteins present in amebic phagosomes (9, 36, 40). Two of three groups using this approach found the protein calreticulin (AmoebaDB accession number EHI_136160) at early time points within E. histolytica phagosomes (9, 36). Although best known as a chaperone protein located in the endoplasmic reticulum (ER) lumen, several studies have implicated cell surface calreticulin as the human receptor for C1q and the collectins (34, 48). Calreticulin functions in complex with CD91 during macrophage phagocytosis of apoptotic cells opsonized with either C1q or MBL (39). In addition, calreticulin can bind directly to apoptotic cells and enable C1q/collectin-independent macrophage phagocytosis (21).
Little is known about E. histolytica calreticulin. Its predicted amino acid sequence is 53% identical and 70% similar to human calreticulin (25). In addition to identification in amebic phagosome preparations (9, 36), E. histolytica calreticulin is known to be immunogenic during human infection and has been shown to colocalize with a resident ER protein, protein disulfide isomerase (PDI) (24, 25). Calreticulin has also been demonstrated within the uroid (i.e., tail region) on the E. histolytica cell surface following capping of surface receptors with the lectin concanavalin A (23). However, the physiologic relevance of this observation remains unclear.
Because calreticulin is present in purified amebic phagosomes and is a known macrophage receptor for C1q and phagocytosis of apoptotic cells, we hypothesized that calreticulin is an E. histolytica receptor that functions in phagocytosis of apoptotic cells and erythrocytes. Here, we present data demonstrating that E. histolytica calreticulin is recruited to the cell surface and localizes to the phagocytic cup during interaction with lymphocytes and erythrocytes. Regulated overexpression of calreticulin significantly increases E. histolytica phagocytosis of apoptotic lymphocytes and Ca2+ ionophore-treated erythrocytes, and E. histolytica calreticulin binds specifically to both apoptotic cells and to human C1q. Our data suggest that cell surface calreticulin is an E. histolytica receptor for C1q that facilitates phagocytosis of host cells.
MATERIALS AND METHODS
Cell lines and culture.
Entamoeba histolytica trophozoites (strain HM-1:IMSS) were grown in TYI-S-33 medium supplemented with 100 U of penicillin/ml and 100 μg/ml streptomycin sulfate (17). For all experiments, amebic trophozoites were harvested from nonconfluent cultures as previously described (30).
The human leukemia T cell line Jurkat (clone E-6) was typically grown in RPMI medium (Gibco BRL) supplemented with 10% fetal bovine serum (FBS), 100 U of penicillin/ml, and 100 μg/ml of streptomycin. For experiments testing calreticulin binding, Jurkat cells were washed with phosphate-buffered saline (PBS) and subcultured for 48 h in serum-free AIM-V and GlutaMAX-I media (Gibco BRL) supplemented with 100 U/ml of penicillin and 100 μg/ml of streptomycin. Jurkat cell apoptosis was induced, where indicated in the text, by exposure to UV light for 15 min, followed by incubation at 37°C for 3 h (29). Fluorescent labeling of cells with carboxyfluorescein succinimidyl ester (CFSE; Invitrogen) was performed according to the manufacturer's protocol. Chinese hamster ovary (CHO) cells were grown in F-12K medium supplemented with 10% FBS, 100 U/ml of penicillin, and 100 μg/ml of streptomycin.
Fresh human erythrocytes were acquired from healthy human volunteers by phlebotomy after informed written consent was obtained. The study was reviewed and approved by the University of Vermont Institutional Review Board. Erythrocytes were isolated from whole blood by Ficoll-Hypaque centrifugation. For phagocytosis assays, erythrocytes were treated with the Ca2+ ionophore A23187 (at 2 μM for 30 min at 37°C) and then labeled with CFSE as above. This ionophore treatment induces phosphatidylserine exposure in >80% of erythrocytes, as determined by annexin V-fluorescein isothiocyanate (FITC) staining and flow cytometry (37; also data not shown).
Production of recombinant E. histolytica calreticulin.
The E. histolytica calreticulin gene was amplified from genomic DNA by PCR using the primers F5′-AAGGTTTATTTCCATGAAACA-3′ and R5′-TTAAAGCTCTTCTTTGTTTTC-3′ and cloned by TOPO-TA cloning into the expression vector pCR-T7/TOPO/NT (Invitrogen), which incorporates a six-histidine N-terminal tag. For protein production, a 500-ml culture of transformed Escherichia coli strain BL21(DE3)pLysS was grown to an optical density at 600 nm (OD600) of 0.5, protein synthesis was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at room temperature, and then the cell pellet was resuspended in buffer containing 20 mM phosphate, 500 mM NaCl, and 20 mM imidazole (pH 7.4) and lysed by sonication. Recombinant calreticulin was purified using nickel resin and a His-Trap kit (GE Healthcare) according to the manufacturer's protocol. Bound protein was washed with 10 ml of lysis buffer and then eluted with 20 mM phosphate, 500 mM NaCl, and 500 mM imidazole (pH 7.4). Protein purity was assessed by image densitometry of Coomassie-stained gels using NIH ImageJ software.
Antibodies.
The anti-E. histolytica calreticulin recombinant monoclonal single-chain Fv (scFv) antibody (O9C clone) was selected using phage display from two phagemid libraries (Tomlinson I and J) provided by the Medical Research Council (MRC) Centre for Protein Engineering (Cambridge, United Kingdom) and using the MRC Centre's protocol (28, 38, 56). Antigen-specific phage were selected by panning against recombinant E. histolytica calreticulin, followed by selection of monoclonal phage with calreticulin binding ability, production of monoclonal O9C scFv peptide in E. coli (strain HB2151), and purification using protein A-agarose (GE Healthcare).
Anti-E. histolytica calreticulin polyclonal rabbit serum was a gift from T. Nozaki (National Institute of Infectious Diseases, Tokyo, Japan) and was affinity purified using recombinant E. histolytica calreticulin immobilized on agarose resin. Rabbit anti-E. histolytica heat shock protein 70 (HSP-70) serum and rabbit anti-inhibitor of cysteine protease 1 (ICP1) polyclonal serum were also gifts from T. Nozaki (45). Rabbit anti-GalNAc lectin polyclonal IgG was a gift from W. A. Petri (University of Virginia, Charlottesville, VA) (35). Rabbit anti-human ADP ribosylating factor 1 (Arf1) polyclonal IgG was a gift from V. Hsu (Brigham and Women's Hospital, Boston, MA) and has been used previously to immuno-stain the E. histolytica Golgi apparatus (22, 53). We described the anti-SREHP mouse monoclonal IgG (10D11) previously (51). The anti-FLAG mouse monoclonal antibody (M2 clone) was purchased from Stratagene, and the goat polyclonal anti-C1q IgG antibody was purchased from Quidel Corporation.
Expression of epitope-tagged proteins in E. histolytica.
The E. histolytica calreticulin (AmoebaDB accession number EHI_136160) and protein disulfide isomerase (PDI) (AmoebaDB accession number EHI_071590) genes were constitutively expressed in E. histolytica under the control of the GalNAc lectin promoter. For this, both the calreticulin and PDI genes beginning distal to the sequence encoding their N-terminal signal peptides were amplified by PCR with addition of 5′ HindIII and 3′ SalI restriction sites. The amplified fragments were then digested and ligated into a pRSETC vector that already contained sequences encoding the GalNAc lectin signal peptide and a FLAG epitope tag between the BamHI and HindIII sites (43). The resultant fusion genes were then subcloned into the neo′ expression vector pGir235 using the BamHI and SalI restriction sites (43). The primers for PCR amplification of calreticulin were F5′-CTACTGAAGCTTAAGGTTTATTTCCATGAAACA-3′ and R5′-CTACTGGTCGACTTAAAGCTCTTCTTTGTTTTC-3′. The primers for PCR amplification of PDI were F5′-CTACTGAAGCTTGATGTAGTATCATTAAATCCA-3′ and R5′-CTACTGGTCGACTTAGAAAACTTCAAGTACATT-3′. For tetracycline-regulated expression of FLAG-calreticulin, the fusion gene was reamplified from pRSETC using the primers F5′-CACCGGTACCATGAAATTATTATTA-3′ and R5′-CTACTGGGATCCTTAAAGCTCTTCTTTGTTTTC-3′ to add 5′ KpnI and 3′ BamHI restriction sites, and the PCR product was ligated into the vector pEhHYG-tetR-O-Cass downstream of the tetracycline-inducible promoter (27).
Entamoeba histolytica trophozoites were transfected as previously described using Attractene transfection reagent (Qiagen) and 20 μg of plasmid DNA (15). Transfected trophozoites were selected with either 1.5 μg/ml G418 (pGir235) or 3.25 μg/ml hygromycin (pEhHYG-tetR-O-Cass) beginning 24 h after transfection, and the drug dosages were doubled weekly to final concentrations of 6 μg/ml G418 or 26 μg/ml hygromycin prior to experimentation. For inducible expression of FLAG-calreticulin, transfected trophozoites were treated overnight with 20 μg/ml of tetracycline.
Immunofluorescence microscopy and flow cytometry.
To prepare microscopy slides, E. histolytica trophozoites in TYI-S-33 medium were allowed to adhere to sterile glass coverslips for 15 min at 37°C. For phagocytosis assays, host cells were then added (2 Jurkat cells/ameba or 1,000 red blood cells [RBC]/ameba), and samples were incubated for an additional 20 min at 37°C. Cells were then fixed, permeabilized or not with 0.2% Triton X-100 in PBS, blocked, and stained with primary and secondary detection reagents diluted as follows: anti-calreticulin scFv monoclonal, 10 μg/ml; rabbit polyclonal anti-GalNAc lectin IgG, 1:100; phalloidin-Alexa Fluor 633, 1:40 (Invitrogen); rabbit polyclonal anti-HSP-70 serum, 1:200; mouse monoclonal anti-FLAG IgG, 1:1,000; and mouse monoclonal anti-SREHP IgG (clone 10D11), 40 μg/ml; All secondary antibodies were purchased from Invitrogen and used at a 1:1,000 dilution. For immunofluorescence microscopy of calreticulin using the scFv monoclonal antibody, a kit for tyramide signal amplification (Invitrogen) was used according to the manufacturer's protocol to enhance the fluorescent signal. After incubation with the scFv monoclonal, protein A-horseradish peroxidase (HRP) (GE Healthcare) was used for secondary detection, and then a tyramide-Alexa Fluor 488 conjugate was used as the substrate. As indicated in the figure legends, images were acquired using either a Zeiss 510 Meta confocal microscope and a 60× (1.4 numerical aperture [NA]) oil immersion objective or a Nikon Eclipse Ti2000 microscope with a 60× (1.4 NA) oil immersion objective, a Z-spacing of 0.26 μm, and a Qimaging ExiBlue digital camera; Autoquant X software (MediaCybernetics) and 45 iterations were used for image deconvolution.
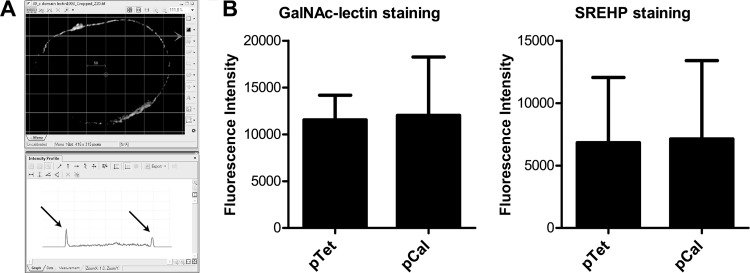
To determine if overexpressing calreticulin altered the quantity of the SREHP and GalNAc lectin per area of cell membrane, trophozoites transfected with either the pEhHYG-tetR-FLAG-calreticulin or empty control plasmid were induced overnight with tetracycline, and then microscopy slides were stained as above for either protein without detergent permeabilization. Z-stack images of at least 10 cells per condition were acquired, and one Z-slice corresponding to the midpoint of each cell was analyzed by measuring the fluorescence intensity at 10 points on the cell membrane with the intensity profile tool in the Nikon NIS Elements software package. Locations to measure the cell membrane fluorescence were arbitrarily selected by overlaying a grid template (8-μm line density) onto the images and measuring the intensity at points where the template crossed the cell membrane (see Fig. 5A).
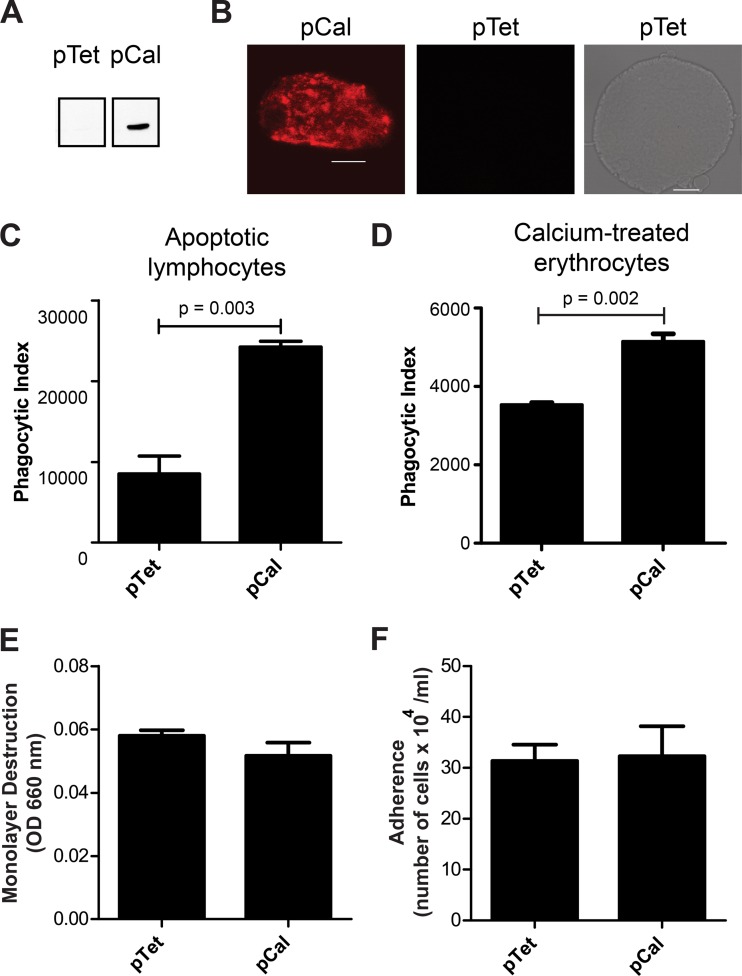
Fig 5.
Tetracycline-regulated overexpression of calreticulin does not alter surface levels of the GalNAc lectin or SREHP. (A) Method for measurement of cell membrane fluorescence intensity. Nonpermeabilized trophozoites were immunostained for the GalNAc lectin or SREHP prior to acquisition of image Z-stacks, image deconvolution, and quantification of fluorescence intensity. Cell membrane locations were arbitrarily selected using an 8-μm density grid template overlaid onto each image (top), and fluorescence intensity was measured where the grid crossed the cell membrane (arrows in the lower screen shot). (B) GalNAc lectin and SREHP surface staining. Each bar graph shows the mean fluorescence and SD for 100 data points measured for cells induced to express calreticulin (pCal) and cells transfected with the empty control vector and treated with tetracycline (10 data points per cell and 10 cells per transfectant).
All flow cytometry was performed using a Coulter EPICS XL-MCL flow cytometer. The FLAG-calreticulin and FLAG-PDI proteins were stained after trophozoites expressing each fusion protein were incubated in medium 199 (Gibco BRL) supplemented with 5.7 mM cysteine, 25 mM HEPES, and 0.5% bovine serum albumin (BSA) at pH 6.8 (M199s medium), either alone or in the presence of Jurkat lymphocytes as described previously. Cells were then washed twice with 111 mM d-galactose in PBS (d-Gal–PBS) to disrupt ameba-host cell rosettes, fixed, and stained with an anti-FLAG mouse monoclonal IgG antibody (diluted 1:1,200 in blocking buffer) and an anti-mouse IgG FITC conjugate (Sigma-Aldrich). To assess calreticulin binding to host cells, recombinant calreticulin was biotinylated using sulfo-NHS-LC-biotin (sulfosuccinimidyl-6-biotinamido-hexanoate; Pierce Biotechnology) according to the manufacturer's protocol and used to stain healthy and apoptotic Jurkat lymphocytes grown in serum-free medium. Healthy and apoptotic cells were fixed with 4% paraformaldehyde, blocked with a streptavidin-biotin blocking reagent (Vector Laboratories), and stained for flow cytometry using 30 μg/ml of biotin-calreticulin and streptavidin-Alexa Fluor 488 (2 μg/ml) (Invitrogen) for secondary detection.
Assays for phagocytosis, tissue culture monolayer destruction, and adherence.
Phagocytosis of apoptotic Jurkat lymphocytes and Ca2+ ionophore-treated erythrocytes was assayed as previously described (29). Trophozoites and either CFSE-labeled apoptotic Jurkat lymphocytes (ameba/Jurkat ratio, 1:5) or Ca2+ ionophore-treated erythrocytes (ameba/erythrocyte ratio, 1:20) were mixed in PBS with 111 mM d-galactose (d-Gal–PBS), centrifuged, and incubated at 37°C for either 15 min (lymphocytes) or 10 min (erythrocytes). The pellets were washed with cold d-Gal–PBS, fixed, and washed two more times, and amebic fluorescence was determined by flow cytometry. Trophozoites with fluorescence above background were scored as phagocytic. The data were expressed as the phagocytic index, defined as the mean fluorescence of phagocytic trophozoites times the percentage of phagocytic trophozoites (19, 29).
The tissue culture cell monolayer destruction assay was described previously (12). A total of 5 × 104 amebic trophozoites in a total volume of 1 ml of M199s medium were added to confluent CHO cell monolayers grown in 24-well plates and incubated for 3 h at 37°C. Detached CHO cells and adherent amebae were then removed by incubation of the plates on ice for 15 min and two washes with PBS. The remaining CHO cell monolayer was stained with 0.1 mg/ml methylene blue in 100 mM borate buffer (pH 8), followed by washing with 100 mM borate buffer. The stain was then extracted with 1 ml of 1N HCl at 37°C for 30 min, and absorbance was measured at 660 nm using a spectrophotometer.
Adherence of trophozoites to fixed CHO cell monolayers was assayed essentially as previously described (41). Confluent CHO cell monolayers grown in 24-well plates were washed two times with PBS, fixed, and treated with 250 mM glycine for 30 min at room temperature. After the monolayers were washed, 2 × 105 trophozoites suspended in 500 μl of M199s medium were added to each well. Amebae were allowed to adhere at 37°C for 30 min, the medium was decanted, and the plates were washed gently two more times with warm M199s medium. Adherent trophozoites were then harvested by addition of 500 μl of ice-cold M199s medium supplemented with 111 mM d-Gal to each well, incubated on ice for 15 min, resuspended in 100 μl, and counted using a hemocytometer.
Biochemical labeling of cell surface calreticulin and coimmunoprecipitation of C1q.
To assess the presence of surface calreticulin biochemically, 5 × 104 E. histolytica trophozoites were resuspended with Jurkat lymphocytes (1:6, ameba/host ratio) in PBS containing 2 mg/ml sulfo-NHS-LC-biotin and 111 mM d-galactose. The suspension was centrifuged and incubated at 37°C for 5 min. The pellets were then washed, lysed using 1% Triton X-100 in 50 mM Tris-HCl (pH 7.4) and 300 mM NaCl containing 800 μM AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride], 50 μM bestatin, 0.4 μM leupeptin, 0.15 μM aprotinin, 0.6 μM E-64, and 400 μM EDTA. Biotinylated proteins were precipitated using streptavidin-agarose beads (Zymed) and analyzed by Western blotting performed using polyclonal rabbit anti-calreticulin IgG (5 μg/ml), rabbit polyclonal anti-GalNAc lectin IgG (1:5,000), and rabbit polyclonal anti-ICP1 IgG (1:1,000) antibodies for primary detection, and developed with an anti-rabbit IgG-HRP (1:10,000) (GE Healthcare) and enhanced chemiluminescence.
For coimmunoprecipitation of FLAG-tagged calreticulin and C1q, apoptotic Jurkat lymphocytes were pretreated with human C1q (Quidel Corporation) (for 30 min at 37°C) and then incubated with either FLAG-calreticulin- or FLAG-PDI-expressing trophozoites, as indicated in Fig. 6D, before lysis and immunoprecipitation with a mouse anti-FLAG monoclonal IgG antibody. Precipitated proteins from an equal number of trophozoites were separated by SDS-PAGE and analyzed by Western blotting, probed first with a goat anti-C1q polyclonal IgG antibody and then stripped and reprobed with the anti-FLAG antibody.
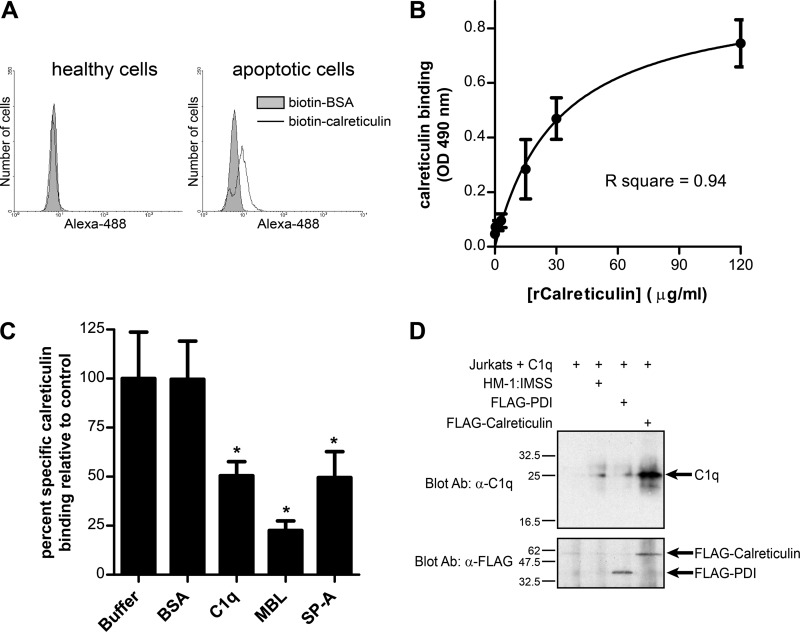
Fig 6.
E. histolytica calreticulin binds to apoptotic lymphocytes and to human C1q. (A) Calreticulin binds specifically to apoptotic lymphocytes, independent of C1q. Healthy and apoptotic Jurkat lymphocytes cultured in serum-free medium were stained for flow cytometry using biotin-calreticulin or biotin-BSA (negative control) and streptavidin-Alexa Fluor 488. Representative histograms of amebic fluorescence are shown. (B) Calreticulin binds directly to human C1q. Recombinant calreticulin (rCalreticulin) binding to immobilized C1q was measured by ELISA using biotinylated anti-calreticulin scFv and streptavidin-HRP. The graph shows a single-site specific binding curve for binding of the indicated concentration of calreticulin, generated by nonlinear regression (mean and SD; n = 3). (C) Specific inhibition of calreticulin-C1q binding by free C1q and collectins. Binding assays were performed in the presence of 3-fold molar excess BSA (negative control), C1q, MBL, and SP-A. Calreticulin binding was expressed as the percentage of specific binding measured in buffer alone (mean and SD; n = 3). *, P < 0.01 compared to buffer control by one-way ANOVA with Bonferroni correction. (D) C1q coimmunoprecipitates with FLAG-calreticulin following interaction of E. histolytica with C1q-coated apoptotic lymphocytes. Apoptotic Jurkat lymphocytes were coated with human C1q and incubated with nontransfected HM-1:IMSS trophozoites or trophozoites expressing FLAG-PDI or FLAG-calreticulin (30 min at 37°C; ameba/host cell ratio, 1.3:1) before lysis and immunoprecipitation with an anti-FLAG antibody. The top panel is an anti-C1q Western blot showing coprecipitated C1q. The bottom panel is the same membrane stripped and reprobed with the anti-FLAG monoclonal antibody. The precipitants from an equal number of amebae were run in each lane.
Calreticulin binding ELISA.
Enzyme-linked immunosorbent assay (ELISA) plates were coated with C1q (1.5 μg/well), followed by blocking with 3% BSA in PBS and addition of recombinant E. histolytica calreticulin at the concentration indicated in Fig. 6B in 20 mM Tris-HCl with 10 mM K2HPO4 and 10 mM CaCl2. Calreticulin was allowed to bind for 1 h at room temperature, after which the plates were washed, and bound calreticulin was detected with biotinylated anti-calreticulin scFv antibody (0.7 μg/ml) and streptavidin-HRP (1:1,000). The ELISAs were developed with o-phenylenediamine dihydrochloride (OPD; 15 min at room temperature), and absorbance was measured at 490 nm. For competition experiments, calreticulin was added to C1q-coated wells at a concentration of 30 μg/ml in the presence of excess (1:3 molar ratio) unbound BSA, C1q, MBL, and surfactant protein A (SP-A). Nonspecific binding was measured as binding of the anti-calreticulin antibody and streptavidin-HRP to immobilized C1q in the absence of calreticulin. Specific binding was calculated as the experimental signal minus the mean nonspecific binding, and all results were expressed as a percentage of calreticulin binding measured in buffer alone.
Figure preparation and statistics.
All data are representative of at least three independent experiments. Images were prepared for publication using Zeiss LSM software for confocal images or Nikon Elements software for deconvolved image stacks. All flow cytometry data were analyzed using WinMDI software (version 2.9). All quantitative data were expressed as the mean and standard deviation (SD). Statistical analyses and graphs were prepared using GraphPad Prism, version 5, software. Statistical significance was determined with an unpaired Student's t test or, in instances with multiple groups for comparison, with a one-way analysis of variance (ANOVA) and Bonferroni correction for multiple comparisons. P values of ≤0.05 were considered statistically significant. Final figures were assembled using Adobe Illustrator CS3.
RESULTS
Calreticulin is concentrated on the cell surface within the phagocytic cup during interaction with Jurkat lymphocytes and erythrocytes.
We began testing if calreticulin might function as an E. histolytica phagocytosis receptor by localizing it during amebic interaction with Jurkat lymphocytes and erythrocytes. First, we selected a recombinant monoclonal single-chain Fv (scFv) antibody specific for calreticulin using an scFv phage library and recombinant E. histolytica calreticulin produced in Escherichia coli as bait (38). We also obtained and affinity purified a rabbit polyclonal antibody specific for E. histolytica calreticulin. Both antibodies were specific for E. histolytica calreticulin, as assessed by Western blotting (Fig. 1A). The scFv reagent worked well for immunofluorescence microscopy, and it stained a reticular network that was enriched near the cell membrane and was consistent with a previously described E. histolytica ER compartment (Fig. 1B) (50). The calreticulin staining colocalized with staining of the resident ER protein BiP. No signal was detected in the absence of the anti-calreticulin scFv antibody (data not shown).
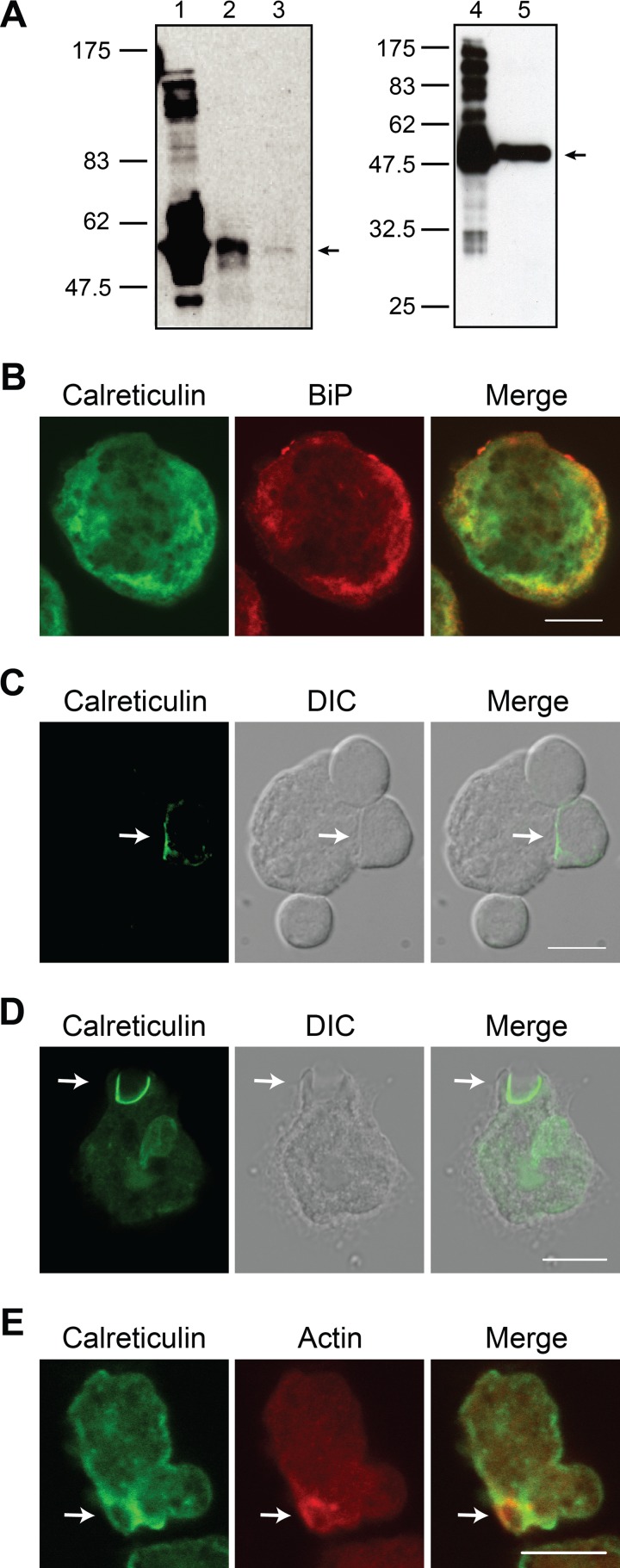
Fig 1.
Calreticulin is present on the cell surface within the phagocytic cup during E. histolytica phagocytosis of Jurkat lymphocytes and erythrocytes. (A) Immunoblots confirming the specificity of an scFv recombinant monoclonal antibody (left) and an affinity-purified rabbit polyclonal IgG antibody (right) for E. histolytica calreticulin. Gel lanes were loaded as follows: lane 1, recombinant calreticulin; lane 2, lysate from FLAG-calreticulin-expressing trophozoites; lane 3, E. histolytica lysate; lane 4, recombinant calreticulin; lane 5, E. histolytica lysate. (B) Confocal microscopy showing localization of calreticulin (green) and the ER marker BiP (red) in unstimulated E. histolytica trophozoites. (C and D) Immunofluorescence microscopy showing cell surface calreticulin in the phagocytic cup of nonpermeabilized amebae during phagocytosis of Jurkat lymphocytes (C) and erythrocytes (D). Trophozoites adhered to coverslips were incubated with either healthy Jurkat cells (10 min, 37°C) or erythrocytes (20 min, 37°C) and then immunostained for calreticulin (green) without membrane permeabilization using anti-calreticulin scFv antibody. Images were acquired using either an epifluorescence microscope and image deconvolution (C) or a confocal microscope (D). The arrows indicate partially phagocytosed host cells. (E) Confocal microscopy showing colocalization of calreticulin and polymerized F-actin during erythrophagocytosis. Trophozoites adhered to coverslips were incubated with erythrocytes (20 min, 37°C), permeabilized with detergent, and then stained for calreticulin using anti-calreticulin scFv antibody (green) and polymerized F-actin using Alexa Fluor 633-phalloidin (red). The arrow indicates a phagocytic cup highlighted by polymerized F-actin. Scale bar, 10 μm. DIC, differential interference contrast.
To assess calreticulin localization during phagocytosis and to determine whether the protein was localized on the cell surface where it might function as a phagocytosis receptor, trophozoites that were adhered to coverslips were incubated with healthy Jurkat lymphocytes and immunostained without membrane permeabilization using the scFv anti-calreticulin antibody. Calreticulin was readily localized to the ameba-Jurkat cell synapse within a subset of phagocytic cups (Fig. 1C). Similarly, intense calreticulin staining was demonstrated at the junction with partially phagocytosed erythrocytes (Fig. 1D). However, calreticulin was not detected at all ameba-host cell interfaces (Fig. 1C), leading us to seek a method to positively identify bona fide phagocytic cups so that we could quantify the frequency of calreticulin recruitment. Phalloidin has been used previously to label polymerized F-actin as a marker for E. histolytica phagocytic cups (5) and was used as a costain for this purpose. Strong calreticulin staining colocalized with the phalloidin staining and further indicated that there was translocation of calreticulin to the site of phagocytosis (Fig. 1E). Presence of polymerized F-actin (i.e., localized phalloidin staining) was used to identify phagocytic cups, which were then scored for the presence or absence of calreticulin; 25 of 33 phagocytic cups scored (76%) were positive for calreticulin. In contrast, calreticulin was rarely identified surrounding fully engulfed lymphocytes or erythrocytes (data not shown).
Calreticulin becomes exposed during E. histolytica interaction with healthy lymphocytes and is also present on the surface of unstimulated trophozoites.
The intense calreticulin staining observed on the cell surface within the phagocytic cup could have been due to directed delivery of calreticulin to the cell membrane during phagocytosis, relocalization of calreticulin already present on the cell surface to the site of host cell contact, or both. Consistent with the possibility of directed delivery, no native calreticulin could be detected on the surface of unstimulated trophozoites by using the anti-calreticulin antibodies for immunofluorescence microscopy or flow cytometry (data not shown). We used flow cytometry and trophozoites engineered to overexpress FLAG epitope-tagged calreticulin to further examine if calreticulin is present on the surface of trophozoites at rest or following stimulation with host cells. Trophozoites overexpressing FLAG epitope-tagged protein disulfide isomerase (PDI), an irrelevant ER protein, were used as control cells. For this, the E. histolytica calreticulin or PDI genes were constitutively expressed as fusion proteins comprised of the N-terminal signal peptide from the GalNAc lectin followed by a FLAG epitope and the protein of interest (Fig. 2A). Expression of the FLAG epitope-tagged proteins was confirmed by Western blotting (Fig. 2B). The overexpressed FLAG-calreticulin and FLAG-PDI proteins both colocalized with the ER marker BiP, and absence of colocalization with the Golgi complex marker ADP-ribosylating factor 1 (Arf1) indicated that both epitope-tagged proteins were appropriately retained within the ER compartment (Fig. 2C and D).
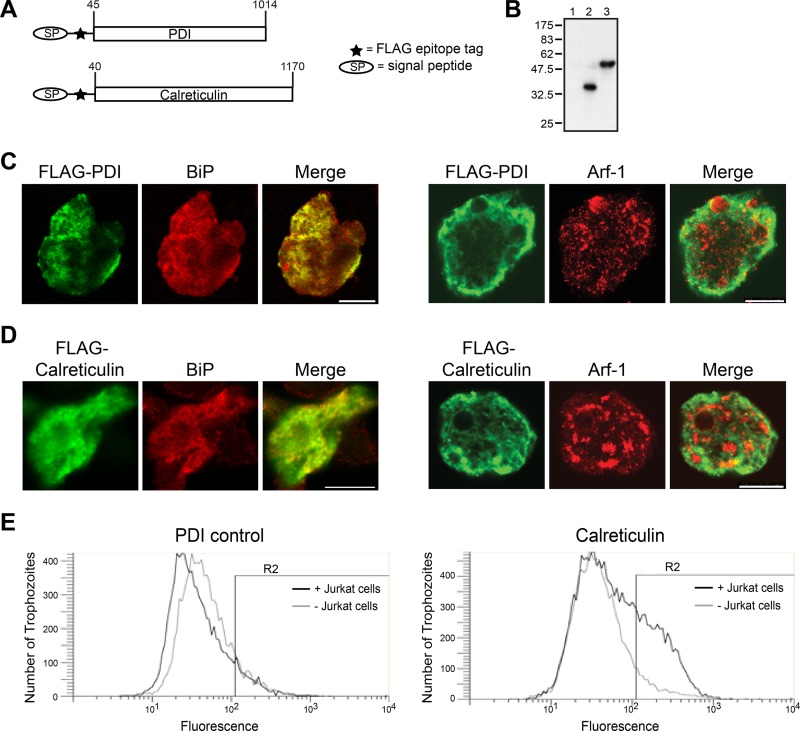
Fig 2.
FLAG epitope-tagged calreticulin becomes exposed on the surface of E. histolytica trophozoites during interaction with healthy T lymphocytes. (A) Schematic describing two FLAG-tagged fusion proteins stably expressed in E. histolytica trophozoites. (B) Immunoblot confirming expression of the FLAG-calreticulin and FLAG-PDI fusion proteins. Whole amebic lysates were probed with an anti-FLAG monoclonal antibody: lane 1, no vector control; lane 2, FLAG-PDI; lane 3, FLAG-calreticulin. (C and D) Confocal microscopy showing localization of the expressed fusion proteins in E. histolytica trophozoites. FLAG-PDI- and FLAG-calreticulin-expressing cells were stained with an anti-FLAG monoclonal antibody (green) and for either the ER marker BiP or the Golgi marker Arf1 (red). Both fusion proteins were retained within the ER. Scale bars, 10 μm. (E) FLAG-calreticulin became exposed on the amebic surface during interaction with healthy T lymphocytes. Amebae expressing FLAG-calreticulin or FLAG-PDI were incubated alone or with healthy Jurkat lymphocytes (45 min at 37°C; ameba/host ratio of 1:2), and exposed FLAG-calreticulin and FLAG-PDI were immunostained and analyzed by flow cytometry. Representative fluorescence-activated cell sorting histograms of amebic fluorescence are shown. The percentage of cells with exposed FLAG-calreticulin increased after incubation with lymphocytes as follows: 14.1% ± 2.2% for unstimulated FLAG-PDI control versus 13.7% ± 5.0% for unstimulated FLAG-calreticulin and 26.2% ± 2.8% for stimulated FLAG-calreticulin. (Values are mean percent and SD [n = 3]; P < 0.01 for stimulated FLAG-calreticulin compared to either control group by one-way ANOVA with a Bonferroni correction).
We next used immunofluorescence staining and flow cytometry to determine if either FLAG epitope-tagged protein could be detected on the surface of rophozoites at rest or following exposure to healthy Jurkat T lymphocytes. Both transfectants were incubated either with or without Jurkat lymphocytes, and after 45 min cells were stained for flow cytometry with an anti-FLAG antibody without membrane permeabilization (Fig. 2E). Nearly twice as many cells expressing the FLAG-calreticulin protein were positive for surface staining following stimulation with healthy Jurkat lymphocytes as either the unstimulated FLAG-calreticulin-expressing trophozoites or the unstimulated or stimulated FLAG-PDI control cells (Fig. 2E, R2 gates). There was no significant difference in surface staining of the FLAG-PDI and unstimulated FLAG-calreticulin cells, and incubation with lymphocytes had no significant effect on FLAG-PDI surface exposure. When cells were detergent permeabilized, essentially all of the FLAG-calreticulin and FLAG-PDI expressing cells stained, confirming the ability to detect both fusion proteins by this method (data not shown).
It was not possible to detect calreticulin on the surface of unstimulated trophozoites by immunofluorescence microscopy or flow cytometry, but it remained possible that calreticulin was present on the surface of unstimulated trophozoites at levels below the limits of detection for these methods. Thus, we employed a more sensitive biochemical approach using the membrane-impermeant biotinylation reagent sulfo-NHS-LC-biotin to test if native calreticulin was present on the cell surface (16). The rationale was that calreticulin would be biotinylated only if it was exposed on the cell surface or secreted. After incubation, biotinylated cell surface proteins were precipitated with streptavidin beads, and Western blotting was performed. The blots were simultaneously probed using antibodies specific for calreticulin, the cytosolic protein inhibitor of cysteine protease 1 (ICP1; negative control), and the GalNAc-specific cell surface lectin (positive control). Biotinylated calreticulin was precipitated by the streptavidin beads regardless of coincubation of amebae with lymphocytes (Fig. 3). As expected, the GalNAc-specific lectin was precipitated with the streptavidin beads, confirming that the surface proteins were biotinylated and that the purification method worked. Although easily detected in the whole-cell lysate, the cytoplasmic protein ICP1 was not precipitated using the streptavidin beads, confirming that the cell membrane remained impermeant to sulfo-NHS-LC-biotin during the incubation. Finally, no bands were present following bead purification in the absence of sulfo-NHS-LC-biotin treatment, confirming that the streptavidin precipitation was specific.
Fig 3.
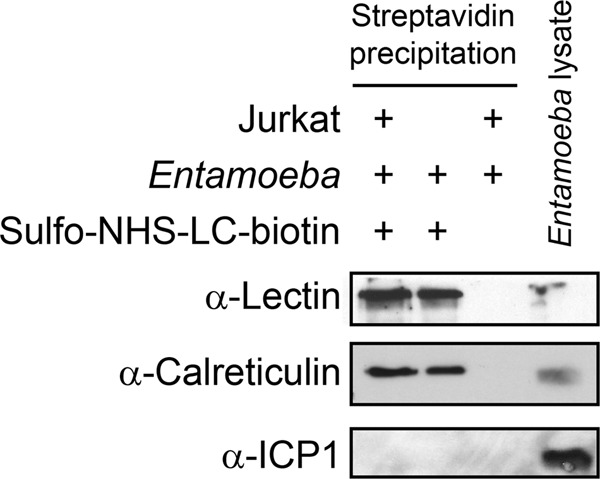
Calreticulin is also present on the surface of unstimulated trophozoites. Amebic trophozoites were incubated with or without healthy Jurkat lymphocytes (37°C for 5 min), and then surface proteins were specifically biotinylated with sulfo-NHS-LC-biotin. Biotinylated proteins were precipitated using streptavidin-coated beads and analyzed by Western blotting using antibodies specific for the GalNAc lectin (positive control), the cytosolic protein ICP1 (negative control), and calreticulin. Nonbiotinylated whole E. histolytica lysates were run as a positive control for the Western blot.
Regulated overexpression of calreticulin increases E. histolytica's phagocytic ability.
Repeated attempts to silence calreticulin gene expression by expression of antisense RNA and by RNA-mediated interference using both endogenous expression of short hairpin small interfering RNAs (siRNAs) and the G3 strain-based method were unsuccessful (data not shown) (1, 13, 33). Therefore, we turned to tetracycline-regulated overexpression of calreticulin in order to test if calreticulin expression affects E. histolytica phagocytosis by subcloning FLAG epitope-tagged calreticulin into the tetracycline-regulated expression vector pEhHYG-tetR-O-Cass (27). Cells transfected with the empty vector (pTet) were used as a control. Inducible expression of the FLAG epitope-tagged fragments was confirmed by Western blotting and immunofluorescence microscopy, which demonstrated a distribution of the recombinant protein that was indistinguishable from that seen for native calreticulin (Fig. 4A and B).
Fig 4.
Inducible overexpression of calreticulin increases phagocytic ability but does not affect tissue culture monolayer destruction or amebic adherence. (A) Immunoblot confirming tetracycline-induced expression of FLAG-calreticulin in E. histolytica. (B) Confocal microscopy showing localization of FLAG-calreticulin following overnight induction of expression with tetracycline. No staining of cells transfected with the empty vector control was detected. Scale bar, 10 μm. (C to F) Phenotypic assays comparing phagocytosis of apoptotic lymphocytes (C), phagocytosis of Ca2+ ionophore-treated erythrocytes (D), tissue culture monolayer destruction (E), and adherence to CHO cell monolayers (F) by trophozoites induced to express FLAG-calreticulin (pCal) and cells transfected with an empty control vector and treated with tetracycline (pTet). All graphs show means and SDs (n = 3). Induced expression of calreticulin significantly increased phagocytosis of both apoptotic lymphocytes and Ca2+ ionophore-treated erythrocytes (P = 0.003 and P = 0.002, respectively, versus empty vector control; Student's t tests).
Following overnight induction with tetracycline, phagocytosis assays using fluorescently labeled apoptotic Jurkat lymphocytes as targets were performed in the presence of 111 mM d-galactose to eliminate confounding effects from the GalNAc lectin. Parasites overexpressing FLAG-calreticulin phagocytosed apoptotic cells more than twice as efficiently as trophozoites transfected with the empty vector control (Fig. 4C). Similarly, Ca2+ ionophore treatment induces erythrocyte phosphatidylserine exposure (37), and trophozoites overexpressing FLAG-calreticulin phagocytosed Ca2+ ionophore-treated erythrocytes more efficiently than the empty vector control parasites (Fig. 4D). We also tested cell culture monolayer destruction and adherence. Overexpression of FLAG-calreticulin did not affect amebic destruction of Chinese hamster ovary (CHO) cell monolayers or adherence to CHO cell monolayers compared to trophozoites transfected with the control vector (Fig. 4E and F, respectively).
Because calreticulin is an ER chaperone protein, overexpression of calreticulin could alter expression of other cell surface proteins. We first assessed GalNAc lectin and SREHP surface staining using flow cytometry, but the analysis was complicated by wide culture-to-culture variability in the staining (data not shown). Therefore, we used immunofluorescence microscopy with image deconvolution to measure the mean fluorescence per area of cell membrane after staining the GalNAc lectin and SREHP on trophozoites overexpressing calreticulin (pCal) or transfected with the control vector (pTet) following incubation with tetracycline. The staining intensity for each antigen was measured at 10 cell membrane locations selected arbitrarily using a grid template for 10 cells per transfectant (Fig. 5A). When intensities were averaged, there was no difference in GalNAc lectin or SREHP surface staining between the transfectants (Fig. 5B). We concluded that expression of calreticulin enhanced E. histolytica phagocytosis but that the phenotype could not be attributed to increased chaperone activity resulting in altered expression of these known phagocytosis receptors on the amebic surface.
E. histolytica calreticulin binds to apoptotic cells and human C1q.
Human calreticulin has been found on the surface of apoptotic cells, where it acts in trans to trigger CD91-dependent macrophage phagocytosis that is independent of C1q (21). Therefore, we wondered if E. histolytica calreticulin binds directly to apoptotic cells. To test this possibility, we produced recombinant E. histolytica calreticulin with an N-terminal six-histidine tag in E. coli and used biotinylated recombinant calreticulin and streptavidin-Alexa Fluor 488 to assay calreticulin-biotin binding to healthy and apoptotic lymphocytes that were grown in serum-free (i.e., collectin- and C1q-free) medium. The recombinant calreticulin used was >90% pure (data not shown). As shown in Fig. 6A, E. histolytica calreticulin bound to apoptotic lymphocytes but not to healthy lymphocytes, while biotinylated bovine serum albumin (BSA) did not bind to either. Thus, E. histolytica calreticulin bound directly to apoptotic cells, using a mechanism that was independent of C1q and the collectins.
Since calreticulin also mediates C1q/collectin-dependent macrophage phagocytosis and since C1q stimulates E. histolytica phagocytosis of apoptotic cells (39, 49), we next tested if E. histolytica calreticulin also binds to human C1q. Recombinant calreticulin binding to immobilized C1q was detected using an ELISA. Calreticulin binding was readily detected at the lowest doses used and was saturable. After subtraction of nonspecific binding, the data fit a single-site ligand binding curve generated by nonlinear regression (Fig. 6B). Soluble C1q and the collectins MBL and surfactant protein A specifically inhibited calreticulin binding to immobilized C1q when included at 3-fold molar excess, further supporting the specificity of the interaction (Fig. 6C).
Finally, we performed coimmunoprecipitation experiments using the FLAG-calreticulin- and FLAG-PDI-expressing trophozoites to determine if cell surface calreticulin might interact with human C1q during interaction with host cells. Anti-FLAG immunoprecipitation was performed after incubation of E. histolytica trophozoites with apoptotic lymphocytes that were coated with human C1q. Roughly equal quantities of FLAG-calreticulin and FLAG-PDI were recovered (Fig. 6D, bottom panel). A small quantity of C1q was recovered with immunoprecipitation from untransfected and FLAG-PDI-expressing amebas. In contrast, much more C1q coprecipitated with FLAG-calreticulin (Fig. 6D, top panel). Collectively, these data indicated that calreticulin and C1q interact both in vitro and during incubation of E. histolytica with apoptotic lymphocytes.
DISCUSSION
C1q and mannose binding lectin (MBL) are chemoattractants for E. histolytica, and C1q stimulates E. histolytica phagocytosis when bound to apoptotic cells (49). The major conclusion of this study is that E. histolytica calreticulin functions as a receptor for C1q during amebic phagocytosis of host cells. Multiple lines of evidence support this conclusion. E. histolytica calreticulin, which is predominantly located in the ER, is enriched on the cell surface within the phagocytic cup during interaction of E. histolytica trophozoites with healthy Jurkat lymphocytes and erythrocytes. Localization of calreticulin to the phagocytic cup is early and transient since it was rarely detected in fully formed phagosomes (data not shown) but was observed in a high proportion of nascent phagosomes marked by actin polymerization. This is in accordance with previously published phagosome proteomics data, in which calreticulin was abundant only at early time points (9). Consistent with a functional role during phagocytosis, regulated overexpression of calreticulin increases amebic phagocytosis of apoptotic lymphocytes and Ca2+-treated erythrocytes. Overexpressing calreticulin does not affect amebic adherence to or destruction of tissue culture monolayers and does not alter expression of the GalNAc lectin or SREHP receptors on the cell surface, and both observations further support direct involvement of calreticulin in amebic phagocytosis rather than a secondary effect due to its chaperone function. Furthermore, E. histolytica calreticulin binds directly to human C1q. Calreticulin binding to C1q is saturable and specifically competed by C1q and the collectin family members SP-A and MBL, suggesting that E. histolytica calreticulin likely binds to the collagenous tail domain that is shared by members of this protein family. Like human calreticulin, E. histolytica calreticulin can also bind to apoptotic lymphocytes grown in serum-free medium in an apparently C1q/collectin-independent manner, but it does not bind to healthy cells.
Calreticulin has been observed on the cell surface in several other species and on the surface of E. histolytica following capping of surface antigens with concanavalin A (2, 4, 23, 54). Our study demonstrated the presence of calreticulin on the cell surface using microscopy, a biochemical method that selectively labeled proteins exposed on the cell surface, and flow cytometry to detect delivery of epitope-tagged protein after prolonged incubation with healthy lymphocytes. Collectively, the data indicate that calreticulin reaches the phagocytic cup both by capping of protein already present on the cell surface and by delivery of additional calreticulin to the cell surface during interaction with host cells. Because recombinant calreticulin binds to apoptotic lymphocytes but not healthy lymphocytes and because E. histolytica is known to induce lymphocyte apoptosis and erythrocyte phosphatidylserine exposure (10, 29, 30), it is likely that calreticulin becomes localized to the phagocytic cup following host cell killing.
Since calreticulin is primarily located in the ER lumen, how it reaches the cell surface is a major unanswered question that needs further investigation. The simplest explanation is that E. histolytica may use ER-dependent phagocytosis under some conditions. Whether ER-dependent phagocytosis occurs remains highly controversial, but several reports suggest that it does and may be favored during sustained phagocytosis or engulfment of large particles (8, 20). In this regard, it is interesting that calreticulin and several other ER proteins were identified in two E. histolytica phagosome proteomics studies but not in a third study (9, 36, 40). Additional work specifically intended to address the possibility of ER-dependent E. histolytica phagocytosis is needed.
Calreticulin is a luminal ER protein with no transmembrane domain. Thus, to function in phagocytosis, it must be retained on the cell surface by interaction with membrane lipids or at least one additional E. histolytica surface protein. CD91 plays this role in macrophages and also transmits signals that initiate particle uptake (39). Identification of such a calreticulin receptor is a major long-term goal of this project. A number of proteins predicted to be type I integral membrane proteins with low-level homology to the extracellular portion of CD91 are present in the E. histolytica genome, but none shows conservation of the intracellular signaling motifs of CD91. Thus, biochemical and/or genetic methods will likely be required to determine if E. histolytica has a specific receptor for calreticulin. Finally, CD91 serves as a receptor for a number of ER chaperones involved in endocytosis and antigen cross-presentation by macrophages, including calreticulin, HSP-70, HSP-90, and GP96 (7). It remains to be determined whether or not homologues of these proteins also function in E. histolytica phagocytosis, but this would be consistent with a model in which calreticulin and other “sticky” ER chaperones function as bridges between apoptotic host cells and E. histolytica scavenger receptors during phagocytosis.
In summary, this study establishes a role for calreticulin on the cell membrane of E. histolytica in amebic phagocytosis of host cells. The data indicate that calreticulin is present on the surface of unstimulated amebic trophozoites and that additional calreticulin is delivered to the cell surface during interaction with host cells. E. histolytica calreticulin binds to apoptotic lymphocytes independently of C1q and the collectins and also binds to C1q. Since calreticulin has no transmembrane domain, it presumably functions as a bridge between host ligands and the ameba. Important questions remain regarding how calreticulin is retained on the amebic surface and how it is brought there during interaction with host cells.
ACKNOWLEDGMENTS
We thank members of our laboratory for their helpful discussions and critique of the manuscript.
This work was supported by NIAID K08 AI053678 and R01 AI072021 and NIAID ARRA supplement R01 AI072021-02S1 to C.D.H.
Footnotes
Published ahead of print 2 April 2012
REFERENCES
- 1. Ankri S, Stolarsky T, Mirelman D. 1998. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol. Microbiol. 28:777–785 [DOI] [PubMed] [Google Scholar]
- 2. Arosa FA, de Jesus O, Porto G, Carmo AM, de Sousa M. 1999. Calreticulin is expressed on the cell surface of activated human peripheral blood T lymphocytes in association with major histocompatibility complex class I molecules. J. Biol. Chem. 274:16917–16922 [DOI] [PubMed] [Google Scholar]
- 3. Arroyo R, Orozco E. 1987. Localization and identification of an Entamoeba histolytica adhesin. Mol. Biochem. Parasitol. 23:151–158 [DOI] [PubMed] [Google Scholar]
- 4. Asgari S, Schmidt O. 2003. Is cell surface calreticulin involved in phagocytosis by insect hemocytes? J. Insect Physiol. 49:545–550 [DOI] [PubMed] [Google Scholar]
- 5. Bailey GB, Day DD, Gasque JW. 1985. Rapid polymerization of Entamoeba histolytica actin induced by interaction with target cells. J. Exp. Med. 162:546–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bailey GB, Gilmour JR, McCoomer NE. 1990. Roles of target cell membrane carbohydrate and lipid in Entamoeba histolytica interaction with mammalian cells. Infect. Immun. 58:2389–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Basu S, Binder RJ, Ramalingam T, Srivastava PK. 2001. CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity 14:303–313 [DOI] [PubMed] [Google Scholar]
- 8. Becker T, Volchuk A, Rothman JE. 2005. Differential use of endoplasmic reticulum membrane for phagocytosis in J774 macrophages. Proc. Natl. Acad. Sci. U. S. A. 102:4022–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boettner DR, et al. 2008. Entamoeba histolytica phagocytosis of human erythrocytes involves PATMK, a member of the transmembrane kinase family. PLoS Pathog. 4:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boettner DR, Huston CD, Sullivan JA, Petri WA. 2005. Entamoeba histolytica and Entamoeba dispar utilize externalized phosphatidylserine for recognition and phagocytosis of erythrocytes. Infect. Immun. 73:3422–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bracha R, Kobiler D, Mirelman D. 1982. Attachment and ingestion of bacteria by trophozoites of Entamoeba histolytica. Infect. Immun. 36:396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bracha R, Mirelman D. 1984. Virulence of Entamoeba histolytica trophozoites. Effects of bacteria, microaerobic conditions and metronidazole. J. Exp. Med. 160:353–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bracha R, Nuchamowitz Y, Anbar M, Mirelman D. 2006. Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog. 2:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brandt H, Perez-Tamayo R. 1970. The pathology of human amebiasis. Hum. Pathol. 1:351–385 [DOI] [PubMed] [Google Scholar]
- 15. Buss SN, et al. 2010. Members of the Entamoeba histolytica transmembrane kinase family play non-redundant roles in growth and phagocytosis. Int. J. Parasitol. 40:833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis PH, et al. 2006. Identification of a family of BspA like surface proteins of Entamoeba histolytica with novel leucine rich repeats. Mol. Biochem. Parasitol. 145:111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diamond LS, Harlow DR, Cunnick C. 1978. A new medium for axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431–432 [DOI] [PubMed] [Google Scholar]
- 18. Fadok VA, Bratton DL, Henson PM. 2001. Phagocyte receptors for apoptotic cells: recognition, uptake, and consequences. J. Clin. Invest. 108:957–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fadok VA, et al. 1992. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148:2207–2216 [PubMed] [Google Scholar]
- 20. Gagnon E, et al. 2002. Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110:119–131 [DOI] [PubMed] [Google Scholar]
- 21. Gardai SJ, et al. 2005. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 123:321–334 [DOI] [PubMed] [Google Scholar]
- 22. Ghosh SK, et al. 1999. Chitinase secretion by encysting Entamoeba invadens and transfected Entamoeba histolytica trophozoites: localization of secretory vesicles, endoplasmic reticulum, and Golgi apparatus. Infect. Immun. 67:3073–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Girard-Misguich F, Sachse M, Santi-Rocca J, Guillen N. 2008. The endoplasmic reticulum chaperone calreticulin is recruited to the uropod during capping of surface receptors in Entamoeba histolytica. Mol. Biochem. Parasitol. 157:236–240 [DOI] [PubMed] [Google Scholar]
- 24. Gonzalez E, et al. 2011. Entamoeba histolytica calreticulin: an endoplasmic reticulum protein expressed in trophozoites into experimentally induced amoebic liver abscesses. Parasitol. Res. 108:439–449 [DOI] [PubMed] [Google Scholar]
- 25. Gonzalez E, et al. 2002. Calreticulin-like molecule in trophozoites of Entamoeba histolytica HM1:IMSS (Swissprot: accession P83003). Am. J. Trop. Med. Hyg. 67:636–639 [DOI] [PubMed] [Google Scholar]
- 26. Griffin JL. 1972. Human amebic dysentery: electron microscopy of Entamoeba histolytica contacting, ingesting, and digesting inflammatory cells. Am. J. Trop. Med. Hyg. 21:895–906 [PubMed] [Google Scholar]
- 27. Hamann L, Buss H, Tannich E. 1997. Tetracycline-controlled gene expression in Entamoeba histolytica. Mol. Biochem. Parasitol. 84:83–91 [DOI] [PubMed] [Google Scholar]
- 28. Hoogenboom HR. 2005. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 23:1105–1116 [DOI] [PubMed] [Google Scholar]
- 29. Huston CD, Boettner DR, Miller-Sims V, Petri WA. 2003. Apoptotic killing and phagocytosis of host cells by the parasite Entamoeba histolytica. Infect. Immun. 71:964–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huston CD, Houpt ER, Mann BJ, Hahn CS, Petri WA. 2000. Caspase 3-dependent killing of host cells by the parasite Entamoeba histolytica. Cell. Microbiol. 2:617–625 [DOI] [PubMed] [Google Scholar]
- 31. Jimenez F. 1981. Pathology of amebiasis. Bull. N. Y. Acad. Med. 57:217–223 [PMC free article] [PubMed] [Google Scholar]
- 32. Leippe M, Ebel S, Schoenberger OL, Horstmann RD, Muller-Eberhard HJ. 1991. Pore-forming peptide of pathogenic Entamoeba histolytica. Proc. Natl. Acad. Sci. U. S. A. 88:7659–7663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Linford AS, et al. 2009. Short hairpin RNA-mediated knockdown of protein expression in Entamoeba histolytica. BMC Microbiol. 9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malhotra R, Willis AC, Jensenius JC, Jackson J, Sim RB. 1993. Structure and homology of human C1q receptor (collectin receptor). Immunology 78:341–348 [PMC free article] [PubMed] [Google Scholar]
- 35. Mann BJ, et al. 1993. Neutralizing monoclonal antibody epitopes of the Entamoeba histolytica galactose adhesin map to the cysteine-rich extracellular domain of the 170-kilodalton subunit. Infect. Immun. 61:1772–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marion S, Laurent C, Guillen N. 2005. Signalization and cytoskeleton activity through myosin IB during the early steps of phagocytosis in Entamoeba histolytica: a proteomic approach. Cell. Microbiol. 7:1504–1518 [DOI] [PubMed] [Google Scholar]
- 37. Nguyen DC, et al. 2011. Regulation of phosphatidylserine exposure in red blood cells. Cell. Physiol. Biochem. 28:847–856 [DOI] [PubMed] [Google Scholar]
- 38. Nissim A, et al. 1994. Antibody fragments from a ‘single pot’ phage display library as immunochemical reagents. EMBO J. 13:692–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ogden CA, et al. 2001. C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J. Exp. Med. 194:781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okada M, et al. 2005. Proteomic analysis of phagocytosis in the enteric protozoan parasite Entamoeba histolytica. Eukaryot. Cell 4:827–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Padilla-Vaca F, Ankri S, Bracha R, Koole LA, Mirelman D. 1999. Down regulation of Entamoeba histolytica virulence by monoxenic cultivation with Escherichia coli O55 is related to a decrease in expression of the light (35-kilodalton) subunit of the Gal/GalNAc lectin. Infect. Immun. 67:2096–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petri WA, Jr, Smith RD, Schlesinger PH, Murphy CF, Ravdin JI. 1987. Isolation of the galactose binding lectin of Entamoeba histolytica. J. Clin. Invest. 80:1238–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramakrishnan G, Lee S, Mann BJ, Petri WA. 2000. Entamoeba histolytica: deletion of the GPI anchor signal sequence on the Gal/GalNAc lectin light subunit prevents its assembly into the lectin heterodimer. Exp. Parasitol. 96:57–60 [DOI] [PubMed] [Google Scholar]
- 44. Ravdin JI, Guerrant RL. 1981. Role of adherence in cytopathic mechanisms of Entamoeba histolytica. Study with mammalian tissue culture cells and human erythrocytes. J. Clin. Invest. 68:1305–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sato D, Nakada-Tsukui K, Okada M, Nozaki T. 2006. Two cysteine protease inhibitors, EhICP1 and 2, localized in distinct compartments, negatively regulate secretion in Entamoeba histolytica. FEBS Lett. 580:5306–5312 [DOI] [PubMed] [Google Scholar]
- 46. Seydel KB, Li E, Zhang Z, Stanley SL. 1998. Epithelial cell-initiated inflammation plays a crucial role in early tissue damage in amebic infection of human intestine. Gastroenterology 115:1446–1453 [DOI] [PubMed] [Google Scholar]
- 47. Sim S, et al. 2005. NADPH oxidase-derived reactive oxygen species-mediated activation of ERK1/2 is required for apoptosis of human neutrophils induced by Entamoeba histolytica. J. Immunol. 174:4279–4288 [DOI] [PubMed] [Google Scholar]
- 48. Stuart GR, Lynch NJ, Day AJ, Schwaeble WJ, Sim RB. 1997. The C1q and collectin binding site within C1q receptor. Immunopharmacology 38:73–80 [DOI] [PubMed] [Google Scholar]
- 49. Teixeira JE, Heron BT, Huston CD. 2008. C1q- and collectin-dependent phagocytosis of apoptotic host cells by the intestinal protozoan Entamoeba histolytica. J. Infect. Dis. 198:1062–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Teixeira JE, Huston CD. 2008. Evidence of a continuous endoplasmic reticulum in the protozoan parasite Entamoeba histolytica. Eukaryot. Cell 7:1222–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Teixeira JE, Huston CD. 2008. Participation of the serine-rich Entamoeba histolytica protein in amebic phagocytosis of apoptotic host cells. Infect. Immun. 76:959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Teixeira JE, Mann BJ. 2002. Entamoeba histolytica-induced dephosphorylation in host cells. Infect. Immun. 70:1816–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vasudevan C, et al. 1998. The distribution and translocation of the G protein ADP-ribosylation factor 1 in live cells is determined by its GTPase activity. J. Cell Sci. 111:1277–1285 [DOI] [PubMed] [Google Scholar]
- 54. White TK, Zhu Q, Tanzer ML. 1995. Cell surface calreticulin is a putative mannoside lectin which triggers mouse melanoma spreading. J. Biol. Chem. 270:15926–15929 [DOI] [PubMed] [Google Scholar]
- 55. WHO 1997. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico 28–29 January, 1997. Epidemiol. Bull. 18:13–14 [PubMed] [Google Scholar]
- 56. Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. 1994. Making antibodies by phage display technology. Annu. Rev. Immunol. 12:433–455 [DOI] [PubMed] [Google Scholar]