Abstract
The enteropathogen Salmonella enterica serovar Typhimurium employs a suite of tightly regulated virulence factors within the intracellular compartment of phagocytic host cells resulting in systemic dissemination in mice. A type VI secretion system (T6SS) within Salmonella pathogenicity island 6 (SPI-6) has been implicated in this process; however, the regulatory inputs and the roles of noncore genes in this system are not well understood. Here we describe four clusters of noncore T6SS genes in SPI-6 based on a comparative relationship with the T6SS-3 of Burkholderia mallei and report that the disruption of these genes results in defects in intracellular replication and systemic dissemination in mice. In addition, we show that the expression of the SPI-6-encoded Hcp and VgrG orthologs is enhanced during late stages of macrophage infection. We identify six regions that are transcriptionally active during cell infections and that have regulatory contributions from the regulators of virulence SsrB, PhoP, and SlyA. We show that levels of protein expression are very weak under in vitro conditions and that expression is not enhanced upon the deletion of ssrB, phoP, slyA, qseC, ompR, or hfq, suggesting an unknown activating factor. These data suggest that the SPI-6 T6SS has been integrated into the Salmonella Typhimurium virulence network and customized for host-pathogen interactions through the action of noncore genes.
INTRODUCTION
Protein secretion is an essential determinant of the virulence of pathogenic bacteria. Multiple systems have evolved in order to secrete proteins into the extracellular environment or across the membranes of target cells (18). In Gram-negative Proteobacteria, these systems include the well-characterized type III secretion system (T3SS) and T4SS that permit the translocation of protein effectors from the bacterial cytoplasm directly into target cells to modulate the host environment (8, 25). An additional type of protein secretion system involved in protein translocation, known as a T6SS, has been found to contribute to interactions between bacteria and both bacteria and eukaryotic cells, including unicellular and multicellular eukaryotes (47). This system employs an assortment of membrane-associated proteins in order to coordinate the localization and assembly of the system; however, the secretory apparatus itself is composed of proteins that are structurally analogous to that of an inverted contractile bacteriophage tail (1, 29, 39). This coordinated contractile tail is utilized to deliver effector proteins to targets including the actin cytoskeleton in eukaryotic cells and peptidoglycan in bacterial cells (41, 45).
Salmonella enterica subsp. enterica serovar Typhimurium (S. Typhimurium) employs two well-characterized T3SSs to manipulate host cells through effector translocation in order to invade gut epithelial cells and disseminate systemically within phagocytic cells (18, 20). Most serovars of S. enterica subsp. enterica also encode a T6SS within Salmonella pathogenicity island 6 (SPI-6), while two other classes of T6SS have been described in S. bongori and S. enterica subsp. arizonae at SPI-2 and SPI-6, respectively (5, 17, 46). The S. enterica subsp. enterica T6SS was first described as part of the S. enterica centisome 7 genomic island (SCI) as a contributing factor in eukaryotic pathogenesis (16). Aside from the T6SS, SCI also contains the adhesin and invasin PagN, the fimbrial gene cluster safABCD, and the transcriptional regulator sinR (15, 26). Deletion of SCI resulted in an approximately 50% reduction in internalization of HEp-2 cells that could not be complemented by PagN expression; however, complementation with safABCD or sinR was not tested (16). Disruption of SPI-6 has also been found to result in a defect in systemic dissemination in orally infected BALB/c mice (19). In support, a number of SPI-6 T6SS genes have been implicated in long-term persistence in macrophages and mice through transposon mutagenesis; however, these transposon mutations have not been verified by precise deletions (9, 24, 28). Contrasting with these reports, another study described a hypervirulence phenotype in BALB/c mice upon deletion of the core structural gene sciS and found an increase in the number of bacteria within macrophages after 6 h, suggesting a time-dependent effect on replication (38). Further supporting a role in interactions with macrophages, genome-wide transcriptional profiling of S. Typhimurium during macrophage infections found that many of the SPI-6 T6SS genes were upregulated during macrophage infection although an increase over time between 4 and 12 h was not observed (21). An earlier gene expression profiling study also observed a trend toward the increased expression of some of these genes; however, statistical significance was not met (13). A mechanism leading to upregulation in this environment is unknown; however, an in vivo host-dependent activation mechanism has been previously described for the T6SS of Vibrio cholerae which requires endocytosis by phagocytic cells (32).
Distinct classes of T6SS maintain a core set of 13 genes required for localization of the system to the bacterial inner membrane and for functional secretion (7). T6SSs employ the proteins encoded by these genes to coordinate the secretion of protein substrates via the assembly, contraction, and disassembly of a phage-like tail (1, 43). For some T6SS-harboring organisms, the core gene encoding the tailspike protein encodes a fusion protein that allows the translocation of an effector domain (43). However, in S. Typhimurium and other T6SS-harboring organisms, no evolved tailspike protein is present (5). In addition to the core genes, organisms that possess a T6SS typically encode less-well-conserved, T6SS-associated genes that differ extensively in their number and location throughout the genome, and some of these genes have been reported to act as effector or regulatory proteins (2, 14, 43). Recently, a set of genes encoding T6SS-secreted toxin proteins were identified in Pseudomonas aeruginosa that are involved in interbacterial competition (45). While S. Typhimurium contains a number of noncore T6SS-associated genes, their role in the pathogenesis of the host small intestine or intracellular environment is poorly understood (5).
Noncore T6SS genes are known to play a role in the integration of T6SS into global regulatory networks. In V. cholerae, some strains constitutively express T6SS-associated proteins while others do not, suggesting niche adaptation (43). Regulatory control at the transcriptional level by the activator of alternative sigma factor 54, VasH, has been observed to occur in V. cholerae (23). In addition, posttranslational regulation by a T6SS-encoded threonine phosphorylation pathway has been described for the T6SS of P. aeruginosa (36). The noncore T6SS-associated gene TagR has been characterized as acting as a posttranslational repressor of this phosphorylation-dependent mechanism (50). Cues from within the host intracellular environment likely play a role in the regulation of the S. Typhimurium T6SS, as it has been determined that transcription of sciS increases approximately 16 h after the invasion of macrophages, and transcript levels are negatively correlated with the activity of the two-component response regulator of the SPI-2 T3SS, SsrB (38). Two-component signaling plays a major role in the regulation of virulence in S. Typhimurium, and these systems may regulate T6SS activity in concert with noncore T6SS genes, as is the case for the sensor kinases LadS and RetS in Pseudomonas syringae (44). A hallmark of T6SS activity has been the stable expression of phage-like proteins Hcp and VgrG, which form the contractile sheath and tailspike, respectively (with the ortholog of the latter annotated as VrgS in S. Typhimurium). The expression of these proteins in some organisms is absent under experimental conditions, likely due to the absence of inducing stimuli. This can be circumvented by the use of strains with constitutive T6SS activity, as in the case of a retS mutant of P. syringae or the V52 V. cholerae strain (43, 44). Whether or not stable expression and secretion of the Hcp and VrgS proteins occur under in vitro or in vivo conditions in S. Typhimurium is unknown. However, plasmid-based overexpression of the hcp ortholog in the SPI-6 T6SS of S. Typhi results in secretion in vitro, suggesting a functional system that can contribute to bacterial pathogenesis (53).
In this study, we determined that both the SPI-6 core T6SS and associated genes contribute to systemic dissemination during in vivo infections of mice and contribute to intracellular replication in macrophages. We used cell infections to determine that the Hcp and VrgS proteins are most highly expressed during late stages of infection of macrophages. We identified six transcriptionally active regions within the SPI-6 T6SS, three of which receive transcriptional input from SsrB, PhoP, and SlyA during growth in vitro. We then showed that expression of the Hcp and VrgS proteins in vitro does not change significantly upon the deletion of these and other key virulence regulatory proteins, suggesting that the induction of this system may rely on an unknown activator specific to the T6SS in vivo.
MATERIALS AND METHODS
Ethics statement.
All experiments with animals were conducted according to guidelines set by the Canadian Council on Animal Care. The Animal Review Ethics Board at McMaster University approved all of the protocols developed for this work.
Informatics.
Genome sequences were retrieved from NCBI GenBank (S. enterica subsp. enterica serovar Typhimurium strain LT2 [accession number AE006468.1], Burkholderia mallei ATCC 23344 [accession number NC_006349.2], and Burkholderia pseudomallei K96243 [accession number NC_006351.1]) and the Wellcome Trust Sanger Institute (S. enterica subsp. enterica serovar Typhimurium strain SL1344). BLASTP searches were performed online using the NCBI BLAST server against the nonredundant protein sequence database, and protein sequences were obtained from the S. Typhimurium strain LT2 genome file NC_003197.faa. The noncore gene search analysis employed an expect threshold of 1, and the number of target sequences was limited to 1,000. A custom Python script was used to parse the BLAST results for organisms with at least two of the proteins queried that did not belong to the genus Salmonella.
Strains and growth conditions.
All of the S. enterica strains used in this study for experimental work were S. enterica subsp. enterica serovar Typhimurium strain SL1344 derivatives. Constructs were designed using the S. Typhimurium LT2 genome and tested in S. Typhimurium SL1344. These genomes differ within the SPI-6 T6SS region by a silent mutation in sciG, an M1175T mutation in sciS, an in-frame 5-amino-acid mutation (A50 to L54) in sciI, and an in-frame 166-amino-acid mutation (V620 to Q786) in rhs1. None of these mutations are expected to have affected the results of experiments. The media employed included LB broth (1% [wt/vol] tryptone, 1% [wt/vol] sodium chloride, 0.5% [wt/vol] yeast extract); acidic, low-phosphate, low-magnesium (LPM) minimal medium [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 80 mM morpholinepropanesulfonic acid, 0.3% glycerol, 0.1% Casamino Acids, 24 μM MgCl2, 337 μM PO43−]; M9 minimal medium (5 mM Na2HPO4, 22 mM KH2PO4, 8.6 mM NaCl, 18.6 mM NH4Cl, 11.1 mM glucose, 2 mM MgSO4, 100 μM CaCl2, 0.1% Casamino Acids, pH 7.4); and Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Life Technologies). Bacteria were grown at 37°C with shaking at 225 rpm.
Construct inserts were amplified from S. Typhimurium SL1344 genomic DNA using platinum Taq high-fidelity DNA polymerase (Life Technologies), purified on PCR purification spin columns (Qiagen), digested using restriction endonucleases (NEB), and ligated using T4 DNA ligase (NEB). Constructs were propagated through Escherichia coli DH5α and electroporated into S. Typhimurium SL1344 competent cells. Integrated chromosomal transcriptional fusion reporters were generated by cloning the intergenic regions of interest into integrative plasmid pIVET5n and conjugating these into S. Typhimurium SL1344 through E. coli SM10 lambda pir (10). Deletion mutants were generated using the lambda red mutagenesis method (12). Hemagglutinin (HA) tags were introduced into the chromosome using a modified lambda red mutagenesis method (52). Lambda red cassettes were amplified using platinum Taq DNA polymerase (Life Technologies) from template plasmid pKD3 (chloramphenicol [Cm] replacement), pKD4 (kanamycin [Kn] replacement), pSUB314 (Cm-marked HA fusion), or pSUB315 (Kn-marked HA fusion), purified on PCR purification spin columns (Qiagen), and electroporated into water- and glycerol-washed S. Typhimurium SL1344 competent cells containing the helper plasmid pKD46 (12, 52). Plasmid-based luciferase transcriptional fusion reporters were generated by cloning the intergenic regions of interest into the pGEN-luxCDABE plasmid (27). Flag epitope expression constructs were generated by cloning the indicated genes into the pFLAG-CTC expression vector (Sigma) and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and secretion assays were performed with LPM minimal medium as previously described (11). Briefly, after 6 h of growth in LPM minimal medium, bacteria were pelleted at 10,000 × g, medium was removed and filtered through a 0.2-μm acrodisc syringe filter (PALL), trichloroacetic acid was added to 10%, and the mixture was incubated on ice at 4°C overnight. Samples were pelleted at 18,000 × g for 30 min at 4°C, washed with 1 ml acetone, and pelleted again at 18,000 × g for 30 min, and the pellet was allowed to air dry. The pellet was then resuspended in an optical density at 600 nm (OD600)-normalized volume of 2× SDS loading dye. For sciG complementation, sciG and its upstream region were cloned into low-copy-number pWSK29 plasmid (54). The following antibiotics were added to the medium when necessary unless otherwise specified: ampicillin (100 μg/ml), Cm (34 μg/ml), Kn (50 μg/ml), streptomycin (50 μg/ml), and gentamicin (10 or 100 μg/ml). All of the strains and plasmids employed here are listed in Table S1 in the supplemental material. All of the primers used to generate constructs are listed in Table S2 in the supplemental material.
Mouse infections.
Bacteria from stationary-phase LB cultures were washed and diluted in inoculation buffer (0.1 M HEPES buffer, 0.9% NaCl), and 10- to 15-week-old female C57BL/6 mice were infected orally with 100 μl of 2 × 107 CFU/ml in groups of three to five. Endpoint analyses and competitive infections (CIs) were performed as previously described (11). For endpoint experiments, mice were monitored for endpoint and sacrificed when they had lost 20% of their initial body weight. For CI experiments, mixed inoculums of wild-type (pseudogene ushA::Cm marked) and mutant strains were used. At 72 h postinfection, mice were sacrificed and their spleens and livers were harvested, homogenized in phosphate-buffered saline (PBS), and plated to determine bacterial numbers, which were compared to initial inoculum plating numbers as CFU ratios ([mutant output/wild-type output]/[mutant input/wild-type input]). For sciG complementation, the wild-type reference strain also possessed the empty pWSK29 plasmid. Statistical analyses were performed in Prism using the nonparametric Wilcoxon signed-rank test to determine whether mutant CI values differed from 1 (P < 0.05 at a 95% confidence interval).
Cell infections.
RAW 264.7 murine macrophage-like cells were seeded at 2 × 105 CFU per well in 24-well plates and grown at 37°C and 5% CO2 in DMEM supplemented with 10% FBS (Life Technologies). Bacteria from overnight LB cultures were pelleted, resuspended in DMEM–10% FBS, and opsonized in human serum, and macrophages were infected at a multiplicity of infection (MOI) of 50 for 30 min. This was followed by two PBS rinses, a 1.5-h incubation in DMEM–10% FBS plus 100 μg/ml gentamicin, two PBS rinses, and incubation to the given lysis time point in DMEM–10% FBS plus 10 μg/ml (11). At the indicated time points, infected cells were washed with PBS, lysed in cell lysis buffer (1 ml PBS, 1% Triton X-100, 0.1% SDS), and plated to establish bacterial counts. For epithelial cell coculture experiments, HeLa and HEp-2 cells were seeded at 1 × 105 CFU per well in 24-well plates and cocultured at an MOI of 10 from pelleted and DMEM–10% FBS-washed 3-h exponential-phase LB cultures. For epithelial cell infections, HeLa or HEp-2 cells were seeded at 1 × 105 CFU per well in 24-well plates and infected at an MOI of 50 from pelleted and DMEM–10% FBS-washed 3-h exponential-phase LB cultures for 15 min, washed twice with PBS, and allowed to complete invasion in fresh DMEM–10% FBS for 20 min. Cells were then washed twice with and incubated in DMEM–10% FBS plus 100 μg/ml gentamicin for 1.5 h, washed twice with PBS, and lysed in cell lysis buffer. Lysates and infection inoculum were plated and counted to calculate invasion efficiency. For sciG complementation, the wild-type reference strain also possessed the empty pWSK29 plasmid. Statistical analyses were performed in Prism using a one sample t test.
Transcriptional reporter assays.
For beta-galactosidase assays, strains were grown overnight in M9 medium, subcultured at 1:50 in LPM minimal medium, and then grown at 37°C with shaking. At defined time intervals, OD600 was read using a spectrophotometer and 200-μl culture samples were pelleted and frozen for analysis. For analysis, pellets were resuspended in 200 μl PBS and lysed by the addition of 50 μl chloroform. A 2-μl volume of the lysate was combined with 100 μl of Tropix Galacto-Star detection reagent (Applied Biosystems), and luminescence in relative light units (RLU) was measured using an Envision plate reader (Perkin-Elmer) (10). For intracellular luminescence assays, macrophages in four sets of wells were identically infected as previously described. At each time point, one set of wells was washed with PBS and luminescence was measured using an Envision plate reader. Cells were then lysed in cell lysis buffer and plated to determine bacterial counts. Statistical analyses were performed in Prism using a one-sample t test.
Immunoblotting.
Strains were grown overnight in LB medium at 37°C. Cultures were pelleted, washed, and diluted in DMEM containing 10% FBS. Inoculum was added to 12-well tissue culture plates with or without eukaryotic cells for coincubation protocol or used for infection as previously described. Plates were incubated at 37°C under 5% CO2. At defined time points, the contents of the well were collected, pelleted, and washed in PBS. Pellets were lysed in 200 μl 2× SDS sample buffer (100 mM Tris-HCl [pH 6.8], 20%% [vol/vol] glycerol, 4% [wt/vol] SDS, 0.002% [wt/vol] bromophenol blue, 4 M urea, 0.2 M dithiothreitol). Samples were boiled for 10 min at 100°C, centrifuged for 1 min at 17,900 × g, and subjected to 15% SDS-PAGE. Protein was transferred to polyvinylidene difluoride membrane and incubated overnight with 1:10,000 mouse anti-DnaK (Stressgen), 1:1,000 mouse anti-HA (Covance), or 1:5,000 mouse anti-FLAG (Sigma) antibodies. Blots were then washed in Tris-buffered saline–Tween 20 (TBST), incubated with 1:5,000 goat anti-mouse–horseradish peroxidase antibodies (Sigma), washed in TBST, and detected using enhanced chemiluminescence (Western Lightning; Perkin-Elmer). Low-signal Western blot assays of samples collected from regulator mutant bacteria were performed with SuperSignal West Femto (Thermo Scientific).
RESULTS
The SPI-6 T6SS noncore genes include T6SS-associated gene pairs.
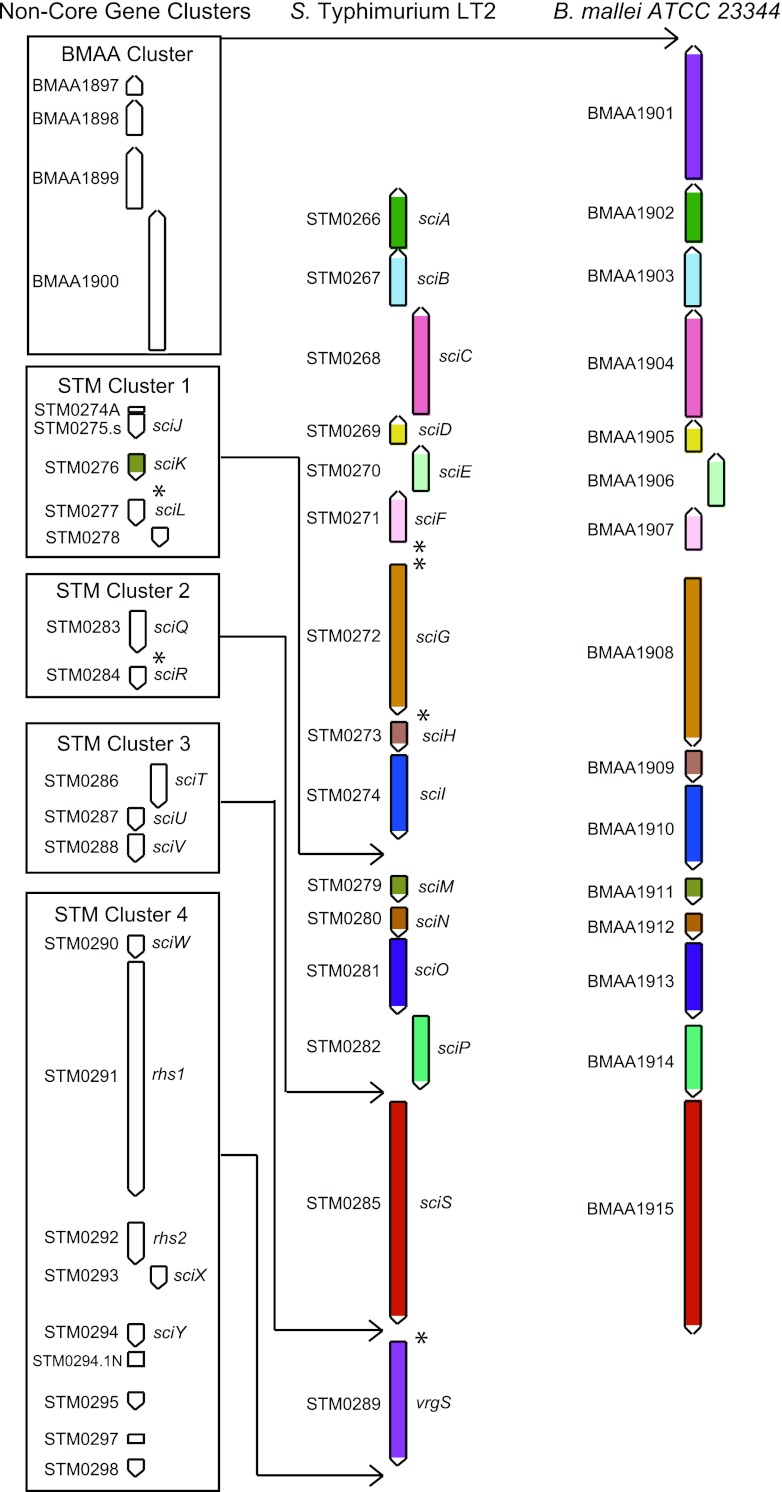
The S. Typhimurium SPI-6 T6SS contains 13 core T6SS genes in addition to other uncharacterized genes (5). Many T6SSs have undergone preliminary characterization in other sequenced bacteria, and we hoped to gain insight into these uncharacterized genes by identifying a similar characterized system. Using the SPI-6 T6SS core protein sequences, we performed a BLASTP search of the NCBI BLAST nonredundant protein sequence database to identify an organism that encodes a similar T6SS. We observed that the T6SS-3 (Tss3) of Burkholderia mallei and B. pseudomallei is highly similar (49) (Fig. 1; see Table S3 in the supplemental material). These systems have been previously reported to be closely related through phylogenetic analyses (4, 5, 7). The Burkholderia sp. system has 15 genes in common with that of S. Typhimurium, representing 13 core genes, and they share synteny in core T6SS genes aside from their VgrG ortholog, which is positioned at opposite sides of the island. The two systems differ in the number of additional nonconserved genes: four genes in a single cluster in the Burkholderia variants (BMAA cluster) and 10 genes within three clusters in the S. Typhimurium T6SS (STM clusters 1 to 3). These clusters contain genes sciJ to sciL (STM0274A to STM0278), sciQ and sciR (STM0283 and STM0284), and sciT to sciV (STM0286 to STM0288), respectively. A fourth cluster of genes (STM cluster 4), sciW to sciY (STM0290 to STM0298), located between the T6SS and safABCD operon contains recombination hot spot (Rhs) elements, poorly conserved genes, and transposases.
Fig 1.
Diagram showing the 15 T6SS genes that the S. Typhimurium LT2 SPI-6 T6SS and the B. mallei ATCC 23344 Tss3 T6SS have in common. Orthologs are indicated by shading with the same color. Nonconserved genes in B. mallei (BMAA cluster) and S. Typhimurium (STM clusters) are displayed in boxes on the left, and their positions relative to the core genes are indicated by black lines. Observed transcriptionally active regions are denoted by asterisks.
In order to better understand the noncore genes that are not conserved between the S. Typhimurium SPI-6 T6SS and the Burkholderia Tss3, we searched for organisms in which orthologs of at least two of these genes exist (see Table S4 in the supplemental material). We did not include sciK, a paralog of sciM, in cluster 1, given its extensive conservation as an Hcp-encoding gene even though it is absent from the B. mallei Tss3. In cluster 1, STM0274A and sciJ were not well conserved outside Salmonella and were found to exist in only Escherichia sp. strain TW09308. In contrast, orthologs of sciL and STM0278 were found to co-occur in 14 other genomes, including those of species of Escherichia, Enterobacter, Erwinia, and Frateuria, in close proximity to an sciS ortholog (suggestive of a T6SS), and in those of species of Advenella, Pantoea, and Pseudomonas, not in close proximity to an sciS ortholog. In cluster 2, orthologs of sciQ and sciR were found to co-occur only in Agrobacterium tumefaciens in close proximity to an sciS ortholog. In cluster 3, orthologs of sciU and sciV were found to co-occur in four genomes, including those of species of Bordetella, in close proximity to an sciS ortholog, and in those of species of Delftia and Vibrio, not in close proximity to an sciS ortholog. While not always in close proximity, all of the organisms indicated here that possessed these gene pairs did contain at least one sciS ortholog. While many genomes were found to contain orthologs of genes within cluster 4, only Enterobacter hormanechei ATCC 49162, Cronobacter sakazakii ATCC BAA-894, Cronobacter turicensis z3032, E. coli TA280, and Pantoea sp. strain SL1_M5 orthologs of these genes were found near orthologs of genes from clusters 1 to 3.
Our search also recovered the previously identified orphan Hcp-encoding gene STM3131, which is not linked with the SPI-6 T6SS and likely has an origin distinct from that of the S. enterica subsp. enterica and arizonae SPI-6 T6SS (5). Recognizing the possibility that STM3131 could contain a gene product functionally relevant to the Salmonella SPI-6 T6SS, we included this gene in downstream experiments.
The SPI-6 T6SS contributes to pathogenesis in a mouse model of typhoid.
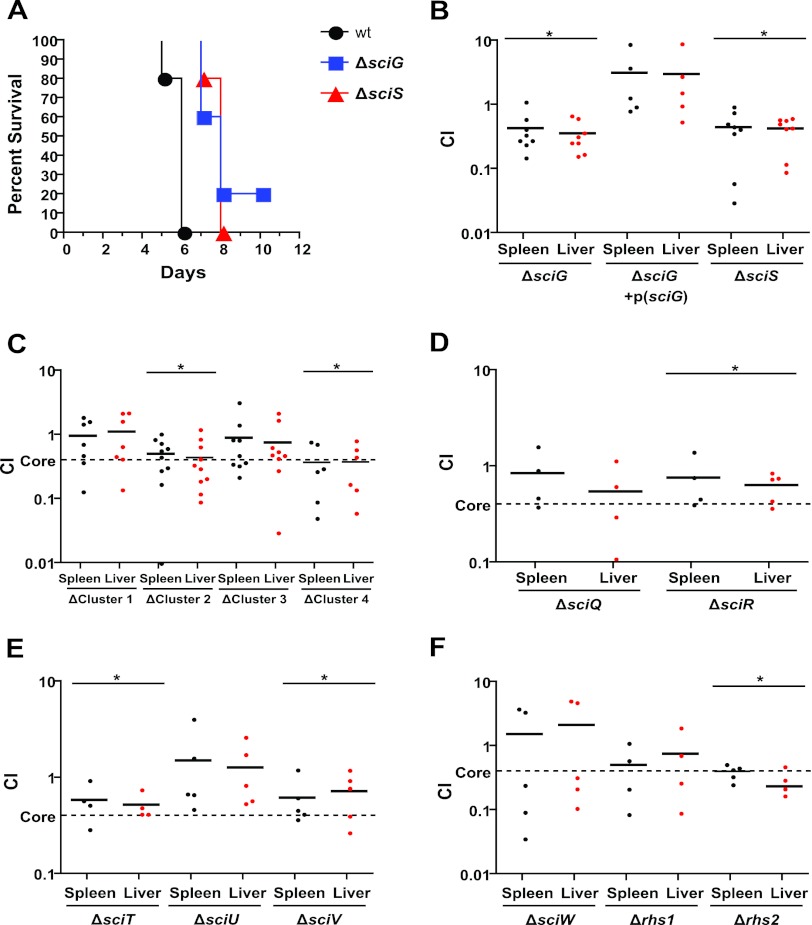
We generated mutants of both sciG (the clpV ortholog), and sciS (the icmF ortholog), genes which have been established as essential for T6SS assembly and functional secretion in other systems (6, 42). When mice were infected with wild-type and mutant strains of S. Typhimurium, mice infected with the mutant strains reached an endpoint approximately 1 to 2 days later than mice infected with wild-type S. Typhimurium (Fig. 2A). The major processes leading to death in this model are systemic dissemination and replication of the bacteria in organs including the spleen and liver. To determine whether the T6SS contributes to these processes, we performed CI experiments with the sciG and sciS mutants against the wild-type strain (Fig. 2B). We obtained combined (liver and spleen) CI values for these mutants of 0.39 and 0.43, respectively (see Table S5 in the supplemental material), that were in agreement with previously reported data for deletion of the entire SPI-6 locus (19). This defect in systemic dissemination was successfully complemented by plasmid-based expression of sciG under the control of its native promoter.
Fig 2.
Contribution of the SPI-6 T6SS to S. Typhimurium pathogenesis in mice. (A) Survival curve of SPI-6 T6SS mutant bacteria in C57BL/6 mice. Mice infected with ΔsciG (ATPase) and ΔsciS (icmF) mutant bacteria reached an endpoint 2 days later than those infected with wild-type (wt) bacteria. (B to F) CI experiments with wild-type and SPI-6 T6SS mutant bacteria and C57BL/6 mice orally infected with 106 CFU. (B) ΔsciG and ΔsciS mutants were less fit in the liver and spleen than wild-type bacteria, a defect that could be resolved by plasmid-based complementation of sciG (*, P < 0.05). (C) Deletion of noncore gene clusters 2 and 4 resulted in CI values similar to those produced by the deletion of essential T6SS genes (core), while deletion of cluster 3 resulted in an intermediate CI value. (D) Deletion of individual genes within cluster 2 could not recapitulate the defect caused by deletion of the entire cluster. (E) Deletion of individual genes within cluster 3 with a nonsignificant trend toward increased replication in the sciU mutant. (F) Deletion of individual genes within cluster 4 showed a strong defect in the Δrhs2 mutant.
The SPI-6 noncore gene clusters contribute to pathogenesis in mice.
Since we were able to measure a virulence effect for the T6SS core genes by oral infection of mice, we employed this model to test the contribution of the SPI-6 noncore gene clusters to systemic dissemination. We generated cluster 1 through 4 deletion mutants. When competed against the wild type in oral infections, cluster 2 and 4 deletion mutants yielded combined organ CI values of 0.47 and 0.37 (see Table S5 in the supplemental material), values similar to that obtained by deletion of the core genes sciG and sciS (Fig. 2C). The combined CI values of cluster 1 and 3 mutants did not differ significantly from that of the wild type, although we observed clustering of values for the cluster 3 deletion near the CI value of the core mutant deletion (average = 0.82, median = 0.49) and therefore included this cluster for further analysis. To identify specific genes contributing to this fitness defect, we generated additional single gene deletion strains for clusters 2, 3, and 4. When competed against the wild type, the individual cluster 2 mutants both had a combined CI value of 0.69 that was not as low as the cluster 2 mutant combined CI value of 0.47 (Fig. 2D). CI experiments with individual cluster 3 mutants showed that the sciT and sciV mutants had combined CI values below that of the wild type (0.55 and 0.67) while the sciU deletion mutant was found to trend toward increased systemic dissemination with a combined CI value of 1.39, albeit not significant (Fig. 2E). CI experiments with selected individual cluster 4 mutants revealed a low combined CI value of 0.32, indicating a strong systemic dissemination defect in the rhs2 mutant (Fig. 2F).
The SPI-6 T6SS contributes to intracellular replication in macrophages.
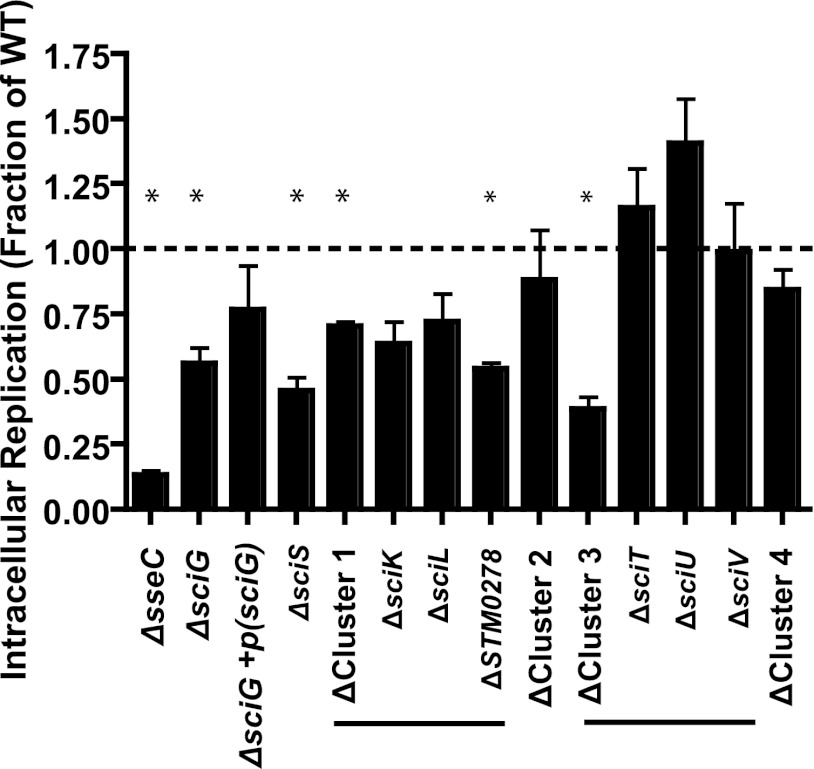
Systemic dissemination is dependent on survival within phagocytic cells. To determine whether the SPI-6 T6SS contributes to intracellular replication, we assessed the intracellular survival/replication rates of T6SS core gene and noncore gene mutants 24 h postinfection in the murine macrophage-like cell line RAW 264.7 (Fig. 3A). Compared to the wild-type S. Typhimurium parent strain, the sciG and sciS core T6SS gene mutants were found to have an intracellular replication ratio of approximately 0.5, compared to the intracellular replication ratio of 0.13 of an sseC SPI-2 T3SS translocon mutant that is defective in intracellular persistence. Plasmid-based expression of sciG was able to complement the sciG mutant to near-wild-type levels of intracellular replication. We observed a significant decrease in replication over the wild-type strain for clusters 1 and 3. Individual deletions within cluster 1 revealed a significant contribution for STM0278, while no individual gene in cluster 3 was found to make a significant contribution (Fig. 3B). While this is not significant, we again observed a trend toward increased replication in the cluster 3 sciU single gene mutant.
Fig 3.
Assessment of SPI-6 T6SS contribution to replication in macrophage cell culture. Intracellular replication of SPI-6 T6SS mutants in RAW 264.7 murine macrophage-like cells expressed as a fraction of that of the wild type (WT) at 24 h postinfection compared with that of a ΔsseC SPI-2 T3SS translocon mutant (*, P < 0.05). Decreased intracellular replication of core gene ΔsciG and ΔsciS mutants was observed, with near-wild-type levels for the sciG complemented strain. Replication defects were also observed in noncore gene cluster 1 and 3 mutants. Decreased intracellular replication for the STM0278 mutant was observed when single gene mutants were tested, while a nonsignificant increase in that of individual noncore gene cluster 3 mutants was observed.
To determine whether this replication phenotype is specific to macrophages, we performed invasion assays with HeLa and HEp-2 epithelial cells. While we observed a clear defect in invasion by our hfq mutant control, which is defective in invasion, we saw no significant effect on invasion in both cell types for any of our core and noncore SPI-6 T6SS mutants (see Fig. S1 in the supplemental material).
Hcp and VrgS protein expression is enhanced during infection of macrophages.
Secretion of Hcp and VgrG orthologs is the hallmark of a functional T6SS. Hcp-1, Hcp-3, and VrgS were cloned and overexpressed as C-terminal FLAG fusion proteins and assayed for expression and secretion following IPTG induction in wild-type and sciG mutant strains (see Fig. S2 in the supplemental material). While all of the proteins were observed in the cytoplasmic fraction, only Hcp-3 was found to be secreted. In addition, it was determined that Hcp-3 secretion into the medium was not dependent on the T6SS ATPase SciG. The Hcp proteins were observed at their predicted sizes of approximately 17 kDa, while the VrgS protein was observed to migrate at approximately 70 kDa rather than the predicted 80 kDa based on the S. Typhimurium LT2 annotation, suggesting a possible misannotated start codon. BLASTP searches indicate low-complexity sequences within the first 100 amino acids, and this region falls outside the conserved Vgr protein domains TIGR03361, COG3501, and TIGR01646 identified by the BLAST integrated conserved domain database (35). Interestingly, trace levels of DnaK were observed only in the secreted fraction of VrgS, suggesting that overexpression of VrgS may be slightly toxic.
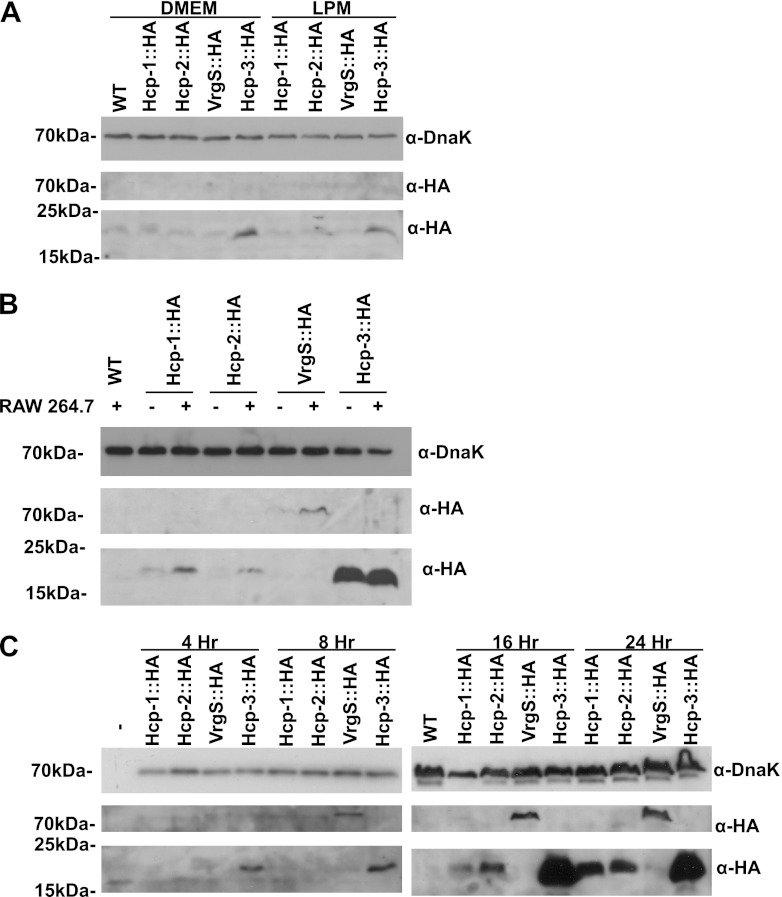
In order to characterize the stable expression of SPI-6 T6SS proteins, we constructed chromosomal HA epitope fusion proteins with the Hcp paralogs SciK (Hcp-1), SciM (Hcp-2), and STM3131 (Hcp-3) and with the VgrG ortholog VrgS such that their expression was controlled by their native chromosomal promoters. Under in vitro growth conditions, only Hcp-3 was detectable after 24 h when bacteria were grown in rich (DMEM–10% FBS) or minimal (LPM) medium (Fig. 4A).
Fig 4.
Expression of Hcp and VrgS C-terminal chromosomal HA epitope fusion proteins assessed by Western blot assay. (A) Following 24 h of growth in DMEM or LPM minimal medium, expression of only Hcp-3::HA was observed. (B) At 24 h postinfection of RAW 264.7 macrophage-like cells with opsonized bacteria, Hcp-1::HA, Hcp-2::HA, and VrgS::HA expression was enhanced compared to that in bacteria grown under identical cell culture conditions in the absence of eukaryotic cells. Expression of Hcp-3::HA was not enhanced. (C) Expression of Hcp-2::HA and VrgS::HA was detected by 16 h postinfection of RAW 264.7 macrophage-like cells with opsonized bacteria. WT, wild type.
Some T6SSs have been shown to require target cell contact for activation (32). To determine whether the absence of chromosomal expression of the Hcp and VrgS proteins in vitro was due to the lack of induction by host cell signals, strains harboring chromosomally tagged genes were either used to infect RAW 264.7 murine macrophage-like cells or grown in tissue culture medium in the absence of macrophages (Fig. 4B). We observed only weak expression of Hcp-1, Hcp-2, and VrgS in the absence of macrophages, whereas the abundance of these proteins was increased following macrophage infection. In contrast, Hcp-3 appears to be constitutively expressed under all of the conditions tested. To determine whether this induction is specific to phagocytic cells or the intracellular environment, HeLa cells were grown in the presence of wild-type or invasion-deficient invA mutant strains. No induction of Hcp-1, Hcp-2, or VrgS expression was observed after 24 h (see Fig. S3 in the supplemental material). Hcp-3 was expressed in both backgrounds. It was noted that Hcp-1 appears to be more strongly expressed than Hcp-2 and VrgS under cell culture conditions both in the absence of macrophages and by wild-type cells in the presence of HeLa cells.
A previous report had shown that transcript levels of the icmF ortholog sciS remain low until late time points (>16 h) in macrophage infections (38). To determine whether Hcp and VrgS expression follows this pattern, we collected the contents of individual wells of infected RAW 264.7 cells at 4, 8, 16, and 24 h postinfection. Hcp-3 expression was evident at early time points, while VrgS, Hcp-1, and Hcp-2 expression was weak. By 16 h, expression of both Hcp-2 and VrgS was observed (Fig. 4C).
The SPI-6 T6SS is not induced by deletion of regulators of intracellular virulence.
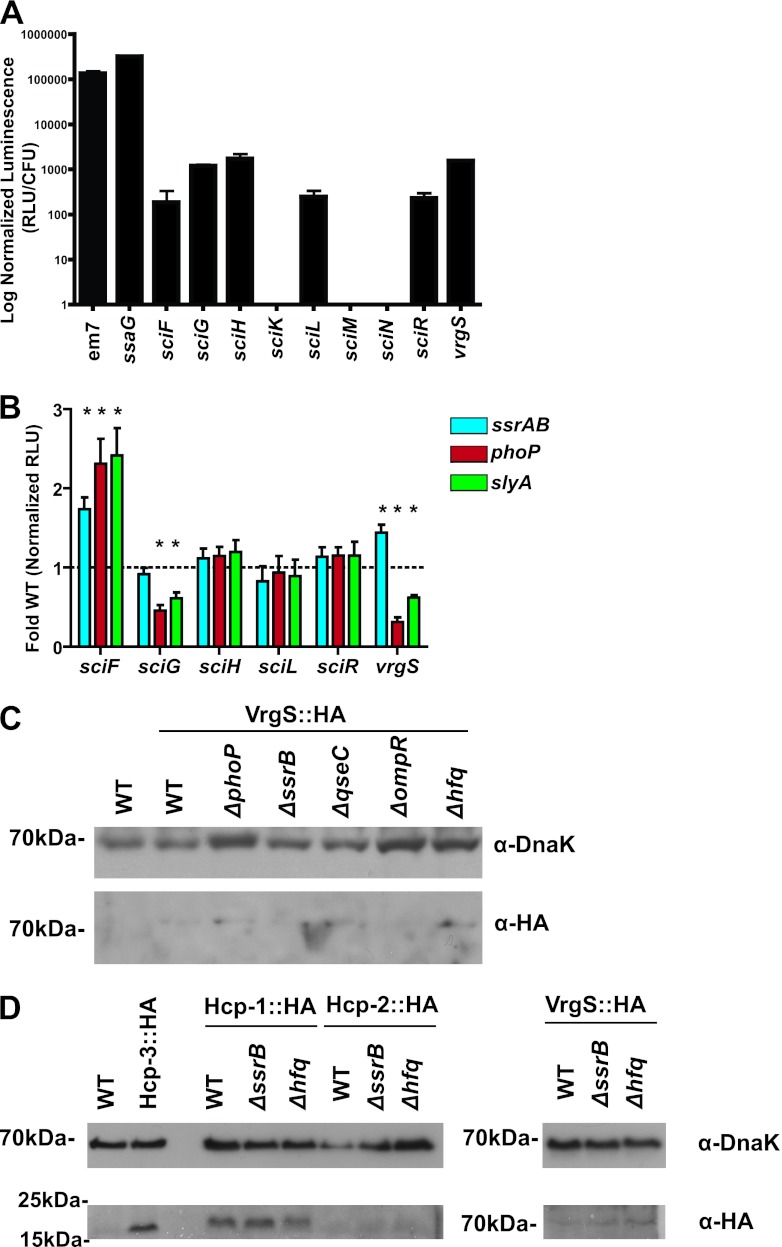
A previous report suggested that the SPI-6 T6SS is induced in response to host cell signaling as a result of regulatory derepression, possibly through the transcription factor SsrB (38). In order to further investigate regulatory contributions, we identified transcriptionally active regions by cloning intergenic regions of greater than 40 bp in the SPI-6 T6SS as transcriptional fusions to luciferase (see Table S6 in the supplemental material). Measurement of luminescence normalized to the number of recovered bacterial cells was performed during RAW 264.7 macrophage-like cell infections to assess transcriptional activity (Fig. 5A). At 24 h postinfection, active transcription from regions upstream of the six genes sciF, sciG, sciH, sciL, sciR, and vrgS was observed. Transcription from regions upstream of sciF, sciL, and sciR was much lower than that from regions upstream of sciG, sciH, and vrgS during exponential growth in vitro (see Fig. S4 in the supplemental material).
Fig 5.
Effect of regulator mutations on SPI-6 T6SS gene expression. (A) Activities of nine SPI-6 T6SS intergenic regions directly upstream of the indicated genes that were cloned as transcriptional fusions with the luxCDABE genes. Luminescence data were collected as RLU and normalized to CFU. At 24 h postinfection of RAW 264.7 macrophage-like cells, six of the nine intergenic regions were transcriptionally active. The constitutive synthetic promoter em7 and the SsrB-dependent promoter of ssaG are also shown as controls. (B) Activities of the six transcriptionally active intergenic regions when cloned as lacZ chromosomal transcriptional fusion reporters in regulator mutant backgrounds and measured during log-phase growth in LPM minimal medium (*, P < 0.05). Luminescence data were collected as RLU and normalized to OD600. The transcriptional activity of the region upstream of sciF is increased in the phoP, slyA, and ssrAB mutant backgrounds, and that of vrgS is increased only in an ssrAB mutant background. Those of sciG and VrgS are decreased in the phoP and slyA mutant backgrounds. (C) Expression of VrgS::HA protein assessed by Western blot assay. Trace levels of the VrgS::HA fusion protein expressed under native promoter control on the chromosome can be detected after 24 h of growth in LB medium, and expression is not strongly upregulated in the phoP, ssrAB, qseC, ompR, and hfq mutant backgrounds. (D) Expression of native-promoter-expressed Hcp-1::HA, Hcp-2::HA, and VrgS::HA chromosomal fusion proteins assessed by Western blot assay in the ssrAB and hfq mutant backgrounds after 24 h of growth in DMEM–10% FBS under cell culture conditions. No significant difference in expression was observed. WT, wild type.
To determine if these transcriptionally active regions are targets of regulatory input from SsrB, lacZ transcriptional reporters were chromosomally integrated downstream of these regions to generate merodiploid reporter strains. The transcriptional activity of these reporters was measured in vitro in defined LPM minimal medium, which provides an environment in which SsrB and upstream regulators of virulence genes, PhoPQ and SlyA, are active (10). We assessed the transcriptional activities of our reporters in backgrounds deficient in these proteins involved in intracellular pathogenesis (56). We observed altered profiles of transcriptionally active regions upstream of sciF, sciG, and vrgS (Fig. 5B). In an ssrAB mutant background, the transcriptional activity of the regions upstream of sciF and vrgS was increased slightly (less than 2-fold). The transcriptional profiles in the phoP and slyA mutant backgrounds were identical and revealed an increase in transcription in the region upstream of sciF by approximately 2.5-fold and a 2-fold decrease in the regions upstream of sciG and vrgS.
To determine whether the expression of the SPI-6 T6SS is repressed by the activity of regulators of intracellular virulence, we assessed the levels of chromosomal fusion protein VrgS::HA expressed under the control of its native promoter in backgrounds deficient in regulators of intracellular pathogenesis (56). These included the two-component systems PhoPQ, OmpR/EnvZ, QseBC, PmrAB, and SsrAB and the RNA binding protein Hfq, regulators important for intracellular pathogenesis (Fig. 5C). Expression of VrgS in these mutant backgrounds in LB medium after 24 h remained barely detectable by an enhanced-sensitivity chemiluminescence system, although expression appeared slightly higher in an hfq mutant background. When expression of Hcp-1, Hcp-2, and VrgS was assessed after 24 h of growth in DMEM under cell culture conditions, we failed to see an increase in expression in either ssrB or hfq mutant background strains over that by the wild type (Fig. 5D). Expression of these proteins was weaker during growth in LPM minimal medium and also failed to show increased expression over that by the wild type (data not shown).
A PmrA box motif was previously identified upstream of the orphan Hcp-3 gene STM3131, although the regulatory input was not confirmed (34). In order to determine whether a regulatory interaction with PmrA occurs at this site, we generated a lacZ transcriptional fusion on the chromosome downstream from STM3131. We also generated lacZ transcriptional fusions on the chromosome for the PmrA-activated promoter of pmrC and the PmrA-repressed promoter of pmrD. Assessment of transcriptional activity indicated that transcription increased 2-fold in a pmrA mutant background, similar to that of the PmrA-repressed promoter of pmrD, suggesting that PmrA acts as a transcriptional repressor of this gene (see Fig. S4 in the supplemental material).
DISCUSSION
T6SS variants have been acquired at least three times within the Salmonella lineage, and little is known about their function and mechanism, although most reports have implicated roles for the S. Typhimurium SPI-6 T6SS in the pathogenesis of mice and infection of macrophages (5, 9, 17, 19, 24, 28, 30, 38). We found that the disruption of noncore T6SS clusters 2 and 4 caused significant defects in systemic dissemination in mice and that disruption of noncore gene clusters 1 and 3 resulted in a significant intracellular replication defect in macrophages. Further supporting this role in intracellular pathogenesis, we showed an increase in Hcp and VrgS protein expression in association with macrophages. Finally, we showed that deletion of a previously identified negative regulator of this system enhances levels of transcription of this system from defined transcriptionally active regions but does not lead to increased levels of Hcp and VrgS protein expression upon deletion.
Distinct T6SSs are encoded by S. bongori, S. enterica subsp. arizonae, and serovars of S. enterica subsp. enterica as the most proximal element to the tRNA genes within SPI-6 or SPI-2, suggesting relatively recent acquisition events (5, 17). In spite of the number of T6SS in Salmonella, an evolved VgrG effector gene has been identified only in S. enterica subsp. arizonae, suggesting that if the S. Typhimurium SPI-6 T6SS functions to deliver effector proteins, they are not encoded as evolved VgrG proteins (5). The SPI-6 T6SS closely resembles the Burkholderia Tss3 (49) present in B. mallei and B. pseudomallei but lacks a reported role in pathogenesis and is absent from B. thailandensis (48). In order to understand the role of the SPI-6 T6SS, we focused on noncore T6SS genes. Noncore T6SS gene transposon mutants of S. Typhimurium have been found to be defective in long-term persistence in mice and macrophages; these include sciR in cluster 2 (9), sciU in cluster 3 (9), and sciW, rhs1, and STM0298 in cluster 4, suggesting that these genes may encode proteins important for T6SS activity (9, 24, 28). By BLAST-based analysis, we found that some of these genes are restricted to Salmonella while others exist as gene pairs in other T6SS-encoding organisms. The sciL and STM0278 genes in cluster 1 are extensively conserved and are particularly interesting candidates for further investigation. Cluster 2 was found to be poorly conserved and carries a conserved gene pair found in only one other non-Salmonella organism. The genes sciU and sciV also exist as a conserved pair in a limited number of organisms, although the association with T6SS is not as strong as that of sciL and STM0278. Many orthologs of genes within cluster 4 were identified, including the sciW and rhs1 genes; however, the extensive conservation of rhs-associated genes and their close similarity made drawing conclusions about these genes difficult. Evidence of conservation of gene pairs within each of these clusters is particularly interesting given the presence of toxin-antitoxin gene pairs found in other T6SS-encoding organisms (22).
In order to quantify the contribution of the SPI-6 T6SS in host pathogenesis, we assessed the systemic dissemination of core and noncore mutants in a murine mouse model of typhoid and found significant contributions for noncore gene clusters 2 and 4. Deletion of individual genes within noncore gene cluster 2 did not decrease systemic dissemination to the same extent as deletion of the entire cluster, suggesting that the transcriptionally active region upstream of sciR may be a contributing element in pathogenesis. Interestingly, the orthologous sciQ and sciR genes in A. tumefaciens are in a different orientation and lack this intergenic region. In the deletions within cluster 3, individual sciT and sciV mutants revealed decreased fitness and a trend toward increased fitness of the sciU mutant. Deletion of all three genes may result in either increased or decreased fitness in vivo, depending on uncontrolled host factors. The sciU and sciV genes may function as a gene pair, given that these two genes exist as an orthologous gene pair in the absence of sciT in four other Proteobacteria member genomes. Deletion of cluster 4 had the greatest effect on systemic dissemination of all of the clusters tested, indicating that it may contain an element essential for T6SS-associated pathogenesis in vivo. This cluster contains Rhs family genes and Rhs-associated genes that are commonly linked with T6SSs and which are extensively distributed in Gram-negative proteobacteria (55). We disrupted sciW, rhs1, and rhs2 and found that disruption of rhs2 produced the same defect in systemic dissemination in mice as disruption of core T6SS genes and the entire SPI-6 locus (19). This Rhs element is located with sciX in a bicistronic operon and may also function as a toxin/immunity gene pair participating in contact-dependent growth inhibition (40).
Survival and replication within host cells permits systemic dissemination in vivo, and disruption of the SPI-6 T6SS resulted in reduced intracellular replication in macrophages. In contrast to systemic dissemination, we found that only noncore gene cluster 1 and 3 mutations had significant effects on intracellular replication in cell culture. Deletion of individual genes within cluster 1 revealed a significant replication defect for STM0278. Like rhs2-sciX, this gene is also part of a bicistronic gene pair with sciL that may also function as a toxin/immunity gene pair. We found that orthologs of these genes are present in close association with T6SS loci in a number of other members of the family Enterobacteriaceae. Given that this gene pair is the most strongly conserved of all of the noncore T6SS-associated genes in clusters 1 to 3, we are further pursuing their characterization. Deletion of individual genes within cluster 3 did not significantly attenuate intracellular replication and in fact produced a nonsignificant increase in intracellular replication. Interestingly, deletion of cluster 3 gene sciU produced a trend toward increased intracellular replication and systemic dissemination. These genes may have redundant functions, or attenuation in the noncore gene cluster 3 mutant may be due to polar effects of the disruption. The difference in mutant fitness between in vivo systemic dissemination and cell culture intracellular replication experiments may be explained by differences in the length of experiment and cell types encountered as assessment of intracellular replication in cell culture is a simplified model of one aspect of systemic dissemination. Oral infection of mice with S. Typhimurium involves passage through the gut environment before systemic dissemination, and it is possible that genes in clusters 2 and 4 are involved in interbacterial competition in this milieu. These multiple gene sets may act to repurpose the general type 6 secretory apparatus for delivery of effector proteins to different cell targets. While other T6SSs, such as that of P. aeruginosa and V. cholerae, have been found to have antibacterial properties, we were unable to observe this phenomenon in the S. enterica T6SS in solid LB agar-based competition assays against E. coli (data not shown) (22, 33). In addition, a previous report found that deletion of the entire SCI genomic island resulted in a 50% reduction of HEp-2 epithelial cells but we were unable to observe an effect on the invasion of both HeLa and HEp-2 epithelial cells by our sciG and sciS mutants, suggesting that gut epithelial cells may not be the target of this system and that this reduction in invasion may be due to a different SCI-encoded factor (16). Interestingly, sciL in cluster 1 and rhs2 and sciX in cluster 4 were found to be upregulated in macrophages and not in HeLa epithelial cells (21). sciL is located in a bicistronic operon with STM0278 in cluster 1, which had the most notable contribution to intracellular replication in macrophages. rhs2 and sciX are located as a bicistronic operon in cluster 4, and rhs2 made the most notable contribution to systemic dissemination in mice. Additionally, the sciL, STM0278, rhs1, and rhs2 genes have the largest number of orthologs of all of the genes in clusters 1 to 4.
Given the number of environments encountered by S. Typhimurium, virulence systems require regulatory control to achieve appropriate situational expression. We observed only weak chromosomal expression and did not observe plasmid-based secretion of Hcp-1, Hcp-2, or VrgS following growth under in vitro conditions after 24 h, which argues against a general role for the SPI-6 T6SS. In contrast, Hcp-3 appears to be constitutively expressed, negatively regulated by PmrA, and secreted under these conditions; however, the secretion of Hcp-3 is, curiously, not dependent on sciG. The ClpV ATPase is encoded by a core T6SS gene, and its abrogation results in the inability of the T6SS to assemble and secrete Hcp in V. cholerae (6). The Hcp-3 coding sequence differs significantly from those of Hcp-1 and Hcp-2, more closely resembling those of the Hcp proteins of S. bongori and Pseudomonas, and may have evolved to perform alternative functions, as is the case for the fourth S. enterica Hcp-encoding gene paralog hilE (16). Hcp-3 may therefore have an unrelated function and may be accessing the extracellular environment through an alternate secretory pathway. sciG may be dispensable for functional secretion in S. Typhimurium; however, the equivalent defects observed in the sciG and sciS mutants in both systemic dissemination in mice and intracellular replication in macrophages make this unlikely. The expression of Hcp-1 and that of Hcp-2 are not identical, as we observed expression of Hcp-1 in the absence of macrophages in DMEM, in contrast to that of Hcp-2 and VrgS, which was barely detectable. Hcp-1 is encoded by sciK within noncore gene cluster 1, and its expression is perhaps a reflection of promoter changes during its acquisition or duplication from sciM.
The absence of expression and secretion of the Hcp-2 and VrgS proteins suggests a system under tight regulatory control. The expression of S. Typhimurium SPI-2 T3SS virulence genes important for replication and survival in macrophages is upregulated upon exposure to the intracellular compartment (31). In a similar manner, we found that the genes for Hcp and VrgS are differentially expressed in the presence or absence of macrophages and between in vitro and tissue culture conditions. Hcp-1 is encoded within noncore gene cluster 1, and like that of Hcp-2 and VrgS, its expression was clearly enhanced during late stages of macrophage infection. The difference in the expression of these proteins between in vitro and cell culture conditions suggests an activating signal specific to growth in the presence of macrophages but may also be in response to general cell stress. Expression of the SPI-6 T6SS may rely on regulatory inputs from the host environment in a manner similar to that of the T6SS of V. cholerae, which requires internalization of bacteria (32). With regard to S. Typhimurium, expression of the T6SS does not appear to occur until after 8 h of infection and has been previously hypothesized to be a result of the loss of repression through an SsrB-dependent regulatory pathway. It was hypothesized that induction may be relevant to host cell death and bacterial escape (38). Another group reported the upregulation of some SPI-6 T6SS genes during macrophage infection but did not observe an increase over time (21). It is possible that this induction relies on the onset of host cell death, which can be affected by differing MOIs and experimental conditions. Indeed, when MOIs were much reduced, we observed that Hcp and VrgS expression was lower (data not shown). An important consideration is that expression levels of this system, even in the context of macrophage infection, remain low and we have not yet been able to observe translocation of tagged proteins to the host cells and therefore further work is necessary to determine whether this system is activated within the intracellular compartment or activated simply in the presence of macrophages.
Induction of the SPI-6 T6SS could be similar to that of the HSI-I T6SS in Pseudomonas spp., where deletion of the sensor kinase RetS leads to enhanced secretory activity of effector proteins (22). We pursued this hypothesis by identifying transcriptionally active regions within the SPI-6 T6SS. We found six transcriptionally active regions, including one within noncore gene cluster 2, active in vitro and in association with host macrophages. A previous report observed increased numbers of transcripts of sciS in ssrB null mutants during late stages of host cell infection (38). We also found increased levels of transcription of the T6SS in an ssrB mutant background, with increased transcriptional activity from regions upstream of sciF and vrgS. The regulatory proteins PhoP and SlyA are involved in the activation of SsrB (3, 37), and we found that transcriptional activity upstream of sciF also increases in this mutant background; however, sciG and vrgS transcriptional activity decreased, suggesting additional regulatory inputs to this system between PhoP and SsrB. The transcriptional profiles of all six reporters in both the phoP and slyA mutant backgrounds were identical, suggesting that they act through a common regulatory input. We previously reported the genome-wide interaction map of SsrB using chromatin immunoprecipitation and found only two interaction sites within the SPI-6 T6SS, upstream of sciF and sciR (51). Given our observation of increased transcriptional activity upstream of sciF and the previously reported increase in sciS transcripts in a ssrB mutant background, we asked whether the loss of these regulatory inputs has a significant effect on VrgS protein expression through regulatory deregulation. Other regulatory proteins aside from PhoP-PhoQ, SsrA-SsrB, and SlyA are necessary for intracellular survival during host infection (56). VrgS levels were not affected in the phoP, ssrB, qseC, ompR, or hfq mutant background, although the disruption of these proteins results in extensive changes in S. Typhimurium gene expression and leads to abrogation of pathogenesis (56). While these results are not extensive, we believe that they argue for the presence of an activating factor that controls T6SS upregulation in response to inducing signals. An unbiased transposon-screening approach is likely to be necessary to identify such a regulator.
In summary, these findings further support a role for the SPI-6 T6SS during host infection, likely mediated through interactions with macrophages and enabled by the SPI-6 T6SS noncore genes STM0278 and rhs2. Two transcriptionally active regions of this system receive negative regulatory input through the SsrB regulatory network; however, the expression of this system appears to be controlled by an as-yet-unknown regulatory factor.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by an operating grant from the Canadian Institutes of Health Research (MOP 82704), an infrastructure grant from the Canada Foundation for Innovation, and the Canada Research Chairs Program from the Government of Canada. D.T.M. is supported by a Canada Graduate Scholarship from the Canadian Institutes of Health Research. C.A.C. was the recipient of a Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada. B.K.C. is the Canada Research Chair in Infectious Disease Pathogenesis.
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 9 April 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernard CS, Brunet YR, Gueguen E, Cascales E. 2010. Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192:3850–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bijlsma JJE, Groisman EA. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57:85–96 [DOI] [PubMed] [Google Scholar]
- 4. Bingle LE, Bailey CM, Pallen MJ. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11:3–8 [DOI] [PubMed] [Google Scholar]
- 5. Blondel CJ, Jiménez JC, Contreras I, Santiviago CA. 2009. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28:315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan K, Kim CC, Falkow S. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect. Immun. 73:5438–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coombes BK, Brown NF, Valdez Y, Brumell JH, Finlay BB. 2004. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J. Biol. Chem. 279:49804–49815 [DOI] [PubMed] [Google Scholar]
- 11. Coombes BK, Wickham ME, Lowden MJ, Brown NF, Finlay BB. 2005. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. U. S. A. 102:17460–17465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JCD. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 14. Filloux A, Hachani A, Bleves S. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154:1570–1583 [DOI] [PubMed] [Google Scholar]
- 15. Folkesson A, et al. 1999. Multiple insertions of fimbrial operons correlate with the evolution of Salmonella serovars responsible for human disease. Mol. Microbiol. 33:612–622 [DOI] [PubMed] [Google Scholar]
- 16. Folkesson A, Löfdahl S, Normark S. 2002. The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res. Microbiol. 153:537–545 [DOI] [PubMed] [Google Scholar]
- 17. Fookes M, et al. 2011. Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathog. 7:e1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerlach RG, Hensel M. 2007. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297:401–415 [DOI] [PubMed] [Google Scholar]
- 19. Haneda T, Ishii Y, Danbara H, Okada N. 2009. Genome-wide identification of novel genomic islands that contribute to Salmonella virulence in mouse systemic infection. FEMS Microbiol. Lett. 297:241–249 [DOI] [PubMed] [Google Scholar]
- 20. Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66 [DOI] [PubMed] [Google Scholar]
- 21. Hautefort I, et al. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell. Microbiol. 10:958–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hood RD, et al. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. 2011. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J. Bacteriol. 193:6471–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klumpp J, Fuchs TM. 2007. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology 153:1207–1220 [DOI] [PubMed] [Google Scholar]
- 25. Kuhle V, Hensel M. 2004. Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell. Mol. Life Sci. 61:2812–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lambert MA, Smith SGJ. 2008. The PagN protein of Salmonella enterica serovar Typhimurium is an adhesin and invasin. BMC Microbiol. 8:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lane MC, Alteri CJ, Smith SN, Mobley HLT. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. U. S. A. 104:16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lawley TD, et al. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leiman PG, et al. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U. S. A. 106:4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Libby SJ, et al. 2010. Humanized nonobese diabetic-scid IL2rgammanull mice are susceptible to lethal Salmonella Typhi infection. Proc. Natl. Acad. Sci. U. S. A. 107:15589–15594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Löber S, Jäckel D, Kaiser N, Hensel M. 2006. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 296:435–447 [DOI] [PubMed] [Google Scholar]
- 32. Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. 2009. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5:234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U. S. A. 107:19520–19524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marchal K, et al. 2004. In silico identification and experimental validation of PmrAB targets in Salmonella typhimurium by regulatory motif detection. Genome Biol. 5:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marchler-Bauer A, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9:797–803 [DOI] [PubMed] [Google Scholar]
- 37. Okada N, et al. 2007. Identification of amino acid residues of Salmonella SlyA that are critical for transcriptional regulation. Microbiology 153:548–560 [DOI] [PubMed] [Google Scholar]
- 38. Parsons DA, Heffron F. 2005. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect. Immun. 73:4338–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. 2009. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U. S. A. 106:4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Poole SJ, et al. 2011. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 7:e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A. 104:15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pukatzki S, et al. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103:1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pukatzki S, McAuley SB, Miyata ST. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12:11–17 [DOI] [PubMed] [Google Scholar]
- 44. Records AR, Gross DC. 2010. Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. J. Bacteriol. 192:3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Russell AB, et al. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sabbagh SC, Forest CG, Lepage C, Leclerc J-M, Daigle F. 2010. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol. Lett. 305:1–13 [DOI] [PubMed] [Google Scholar]
- 47. Schwarz S, Hood RD, Mougous JD. 2010. What is type VI secretion doing in all those bugs? Trends Microbiol. 18:531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwarz S, et al. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6:e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shalom G, Shaw JG, Thomas MS. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153:2689–2699 [DOI] [PubMed] [Google Scholar]
- 50. Silverman JM, et al. 2011. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol. Microbiol. 82:1277–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tomljenovic-Berube AM, Mulder DT, Whiteside MD, Brinkman FSL, Coombes BK. 2010. Identification of the regulatory logic controlling Salmonella pathoadaptation by the SsrA-SsrB two-component system. PLoS Genet. 6:e1000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 98:15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang M, et al. 2011. Molecular characterization of a functional type VI secretion system in Salmonella enterica serovar Typhi. Curr. Microbiol. 63:22–31 [DOI] [PubMed] [Google Scholar]
- 54. Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
- 55. Wang YD, Zhao S, Hill CW. 1998. Rhs elements comprise three subfamilies which diverged prior to acquisition by Escherichia coli. J. Bacteriol. 180:4102–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yoon H, McDermott JE, Porwollik S, McClelland M, Heffron F. 2009. Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium. PLoS Pathog. 5:e1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.