Abstract
Urogenital Chlamydia serovars replicating in reproductive epithelium pose a unique challenge to host immunity and vaccine development. Previous studies have shown that CD4 T cells are necessary and sufficient to clear primary Chlamydia muridarum genital tract infections in the mouse model, making a protective CD4 T cell response a logical endpoint for vaccine development. Our previous proteomics studies identified 13 candidate Chlamydia proteins for subunit vaccines. Of those, PmpG-1 is the most promising vaccine candidate. To further that work, we derived a PmpG303-311-specific multifunctional Th1 T cell clone, designated PmpG1.1, from an immune C57BL/6 mouse and used it to investigate the presentation of the PmpG303-311 epitope by infected epithelial cells. Epithelial presentation of the PmpG303-311 epitope required bacterial replication, occurred 15 to 18 h postinfection, and was unaffected by gamma interferon (IFN-γ) pretreatment. Unlike epitopes recognized by other Chlamydia-specific CD4 T cell clones, the PmpG303-311 epitope persisted on splenic antigen-presenting cells (APC) of mice that cleared primary genital tract infections. PmpG1.1 was activated by unmanipulated irradiated splenocytes from immune mice without addition of exogenous Chlamydia antigen, and remarkably, activation of PmpG1.1 by unmanipulated immune splenocytes was stronger 6 months postinfection than it was 3 weeks postinfection. Enhanced presentation of PmpG303-311 epitope on splenic APC 6 months postinfection reflects some type of “consolidation” of a protective immune response. Understanding the antigen-presenting cell populations responsible for presenting PmpG303-311 early (3 weeks) and late (6 months) postinfection will likely provide important insights into stable protective immunity against Chlamydia infections of the genital tract.
INTRODUCTION
Public health measures to control Chlamydia trachomatis genital tract infections, combining case identification with partner tracing and treatment programs, have had some success in decreasing the incidence of pelvic inflammatory disease (PID), but not the incidence and prevalence of sexually transmitted infections (2, 3, 23). Development of a protective vaccine for prevention of C. trachomatis urogenital tract infections will be challenging, as antibody has no discernible role in clearing primary infections (21, 27); therefore, the critical components of a Chlamydia vaccine will likely be its T cell epitopes. A wealth of data from the Chlamydia muridarum mouse model for Chlamydia genital tract infections suggests the critical T cell epitopes are presented by major histocompatibility complex (MHC) class II (human HLA-DP, -DQ, and DR) molecules to Chlamydia-specific CD4 T cells (18, 19). To date, there are no examples of a protective human subunit vaccine based on T cell immunity or against an intracellular bacterial pathogen.
To facilitate rational vaccine development, we have focused on identifying Chlamydia peptides loaded onto MHC class II molecules using the C. muridarum mouse model (14, 32). Chlamydia peptides loaded onto MHC class II molecules have the potential to be recognized by T cell receptors on Chlamydia-specific T cells and, by definition, are derived from Chlamydia proteins whose biology within infected cells makes them susceptible to host cell antigen-processing and presentation machinery. Identification of Chlamydia proteins processed and presented by infected cells is critical for rational vaccine development, as a large fraction of Chlamydia proteins are likely sequestered apart from processing and presentation machinery by exclusive residence in the inclusion body. We and others have shown that protective immunity against C. muridarum genital tract infections can be induced by adoptive transfer of antigen-pulsed dendritic cells (7, 12, 28, 30); therefore, our initial efforts focused on that cell type. We previously identified a panel of CD4 and CD8 T cell epitopes by immunoprecipitation of MHC class II and class I molecules from infected C57BL/6-derived dendritic cells, eluting the resident peptides, and identifying those peptides using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (14, 32). We found that of the identified epitope source proteins, immunization with a PmpG-1 fusion protein, the source protein for the I-Ab-presented epitope PmpG303-311, provided the greatest protection against C. muridarum infectious challenge in the genital tract (30, 31).
In this study, we advance that research by investigating the characteristics of the vaccine-protective T cell epitope PmpG303-311. To that end, we derived a CD4 T cell clone specific for PmpG303-311 from an immune mouse that had previously cleared a genital tract infection. The resulting CD4 T cell clone, designated PmpG1.1, was a useful tool for investigating the presentation of the PmpG303-311 epitope in vitro and in vivo. We present interesting results of those studies here.
MATERIALS AND METHODS
Mice.
Female C57BL/6 mice were purchased from Harlan Laboratories (Indianapolis, IN). All mice were housed in Indiana University Purdue University-Indianapolis (IUPUI) specific-pathogen-free (SPF) facilities. The IUPUI Institutional Animal Care and Utilization Committee approved all experimental protocols.
Cells and bacteria.
C57epi.1 is a cloned oviduct epithelial cell line derived from a C57BL/6 mouse (H-2b) (11). C57epi.1 cells are grown at 37°C in a 5% CO2 humidified incubator in epithelial cell medium (1:1 Dulbecco's modified Eagle medium-F12K [Sigma, St. Louis, MO]) supplemented with 10% characterized fetal bovine serum (FBS) (HyClone, Logan, UT), 2 mM l-alanyl-l-glutamine (Glutamax I; Gibco/Invitrogen), 5 μg of bovine insulin/ml, and 12.5 μg/ml of recombinant human FGF-7 (keratinocyte growth factor; Sigma).
Mycoplasma-free C. muridarum (Nigg), previously known as C. trachomatis strain MoPn, was grown in McCoy cells (ATCC). The titers of mycoplasma-free C. muridarum stocks were determined on McCoy cells by centrifugation, as previously described (13). UV-inactivated C. muridarum stocks were made by diluting concentrated stocks in phosphate-buffered saline (PBS) and then exposing roughly 3 to 4 ml of diluted stock in a sterile petri dish to 1,200 J/cm2 twice in a UV-cross-linking cabinet (Spectralinker; Spectronics Corporation, Westbury, NY).
Infection of mice.
C57BL/6 mice were treated with 2.5 mg of depoprogesterone (Depo-Provera; Pfizer, New York, NY) injected subdermally 1 week prior to infection. Vaginal infections were accomplished with 5 × 104 inclusion-forming units (IFU) of C. muridarum in 10 μl of sucrose-phosphate-glutamic acid (SPG) buffer. The mice were swabbed 7 days later to confirm infection. Vaginal swab IFU were recovered in SPG buffer and quantified using McCoy cell monolayers as previously described (25).
CD4 T cell clones.
Chlamydia-specific CD4 T cell clones (uvm-2, uvmo-3, uvmo-4, and spl4-10) were grown in RPMI 1640 (Sigma) supplemented with 10% characterized fetal bovine serum (HyClone), 2 mM l-alanyl-l-glutamine (Glutamax I; Gibco/Invitrogen), 25 μg/ml gentamicin (Sigma), 5 × 10−5 M 2-mercaptoethanol (RPMI CM) (Sigma), and secondary mixed lymphocyte culture (MLC) supernatant with recombinant cytokines as previously described (11).
Chlamydia-specific CD4 T cell clone PmpG1.1 recognizing PmpG303-311 was derived from an immune C57BL/6 (H-2b) female mouse that cleared a primary C. muridarum genital tract infection without development of hydrosalpinx. Immune splenocytes were plated at 4 × 106 cells per well in tissue culture-treated 24-well plates containing 1.5 ml RPMI CM with murine recombinant interleukin 1α (IL-1α) (2 ng/ml), IL-6 (2 ng/ml), IL-7 (3 ng/ml), IL-15 (4 ng/ml), human recombinant IL-2 (100 units/ml), 20% secondary MLC supernatant, and 1.5 μg of recombinant PmpG-1 (amino acids [aa] 25 to 500 with an N-terminal His tag) (30). The resulting polyclonal T cell population was purified with Ficoll-Hypaque (Histopaque 1083; Sigma) and restimulated under the same conditions using 5 × 106 irradiated immune splenocytes as antigen-presenting cells (APC). T cells resulting from the secondary PmpG-1-stimulated passage were cloned by limiting dilution using irradiated immune splenocytes pulsed with UV-inactivated C. muridarum as a specific antigen. Five clones resulting from the limiting dilution were screened for activation by PmpG303-311. One clone was specific for PmpG303-311; it was designated PmpG1.1 and kept for further study.
For routine passage, 1 × 105 PmpG1.1 T cells were plated in 24-well tissue culture-treated wells containing 1.5 ml of RPMI CM-15% MLC supernatant supplemented with cytokines as previously described using 5 × 106 γ-irradiated naïve C57BL/6 splenocytes (1,200 rads) that had been prepulsed at 37°C with 2.5 IFU equivalents of UV-inactivated C. muridarum per splenocyte for 20 min (11).
T cell activation.
T cells were activated by irradiated splenic APC with or without antigen or immobilized anti-CD3 antibody. For activation with immobilized anti-CD3 (145-2C11; no azide/low endotoxin [NA/LE]; BD Pharmingen), 96-well tissue culture plates were prepared by incubating 0.5 μg/ml antibody in PBS, 50 μl per well, overnight at 4°C. The wells were washed once with 150 μl of medium prior to use.
T cell activation by irradiated (1,200 rad) splenic APC was performed with 5 × 105 irradiated splenocytes and 2.5 × 104 T cell clone cells in a 150-μl total volume in 96-well round-bottom plates. Proliferation was quantified by the addition of 0.5 μCi 3H-thymidine to each well at 36 h; the wells were harvested 12 h later, and counts-per-minute were quantified with a TopCount Beta Counter (Perkin Elmer, Waltham, MA).
Epithelial cell infections.
C57epi.1 cells were plated in 48-well tissue culture plates and infected with 3 IFU of C. muridarum per cell in 0.3 ml of culture medium. The 48-well plates were centrifuged at 1,200 rpm (300 × g) in a tabletop centrifuge for 30 min and then incubated at 37°C in a 5% CO2 humidified incubator. Mock-infected wells received an equivalent volume of SPG buffer lacking C. muridarum.
For antibody inhibition experiments, monolayers were washed, and then 2 μg (10 μg/ml) of sterile azide-free anti-CD4 monoclonal (GK1.5; eBioscience, San Diego, CA) or isotype control (Ig2b k; eBioscience) antibody was added to each well prior to addition of T cell clones.
Flow cytometry.
Cells were stained for 20 min on ice in RPMI CM-10% FBS with phycoerythrin (PE)-coupled GK1.5 (CD4) or PE-coupled 53-6.7 (CD8α). The cells were fixed with 1% paraformaldehyde after staining and analyzed using a FACSCalibur cytometer (BD Biosciences, San Diego, CA).
ELISA determination of IFN-γ.
Relative IL-2 (1A12 and 5H4; Thermo Scientific, Rockford, IL), gamma interferon (IFN-γ) (XMG1.2; Thermo Scientific), tumor necrosis factor alpha (TNF-α) (ΤΝ3-19.12 and rabbit anti-TNF-α; BD Pharmingen, San Diego, CA), IL-6 (32C11 and 20F; Thermo Scientific), IL-10 (JES5-2A5 and SXC-1; BD Pharmingen), IL-17a (TC11-18H10.1 and TC11-8H4; Biolegend, San Diego, CA), and IL-21 (IL-21 enzyme-linked immunosorbent assay [ELISA] kit; Ebioscience, San Diego, CA) levels were determined by ELISA using monoclonal antibodies according to the manufacturer's protocols. Recombinant murine IL-2, IL-6, IL-10 (Thermo Scientific), TNF-α, IFN-γ (R&D Systems, Minneapolis, MN), IL-17a (Biolegend), and IL-21 (Ebioscience) were used as standards.
Statistical analysis.
Summary figures for each experimental investigation are presented as means and standard deviations (SD) for single experiments or as “pooled” means and standard errors of the mean (SEM) for aggregate data from 2 or more independent experiments. All experiments were performed with a minimum of triplicate wells for each data point. The figure legends indicate the number of independent experiments pooled to generate each figure; for single experiments, the number of replicate wells for each data point is indicated. Student's two-tailed t tests were used to assess the significance of experimental data. The homogeneity of variances was assessed using a folded F test. All data were verified to meet analytic assumptions. Nonparametric tests were also performed and showed similar results. Analyses were performed using SAS 9.2 (SAS Institute, Cary, NC).
RESULTS
Derivation of a PmpG303-311-specific CD4 T cell clone.
We previously identified C. muridarum peptides bound to MHC class II molecules of infected dendritic cells and tested whether vaccination with their epitope source proteins could protect mice from C. muridarum genital tract infections. Our most protective epitope source protein in that study was PmpG-1, containing the T cell epitope PmpG303-311. Studying presentation of T cell epitopes by antigen-presenting cells is experimentally difficult. One approach is to derive monoclonal antibodies specific for peptide epitopes complexed with MHC/HLA molecules that can be used to quantify epitope levels on the cell surfaces by flow cytometry (5, 6, 15). Alternatively, epitope-specific T cell clones can be used as a tool to study presentation of epitope-MHC complexes on the cell surface at or above the threshold level for T cell activation. To study presentation of the protective PmpG303-311 CD4 T cell epitope in vitro and in vivo, we derived a PmpG303-311-specific CD4 T cell clone.
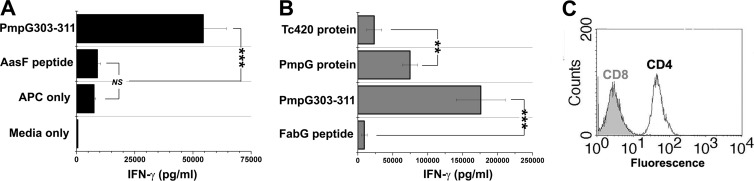
Initially, immune splenocytes from C57BL/6 mice that previously self-cleared primary C. muridarum genital tract infections were stimulated and restimulated in vitro with PmpG303-311 peptide. Polyclonal T cell populations generated by this approach did not show specificity for the PmpG303-311 peptide, even though freshly isolated immune splenocytes had strong and specific IFN-γ responses to PmpG303-311 (data not shown). An alternative strategy was adopted. Immune splenocytes from a C57BL/6 mouse that self-cleared a primary C. muridarum genital tract infection were stimulated ex vivo with recombinant PmpG-1 amino acids 25 to 500, containing the PmpG303-311 epitope and an N-terminal His tag, in primary culture and then restimulated 7 days later under the same culture conditions using irradiated immune splenocytes as the APC. Seven days after the secondary stimulation, the polyclonal population was tested for PmpG303-311 specificity (Fig. 1A), and limiting diluted on irradiated immune splenocytes pulsed with UV-inactivated C. muridarum to generate T cell clones. Five clones resulting from the limiting dilution of the PmpG-1 polyclonal population were screened for specificity; one clone, designated PmpG1.1, was specific for recombinant PmpG-1 aa 25 to 500 and recognized the PmpG303-311 epitope previously identified in proteomic studies (Fig. 1B). PmpG1.1 is a CD4+ CD8-negative (CD8neg) T cell clone (Fig. 1C).
Fig 1.
Derivation of PmpG303-311-specific CD4 T cell clone PmpG1.1. (A) The polyclonal T cell population resulting from the secondary in vitro stimulation of immune splenocytes with PmpG-125-500 was tested for specificity for the PmpG303-311 epitope. The T cells were activated with irradiated naïve splenocyte APC plus 2 μg/ml of the indicated peptides or the medium control, IFN-γ in 48-h culture supernatants quantified by ELISA. The data presented are from a single experiment done in triplicate. The error bars indicate SD. (B) Specificity of the PmpG1.1 T cell clone. The PmpG1.1 T cell clone was activated with immune irradiated splenocyte APC plus 1.5 μg/ml of the indicated proteins or 2 μg/ml of the indicated peptides. IFN-γ in 48-h culture supernatants was quantified by ELISA; aggregate data from two independent experiments are shown. (C) PmpG1.1 was stained for CD8a and CD4 and analyzed by flow cytometry. **, P < 0.005; ***, P < 0.0005; NS, not statistically significant. The error bars indicate SEM.
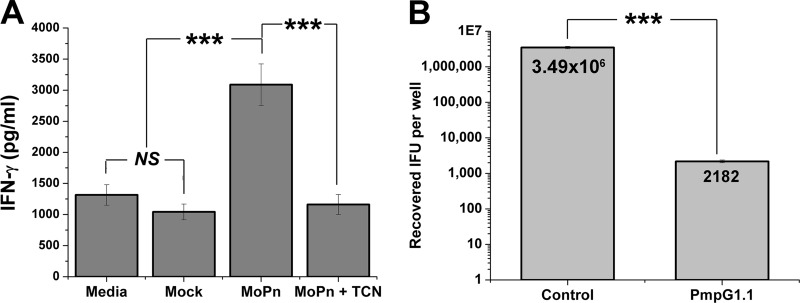
We tested whether PmpG1.1 recognized infected epithelial cells and whether recognition was replication dependent. C57epi.1 epithelial cells were pretreated with 10 ng/ml IFN-γ for 12 h and then mock infected and infected with C. muridarum in the absence or presence of 10 μg/ml tetracycline (which stops bacterial protein synthesis and epitope generation). C57epi.1 cells were harvested 18 h postinfection and then cocultured 1:1 with PmpG1.1 for 24 h in the presence of 10 μg/ml tetracycline (all wells). Culture supernatants were collected after 24 h, and IFN-γ levels were determined by ELISA (Fig. 2A). PmpG1.1 recognized infected epithelial cells, and recognition of infected epithelial cells required bacterial replication, as it was blocked by the presence of tetracycline during the infection.
Fig 2.
PmpG1.1 recognition of infected epithelial cells and termination of C. muridarum replication within them. (A) PmpG1.1 cells were cocultured 1:1 with C57epi.1 cells that were mock infected or 18 h postinfection with 5 IFU per cell (MoPn), without and with tetracycline (TCN) cotreatment; IFN-γ in 24-h culture supernatants was quantified by ELISA. (B) C57epi.1 monolayers, pretreated for 12 h with 10 ng/ml IFN-γ, were infected with C. muridarum (3 IFU per cell). Four hours later, the monolayers were washed and then cocultured with PmpG1.1 T cells at an effector-to-target ratio of 0.75:1. The contents of the wells were harvested 32 h postinfection, and the recovered IFU were enumerated on McCoy monolayers. The mean number of IFU recovered for each condition is shown within the bars. Panels A and B represent single experiments done in quadruplicate. ***, P < 0.0005; NS, not statistically significant. The error bars indicate SD.
A previous study showed that in vivo protection by CD4 T cell clones in adoptive transfer correlated with a clone's ability to control C. muridarum replication in epithelial cells (9). We tested whether PmpG1.1 could control C. muridarum replication in C57epi.1 epithelial cells. C57epi.1 epithelial cell monolayers in 48-well plates were pretreated with 10 ng/ml IFN-γ for 12 h and then infected with C. muridarum (3 IFU per cell). Four hours later, the inocula were removed, the cell monolayers were washed and then cultured in T cell medium with or without 1.5 × 105 PmpG1.1 T cells at an effector-to-target ratio of ∼0.75:1. Twenty-eight hours later (32 h postinfection), the cell monolayers were harvested by adding SPG buffer and scraping. The number of IFU recovered per well was determined on McCoy monolayers (Fig. 2B). PmpG1.1 terminates C. muridarum replication in epithelial cells with an efficiency comparable to those of other potent previously described Chlamydia-specific CD4 T cell clones (10).
Cytokine profile of PmpG1.1.
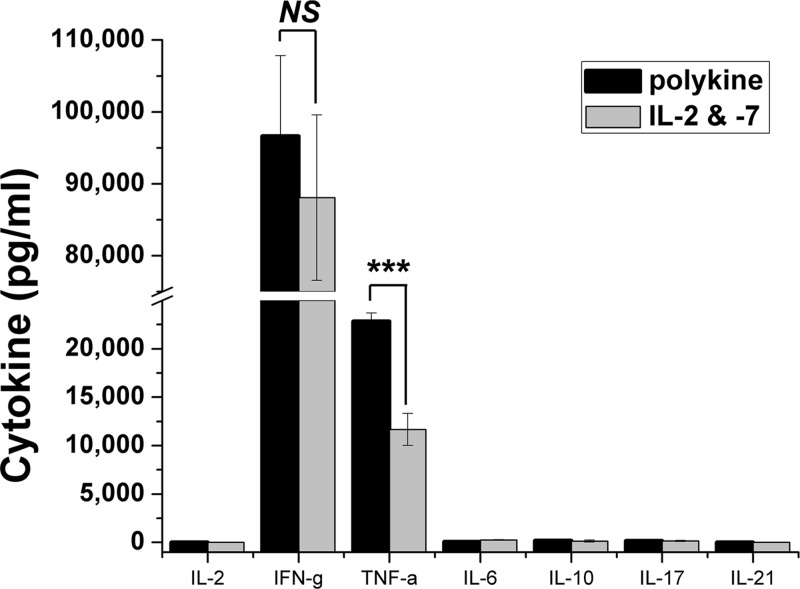
We have shown that multifunctional Th1 T cells secreting both IFN-γ and TNF-α correlate strongly with protective genital tract immunity in mice immunized with recombinant Chlamydia antigens or by C. muridarum nasal infection (31, 32). Accordingly, we analyzed the cytokine profile of PmpG1.1 and investigated whether that profile was influenced by culture conditions ex vivo. Over a 6-month period, PmpG1.1 T cells were passed in parallel using the standard protocol (multiple cytokines plus secondary MLC supernatant [see Materials and Methods]) and the same conditions, except that exogenous cytokines were limited to the IL-2 and IL-7 components of the growth media. PmpG1.1 T cells grown in parallel under these two different conditions for 6 months were activated by immobilized anti-CD3 antibodies at the end of their usual culture cycle. Twenty hours postactivation, the culture supernatants were collected and analyzed for IL-2, IFN-γ, TNF-α, IL-6, IL-10, IL-17, and IL-21 (Fig. 3). Activated PmpG1.1 produced significant levels of IFN-γ and TNF-α. Low levels of IL-6 and IL-17 were close to, and IL-10 overlapped, 0 pg/ml within the 95% confidence intervals. IL-17 production was not induced by addition of transforming growth factor beta (TGF-β), IL-23, and IL-6 to the culture medium (data not shown). No IL-2 or IL-21 was detectable in the culture supernatants. There was an approximately 2-fold difference in the levels of TNF-α production between the two culture conditions but no meaningful difference in the overall cytokine profiles. The lack of a significant difference between PmpG1.1's cytokine profile over 6 months of culture in dramatically different cytokine milieus suggests that the diverse intracellular cytokine patterns seen in activated immune T cells from vaccinated and infected animals reflect stable T cell cytokine differentiation patterns. PmpG1.1 has the multifunctional IFN-γ and TNF-α cytokine profile associated with protective immunity in C. muridarum vaccination studies. We attempted to test PmpG1.1's protective capability by adoptive transfer into Rag1 knockout (KO) mice followed by C. muridarum infectious challenge. Those studies were inconclusive, as PmpG1.1 T cells were not found (zero CD4+ cells) in the spleen, lymph nodes, or genital tracts of adoptively transferred Rag1 KO mice, even with pretransfer natural killer cell depletion using monoclonal antibody NK1.1 (data not shown).
Fig 3.
Cytokine profile of PmpG1.1 maintained in different cytokine milieus. PmpG1.1 T cells (5 × 104) were activated by immobilized anti-CD3 antibody 145-2c11 in medium containing 1 ng/ml IL-7, and 20-h culture supernatants were collected and analyzed for the indicated cytokines by ELISA. Shown are aggregate data from six independent experiments determining IFN-γ levels, with two independent experiments for each of the other cytokines. For IL-2, IL-6, IL-10, and IL-21, the measured level of the cytokine for one or both conditions overlapped 0 pg/ml within the 95% confidence interval. ***, P < 0.0005; NS, not statistically significant. The error bars indicate SEM.
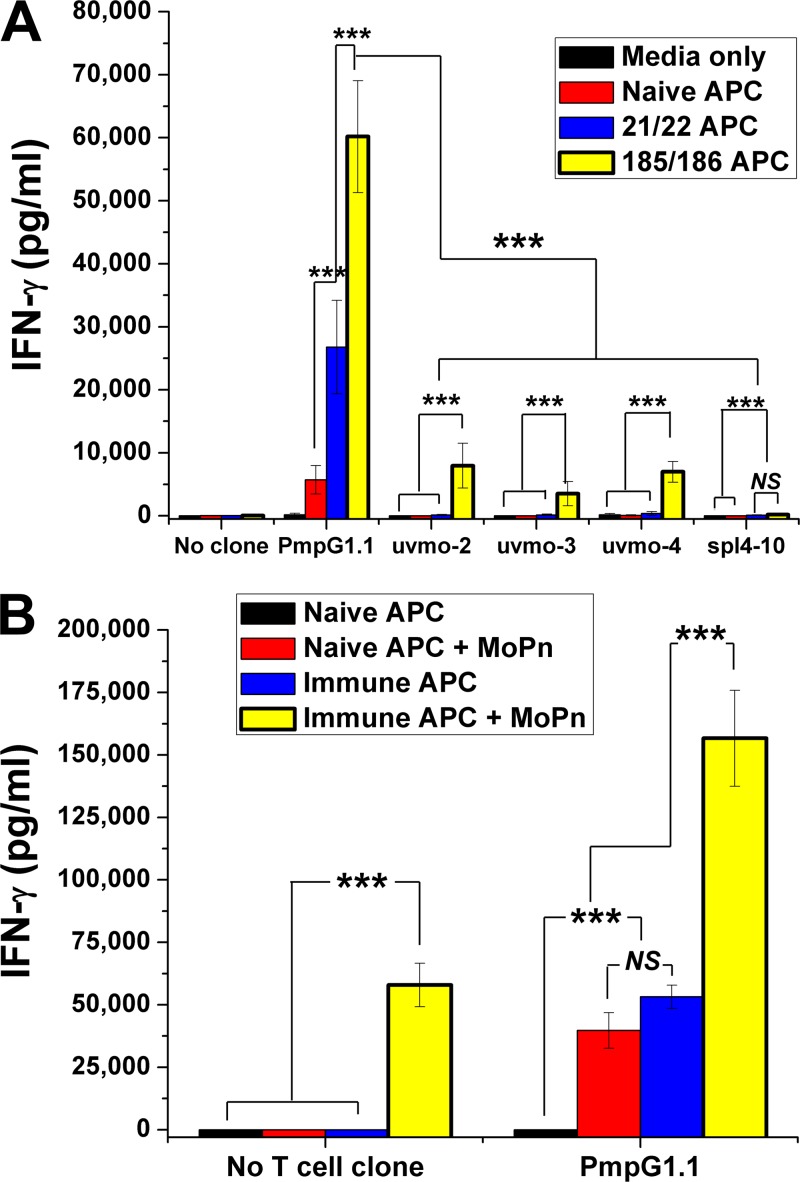
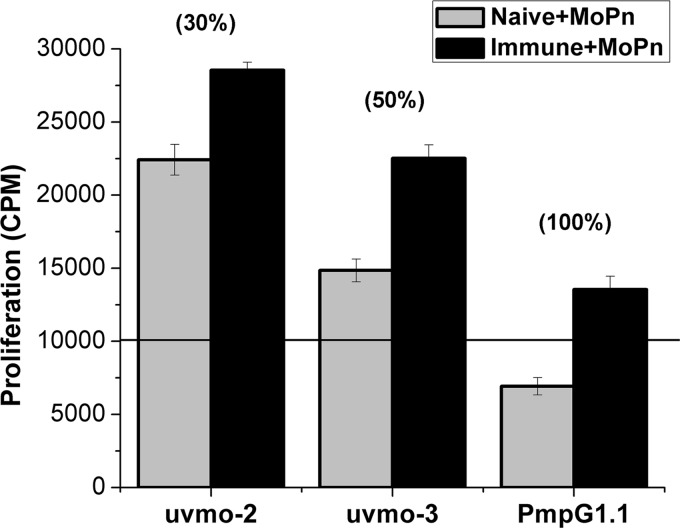
A reasonable hypothesis for the failure to find PmpG1.1 after adoptive transfer into Rag1 knockout mice followed by C. muridarum infection is that PmpG1.1 is a helper-dependent CD4 T cell clone, i.e., that without an ability to make IL-2, PmpG1.1 is dependent on other T cells to proliferate. The recipient Rag1 knockout mice have no endogenous T cells. To investigate this possibility in vitro, we compared the ability of antigen-pulsed irradiated naïve (no T cell help; referred to hereafter as “no help”) versus immune splenocytes (T cell help; referred to hereafter as “help”) to activate uvmo-2, uvmo-3, and PmpG1.1. Irradiated splenocytes pulsed with UV-inactivated C. muridarum do not proliferate significantly; however irradiated immune but not naïve splenocytes pulsed with C. muridarum produce cytokines (data not shown; see Fig. 6B, “No T cell clone” control). uvmo-2 and uvmo-3, clones that make IL-2, were able to proliferate to both antigen-pulsed-irradiated naïve and immune splenocytes, while PmpG1.1 significantly proliferated only when “help” was available (irradiated immune splenocytes plus UV-inactivated C. muridarum) (Fig. 4).
Fig 6.
Presentation of the PmpG303-311 epitope persists in vivo >6 months postinfection. PmpG1.1 was activated by naïve splenocyte APC or immune splenocyte APC from mice 3 weeks (21/22) and 6 months (185/186) postinfection, without or with exogenous UV-inactivated C. muridarum antigen (MoPn). (A) T cell clones were cultured in medium (control) or cocultured with naïve and immune irradiated splenocytes without exogenous antigen. The culture supernatants were collected at 48 h, and IFN-γ was quantified by ELISA; aggregate data from two independent experiments are shown. Error bars indicate SEM. (B) PmpG1.1 cells were cocultured with naïve or immune (6 months postinfection) irradiated splenocytes in the absence and presence of UV-inactivated C. muridarum (MoPn). The culture supernatants were collected at 48 h, and IFN-γ was quantified by ELISA. The data presented are from one experiment done in triplicate. ***, P < 0.0005; NS, not statistically significant. The error bars indicate SD.
Fig 4.
Proliferation of individual T cell clones to naïve and immune APC pulsed with UV-inactivated C. muridarum (MoPn). T cell clones (2.5 × 104) were cocultured with 5 × 105 irradiated naïve or immune (4 months postinfection) splenocytes with UV-inactivated C. muridarum in the presence of 10 μg/ml tetracycline. Wells were pulsed with [3H]thymidine between 36 and 48 h to score T cell proliferation. The values in parentheses are the percent enhancement of proliferation afforded by the immune APC over the naïve APC for each clone. The data presented are from a single experiment performed in triplicate. For accounting purposes, the background proliferation of the irradiated APC was subtracted from each experimental well. None of the T cell clones proliferated to naïve APC without antigen (all <1,000 cpm). For each T cell clone, the immune APC were superior to the naïve APC (all P < 0.0005). uvmo-2 and uvmo-3 had greater proliferation than PmpG1.1 for each of the two experimental conditions, naïve APC or immune APC (all P < 0.0005). The error bars indicate SD.
Timing of presentation of the PmpG303-311 epitope on infected epithelial cells.
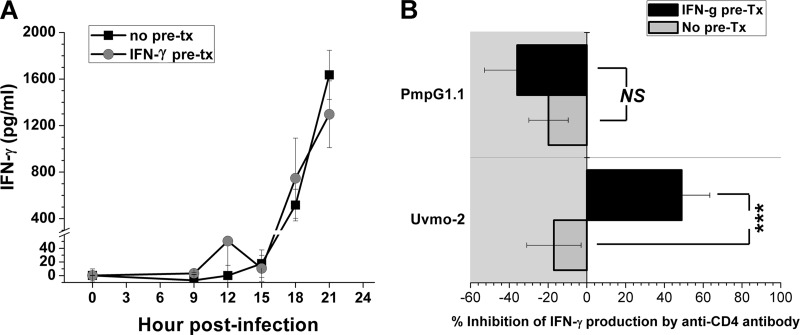
Derivation of a PmpG303-311-specificCD4 T cell clone provided an opportunity to investigate the mechanism and timing of infected-epithelial-cell presentation of a protective Chlamydia epitope. C57epi.1 epithelial cell monolayers in 96-well plates, untreated or pretreated with IFN-γ, were infected at staggered intervals over 12 h to generate mock-infected (0 h) and infected monolayers 9, 12, 15, 18, and 21 h postinfection. At endpoint, the inocula were removed and the monolayers were washed and then cocultured with PmpG1.1 T cells in the presence of tetracycline, an antibiotic that halts bacterial protein synthesis and therefore epitope generation. The culture supernatants were harvested at 48 h, and the levels of IFN-γ were determined by ELISA (Fig. 5A). The PmpG303-311 epitope reaches the cell surface in levels sufficient to activate PmpG1.1 T cells between 15 and 18 h postinfection. IFN-γ pretreatment of epithelial cells did not affect the timing of recognition or the magnitude of PmpG1.1 activation as measured by IFN-γ production. Activation of three Chlamydia-specific CD4 T cell clones that we previously investigated (uvmo-1, uvmo-2, and uvmo-3) was positively affected by pretreatment of epithelial cells with IFN-γ: either earlier recognition during the time course of infection, greater activation, or both (11). A straightforward explanation for the lack of an IFN-γ pretreatment effect for PmpG1.1 T cells would be a high-affinity T cell receptor less dependent on the CD4 coreceptor. To test that hypothesis, we compared PmpG1.1 to uvmo-2, a clone that we have previously shown to be significantly inhibited by monoclonal antibody specific for CD4 (GK1.5) (11). C57epi.1 epithelial cell monolayers, untreated or pretreated with IFN-γ, were mock infected or infected with C. muridarum for 24 h. Twenty-four hours postinfection, the inocula were removed, and the monolayers were washed and then cocultured with T cell clones in the presence of 10 μg/ml GK1.5 or isotype control antibody. The culture supernatants were collected after 24 h of coculture, and IFN-γ was quantified. Interestingly, neither clone could be blocked by GK1.5 under low MHC class II expression conditions (no IFN-γ pretreatment). IFN-γ pretreatment of the epithelial cells enhanced uvmo-2 production of IFN-γ, and that enhanced production of IFN-γ was blocked by GK1.5. IFN-γ pretreatment had no effect on PmpG1.1 IFN-γ production, and PmpG1.1 was not inhibited by GK1.5 under either experimental condition (Fig. 5B). The inability of anti-CD4 antibody to block PmpG1.1 activation is consistent with its having a high-affinity T cell receptor.
Fig 5.
Processing and presentation of the PmpG303-311 epitope by infected epithelial cells. (A) C57epi.1 monolayers, untreated (no pre-tx) or pretreated (pre-tx) with IFN-γ (10 ng/ml for 14 h) in 96-well plates, were infected with C. muridarum (3 IFU per cell) over staggered intervals to generate mock (0-h)-infected and 9-, 12-, 15-, 18-, and 21-h-infected monolayers at endpoint. The infected monolayers were cocultured with 5 × 104 PmpG1.1 T cells in the presence of 10 μg/ml tetracycline; IFN-γ in 48-h culture supernatants was quantified by ELISA. Shown are aggregate data from two independent experiments; the two curves are not statistically different at any time point. Note the break in the scale. (B) C57epi.1 monolayers in 96-well plates, untreated or pretreated with IFN-γ (10 ng/ml for 14 h), were infected with C. muridarum (3 IFU per cell) for 24 h. At 24 h, the monolayers were washed and then cocultured with 5 × 104 T cells in the presence of 10 μg/ml anti-CD4 monoclonal antibody (GK1.5) or 10 μg/ml isotype control monoclonal antibody (mouse Ig2b). Twenty-four hours later, the supernatants were collected and IFN-γ was quantified by ELISA. The percent inhibition of IFN-γ production was calculated by comparing IFN-γ production in the presence of GK1.5 to that in the presence of the isotype control antibody. The data presented are aggregate data from three independent experiments. ***, P < 0.0005; NS, not statistically significant. The error bars indicate SEM.
The PmpG303-311 epitope persists in vivo after clearance of a primary genital tract infection.
During characterization of PmpG1.1, a “high background” was noted when irradiated immune splenocytes were used as antigen-presenting cells. Irradiated immune splenocyte APC were included in the PmpG1.1 derivation strategy because they are more efficient antigen-presenting cells, lowering both the level of antigen required and the magnitude of the T cell response (16, 17, 24). On reflection, the only likely difference between immune and naïve splenocytes is exposure of immune splenocytes to Chlamydia antigens during the primary infection. During the first 10 days of a primary genital tract infection in wild-type C57BL/6 mice, there is low-level dissemination of C. muridarum, as demonstrated by recoverable IFU from lung and spleen (4, 8). To formally investigate the irradiated immune splenocyte phenomenon, C57BL/6 mice in two experimental groups were vaginally infected with C. muridarum ∼6 months apart and used as the source of immune splenocyte APC at days 21/22 and days 185/186 postinfection. Unmanipulated immune splenocytes were compared to naïve splenocyte controls for their ability to activate PmpG1.1 and 4 other previously characterized Chlamydia-specific CD4 T cell clones, uvmo-2, uvmo-3, uvmo-1, and spl4-10 (10, 11). The antigen specificity of the non-PmpG1.1 Chlamydia-specific CD4 T cell clones is unknown, but it is not PmpG303-311 or any of the other 7 C. muridarum CD4 T cell epitopes we previously identified (reference 30 and data not shown). The T cell clones were cocultured with irradiated naïve and irradiated immune splenocytes for 48 h; then, culture supernatants were collected and IFN-γ was quantified by ELISA (Fig. 6A). APC presentation of PmpG303-311 is uniquely preserved in vivo compared to epitopes recognized by the 4 other T cell clones, and remarkably, presentation of PmpG303-311 was stronger 6 months postinfection than it was 3 weeks postinfection. These results suggest some type of consolidation of PmpG303-311 epitope presentation by splenic APC. Three of the other four CD4 T cell clones' epitopes showed a similar, but much lower magnitude (∼10-fold lower), consolidation of epitope presentation. Conversely, the Chlamydia-specific T cell clone spl4-10 showed almost no activation by either naïve or immune APC (<200 pg/ml IFN-γ), largely ruling out a nonspecific activation of T cell clones by immune APC due to unknown cytokine or accessory molecule interactions.
To get a sense of how robust persistent presentation of the PmpG303-311 epitope was on immune splenocytes, we compared the ability of Chlamydia-pulsed naïve splenocytes to activate PmpG1.1 with that of unpulsed and antigen-pulsed immune irradiated splenocytes (Fig. 6b). Six months postinfection, unpulsed immune irradiated splenocytes activated PmpG1.1 to the same degree as antigen-pulsed naïve irradiated splenocytes. Presentation of PmpG303-311 on immune irradiated splenocytes was not saturated, as addition of UV light-inactivated C. muridarum to immune irradiated splenocytes increased PmpG1.1 IFN-γ production by roughly 3-fold.
DISCUSSION
Previous identification of PmpG303-311 as a protective C. muridarum CD4 T cell epitope in C57BL/6 (H-2b) mice provided an opportunity to investigate the immunomechanics of protective immunity. For this report, we derived a CD4 T cell clone specific for the PmpG303-311 epitope in order to investigate PmpG-1 antigen processing and presentation by infected epithelial cells. Understanding how protective versus nonprotective T cell epitopes are processed and presented by infected epithelial cell targets may reveal antigen characteristics associated with protective immunity and may contribute to rational selection of Chlamydia proteins for inclusion in subunit vaccines.
A previous study showed that a CD4 T cell clone's ability to terminate C. muridarum replication in epithelial cells in vitro was correlated with its ability to protect against genital tract C. muridarum infections in adoptive-transfer experiments (9). Consistent with those results, the PmpG1.1 CD4 T cell clone, specific for the protective epitope PmpG303-311, was able to recognize infected epithelial cells and terminate infection in vitro. We attempted to directly test whether PmpG1.1 could protect mice in vivo using adoptive-transfer experiments. The experiments were inconclusive, because we could not find any CD4 T cells in Rag1 knockout mice after adoptive transfer of the PmpG1.1 CD4 T cell clone. This may be related to the fact that the activated PmpG1.1 CD4 T cell clone produces IFN-γ and TNF-α, but not IL-2. In humans, IFN-γ+/TNF-α+/IL-2neg CD4 T cells are found in a perforin+/CD28neg CD4 cell subset that has little or no proliferative capacity with T cell activation without exogenous cytokines and may undergo activation-induced cell death (AICD) under those conditions (1). The PmpG1.1 CD4 T cell clone had a very limited ability to proliferate in response to an antigenic stimulus delivered by naïve splenocytes and may belong to such a CD4 subset. Recipient Rag1 knockout mice lack endogenous T cells, i.e., they had no potential helper T cells to support PmpG1.1 during the C. muridarum genital tract infectious challenge.
The PmpG303-311 epitope appears on the surfaces of infected epithelial cells at levels sufficient to activate PmpG1.1 between 15 and 18 h postinfection. This is the same window of time postinfection in which epitopes for three other C. muridarum-specific CD4 T cell clones appear on the cell surface (11). Unlike the epitopes for the previously characterized CD4 T cell clones, recognition of PmpG303-311 is completely unaffected by pretreatment of the epithelial cells with IFN-γ prior to infection, suggesting that neither alternations in the levels of MHC class II expression nor IFN-γ-triggered changes in antigen processing, such as the transition to immunoproteosomes (29), had any significant effect on processing of PmpG-1 in infected epithelial cells. Of the four epitopes characterized to date, PmpG303-311 is the only epitope whose time to presentation and level on infected epithelial cells, as measured by T cell activation, is not affected by IFN-γ pretreatment of epithelial cell monolayers. The lack of IFN-γ pretreatment effect on the magnitude of PmpG1.1 activation is likely due to a high-affinity T cell receptor, based on the inability of anti-CD4 monoclonal antibody to inhibit PmpG1.1.
Only one of the four C. muridarum CD4 T cell clones characterized to date could recognize an infected epithelial cell earlier than 15 h postinfection. That recognition was very modest at 12 h postinfection and required IFN-γ pretreatment of the epithelial cell monolayer. In the current study, PmpG303-311, a protective T cell epitope, was not visible to CD4 T cells until ≥15 h postinfection. That time point occurs after the C. muridarum elementary body (EB)-to-reticulate body (RB) transition is well under way and supports our previous hypothesis that protective immunity is not likely based on nonspecific disruption of epithelial cell physiology or viability during the “eclipse phase” ∼2 to 15 h postinfection, when noninfectious RB predominate in the infected cell. Rather, protective CD4-mediated T cell immunity is more likely a direct attack on intracellular infectious EB (10).
An incidental finding during the course of this investigation was that the PmpG303-311 epitope persists on splenic APC surfaces 6 months postinfection. This finding suggests establishment of durable protective CD4 T cell immunity and is consistent with previous work showing adoptive transfer of protective immunity with splenic CD4 T cells isolated from mice 5 months after C. muridarum genital tract infection (26). Curiously, in our experiments, there was greater activation of PmpG1.1 with unmanipulated/unpulsed immune irradiated splenocytes taken from mice 6 months postinfection than there was from immune irradiated splenocytes taken from mice 3 weeks postinfection. Unmanipulated/unpulsed immune splenocytes from the 3-week and 6-month time points were, respectively, equal and superior to naïve irradiated splenocytes pulsed with UV-inactivated C. muridarum for activation of PmpG1.1. Early in the course of genital tract infections, C. muridarum transiently disseminates to other organs, including the spleen (<100 IFU), and is cleared from them prior to resolution of the genital tract infection (4, 8). One might have predicted that 3-week-postinfection splenocytes recently exposed to live C. muridarum would have been superior to 6-month-postinfection splenocytes for activating PmpG1.1. Alternatively, C. muridarum DNA persists in the mouse genital tract for ≥3 months after viable bacteria can no longer be recovered from the genital tract (20). The same persistence of DNA could be occurring in the spleen. The possibility that viable but uncultivatable bacteria contribute to ongoing generation of PmpG-1 antigen is somewhat diminished by the finding that doxycycline treatment of infected mice did not have an effect on persistence of C. muridarum DNA in the genital tract (22). In addition, persistence of viable bacteria in the spleen would not likely favor presentation of any single antigen epitope. PmpG1.1 and uvmo-2 activated by naïve or immune irradiated splenocytes pulsed with UV-inactivated C. muridarum produce nearly identical levels of IFN-γ (43,000 ± 6,000 versus 40,000 ± 2,000, and 197,000 ± 30,000 versus 284,000 ± 13,000, respectively). Conversely, unmanipulated/unpulsed immune splenocytes activate PmpG1.1 to produce roughly 10 times as much IFN-γ as uvmo-2 (Fig. 6A). Significantly greater activation of PmpG1.1 by the 6-month-postinfection splenocytes argues either that the level of PmpG303-311 epitope increased over 6 months or, more likely, that the available PmpG303-311 on splenic APC 6 months postinfection resides on a more effective antigen-presenting cell; our current data do not rule out the possibility that the relatively poor APC performance of unpulsed/unmanipulated splenocytes 3 weeks versus 6 months postinfection is the result of a suppressive T cell or APC population present at week 3 that wanes over time. Determining which antigen-presenting cell subset presents the PmpG303-311 epitope 6 months postinfection, the physical state of PmpG-1 antigen/epitope, and whether vaccination can reproduce those conditions will likely provide important insights into durable protective immunity against Chlamydia genital tract infections useful for rational development of a subunit vaccine.
ACKNOWLEDGMENTS
We have no conflicts of interest related to this report.
This research was supported by NIH grants R01AI070514 (R.M.J.) and R01AI076483 (R.C.B.).
Footnotes
Published ahead of print 19 March 2012
REFERENCES
- 1. Appay V, et al. 2002. Characterization of CD4(+) CTLs ex vivo. J. Immunol. 168:5954–5958 [DOI] [PubMed] [Google Scholar]
- 2. Brunham RC, Pourbohloul B, Mak S, White R, Rekart ML. 2005. The unexpected impact of a Chlamydia trachomatis infection control program on susceptibility to reinfection. J. Infect. Dis. 192:1836–1844 [DOI] [PubMed] [Google Scholar]
- 3. Brunham RC, Rekart ML. 2008. The arrested immunity hypothesis and the epidemiology of chlamydia control. Sex. Transm. Dis. 35:53–54 [DOI] [PubMed] [Google Scholar]
- 4. Cotter TW, Ramsey KH, Miranpuri GS, Poulsen CE, Byrne GI. 1997. Dissemination of Chlamydia trachomatis chronic genital tract infection in gamma interferon gene knockout mice. Infect. Immun. 65:2145–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dadaglio G, Nelson CA, Deck MB, Petzold SJ, Unanue ER. 1997. Characterization and quantitation of peptide-MHC complexes produced from hen egg lysozyme using a monoclonal antibody. Immunity 6:727–738 [DOI] [PubMed] [Google Scholar]
- 6. Duc HT, Rucay P, Righenzi S, Halle-Pannenko O, Kourilsky P. 1993. Monoclonal antibodies directed against T cell epitopes presented by class I MHC antigens. Int. Immunol. 5:427–431 [DOI] [PubMed] [Google Scholar]
- 7. He Q, et al. 2005. Molecular basis for the potency of IL-10-deficient dendritic cells as a highly efficient APC system for activating Th1 response. J. Immunol. 174:4860–4869 [DOI] [PubMed] [Google Scholar]
- 8. Igietseme JU, et al. 1998. Chlamydial infection in inducible nitric oxide synthase knockout mice. Infect. Immun. 66:1282–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Igietseme JU, Wyrick PB, Goyeau D, Rank RG. 1994. An in vitro model for immune control of chlamydial growth in polarized epithelial cells. Infect. Immun. 62:3528–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jayarapu K, Kerr M, Ofner S, Johnson RM. 2010. Chlamydia-specific CD4 T cell clones control Chlamydia muridarum replication in epithelial cells by nitric oxide-dependent and -independent mechanisms. J. Immunol. 185:6911–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jayarapu K, Kerr MS, Katschke A, Johnson RM. 2009. Chlamydia muridarum-specific CD4 T-cell clones recognize infected reproductive tract epithelial cells in an interferon-dependent fashion. Infect. Immun. 77:4469–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang X, Shen C, Rey-Ladino J, Yu H, Brunham RC. 2008. Characterization of murine dendritic cell line JAWS II and primary bone marrow-derived dendritic cells in Chlamydia muridarum antigen presentation and induction of protective immunity. Infect. Immun. 76:2392–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson RM. 2004. Murine oviduct epithelial cell cytokine responses to Chlamydia muridarum infection include interleukin-12-p70 secretion. Infect. Immun. 72:3951–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karunakaran KP, et al. 2008. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J. Immunol. 180:2459–2465 [DOI] [PubMed] [Google Scholar]
- 15. Krogsgaard M, et al. 2000. Visualization of myelin basic protein (MBP) T cell epitopes in multiple sclerosis lesions using a monoclonal antibody specific for the human histocompatibility leukocyte antigen (HLA)-DR2-MBP 85-99 complex. J. Exp. Med. 191:1395–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moore T, et al. 2002. Fc receptor regulation of protective immunity against Chlamydia trachomatis. Immunology 105:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moore T, et al. 2003. Fc receptor-mediated antibody regulation of T cell immunity against intracellular pathogens. J. Infect. Dis. 188:617–624 [DOI] [PubMed] [Google Scholar]
- 18. Morrison RP, Caldwell HD. 2002. Immunity to murine chlamydial genital infection. Infect. Immun. 70:2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morrison RP, Feilzer K, Tumas DB. 1995. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect. Immun. 63:4661–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramsey KH, Miranpuri GS, Sigar IM, Ouellette S, Byrne GI. 2001. Chlamydia trachomatis persistence in the female mouse genital tract: inducible nitric oxide synthase and infection outcome. Infect. Immun. 69:5131–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramsey KH, Soderberg LS, Rank RG. 1988. Resolution of chlamydial genital infection in B-cell-deficient mice and immunity to reinfection. Infect. Immun. 56:1320–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reeves DM, Nagarajan U, O'Connell C, Andrews CW, Jr, Darville T. 2007. Lack of an effect of antibiotic treatment on prolonged detection of chlamydial DNA in murine genital tract infection. Antimicrob. Agents Chemother. 51:2646–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rekart ML, Brunham RC. 2008. Epidemiology of chlamydial infection: are we losing ground? Sex. Transm. Infect. 84:87–91 [DOI] [PubMed] [Google Scholar]
- 24. Rock KL, Benacerraf B, Abbas AK. 1984. Antigen presentation by hapten-specific B lymphocytes. I. Role of surface immunoglobulin receptors. J. Exp. Med. 160:1102–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schachter J. (ed). 1980. Chlamydiae (Psittacosis-lymphogranuloma venereum-trachoma group), 3rd ed American Society for Microbiology, Washington, DC [Google Scholar]
- 26. Su H, Caldwell HD. 1995. CD4+ T cells play a significant role in adaptive immunity to Chlamydia trachomatis infection of the mouse genital tract. Infect. Immun. 63:3302–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Su H, Feilzer K, Caldwell HD, Morrison RP. 1997. Chlamydia trachomatis genital tract infection of antibody-deficient gene knockout mice. Infect. Immun. 65:1993–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su H, et al. 1998. Vaccination against chlamydial genital tract infection after immunization with dendritic cells pulsed ex vivo with nonviable Chlamydiae. J. Exp. Med. 188:809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tanaka K. 1994. Role of proteasomes modified by interferon-gamma in antigen processing. J. Leukoc. Biol. 56:571–575 [DOI] [PubMed] [Google Scholar]
- 30. Yu H, Jiang X, Shen C, Karunakaran KP, Brunham RC. 2009. Novel Chlamydia muridarum T cell antigens induce protective immunity against lung and genital tract infection in murine models. J. Immunol. 182:1602–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu H, et al. 2010. Chlamydia muridarum T-cell antigens formulated with the adjuvant DDA/TDB induce immunity against infection that correlates with a high frequency of gamma interferon (IFN-gamma)/tumor necrosis factor alpha and IFN-gamma/interleukin-17 double-positive CD4+ T cells. Infect. Immun. 78:2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu H, et al. 2011. Immunization with live and dead Chlamydia muridarum induces different levels of protective immunity in a murine genital tract model: correlation with MHC class II peptide presentation and multifunctional Th1 cells. J. Immunol. 186:3615–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]