Abstract
Prion diseases, a group of fatal neurodegenerative disorders, are characterized by the presence of the abnormal scrapie isoform of prion protein (PrPSc) in affected brains. A conformational change is believed to convert the normal cellular prion protein into PrPSc. Detection of PrPSc for diagnosis and prophylaxis is impaired because available Abs recognizing epitopes on PrP fail to distinguish between PrPSc and normal cellular prion protein. Here, we report that an anti-DNA Ab, OCD4, as well as gene 5 protein, a well established DNA-binding protein, capture PrP from brains affected by prion diseases in both humans and animals but not from unaffected controls. OCD4 appears to immunoreact with DNA (or a DNA-associated molecule) that forms a conformation-dependent complex with PrP in prion diseases. Whereas PrP immunocaptured by OCD4 is largely protease-resistant, a fraction of it remains protease-sensitive. Moreover, OCD4 detects disease-associated PrP >10 times more efficiently than a widely used Ab to PrP. Our finding that anti-DNA Abs and gene 5 protein specifically target disease-associated DNA–PrP complexes in a wide variety of species and disease phenotypes opens new avenues in the study and diagnosis of prion diseases.
Human prion diseases include sporadic and familial forms (1), such as Creutzfeldt–Jakob disease (CJD) and Gerstmann–Sträussler–Scheinker disease (GSS), as well as acquired forms such as variant CJD (vCJD) transmitted through the consumption of contaminated beef (2, 3). Scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) in cows, and chronic wasting disease (CWD) in deer and elk are the most common prion diseases in animals. According to the prevailing prion hypothesis (4), all forms of prion diseases share a pathogenic mechanism whereby the host-encoded normal cellular prion protein (PrPC) is converted into the abnormal scrapie isoform of prion protein (PrPSc) isoform that is insoluble, pathogenic, and infectious. The PrPC to PrPSc conversion is believed to take place posttranslationally and to involve an α-helix to β-sheet structural transition (5–8). The presence and location of the β-sheet structure is thought to make PrPSc resistant to proteases and to result in a variety of PrPSc conformers known as prion strains (9). How this conversion takes place, the precise physicochemical characteristics of the converted PrPSc conformer, and whether additional molecules, including nucleic acid (10, 11), are also components of the infectious agent are issues that have never been fully clarified. The possibility of the spread of prion diseases in animals and from animals to humans (12, 13) has prompted the generation of many Abs against PrP sequences as possible diagnostic reagents. However, most of them recognize both the PrPC and PrPSc isoforms. A previous report of an Ab immunoreacting specifically with PrPSc has not been confirmed in subsequent studies (14).
Recent evidence suggests that PrP may form a macromolecular complex with nucleic acid (15). To explore whether nucleic acid represents a possible target for PrPSc detection, we screened anti-DNA Abs for their ability to capture PrPSc from homogenates of diseased brains. Surprisingly, several Abs to DNA readily identified PrP in the immunocapture assay. Moreover, like anti-DNA Abs, Ff gene 5 protein (g5p), a single-stranded DNA-binding protein is also capable of capturing PrPSc. OCD4, a mAb raised against a nuclear DNA preparation from human lymphoma cells, was selected for detailed analyses.
Materials and Methods
Reagents and Anti-PrP Abs. Magnetic beads (M-280 tosyl-activated Dynabeads) were from Dynal (Oslo). Salmon testes single-stranded DNA and proteinase K (PK) were purchased from Sigma. Nuclease (benzonase) was from Roche Diagnostics (Indianapolis). Mouse mAb 6H4 from Prionics (Zurich) was used to recognize the sequence of human PrP residues 144–152 (16). Mouse mAb 3F4 from Signet Laboratories (Dedham, MA) was used to recognize an epitope within human PrP residues 109–112 (17). The rabbit anti-C antiserum immunoreacted with human PrP residues 220–231 (18). Horseradish peroxidase-conjugated Ab was purchased from Amersham Biosciences (Piscataway, NJ).
Brain Tissues. Human brain tissues were obtained at autopsy from patients with or without prion diseases and were kept frozen at –80°C. The diagnosis of various disease phenotypes of sporadic CJD (sCJD), familial CJD (fCJD), vCJD, and GSS was confirmed by standard criteria, including histological examination, immunohistochemistry, immunoblotting, and DNA typing. Animal brain tissues with or without prion disease were confirmed by immunohistochemistry and immunoblotting as well.
Production of Anti-DNA Ab. Nuclear DNA extracted (19) from Raji Burkitts lymphoma cells was used as an immunogen to generate OCD4. The production of mAb was carried out according to a standard protocol (20). Screening of mAb by enzyme-linked immunoabsorbent assay was performed by using 96-well plates coated with calf thymus DNA (Sigma).
Preparation of g5p. The g5p (PDB ID code 1VQB) was isolated from Escherichia coli transformed with an Ff gene 5-containing plasmid and purified by using DNA cellulose affinity plus Sephadex G75 sizing columns as described (21, 22). The purity of g5p was >99% as determined by quantitation of Coomassie blue-stained bands on SDS-polyacrylamide gels.
Immunocapture Assay. OCD4 (100 μg of purified IgG) was conjugated to 7 × 108 tosyl-activated superparamagnetic beads (M-280 Dynabeads, Dynal) in 1 ml of PBS at 37°C for 20 h. The OCD4-conjugated beads were incubated with 0.1% BSA in PBS to block nonspecific binding. The prepared OCD4 beads were stable for at least 3 months at 4°C. Brain homogenate (BH) (10%, wt/vol) was prepared at 4°C in lysis buffer (100 mM NaCl/10 mM EDTA/0.5% Nonidet P-40/0.5% sodium deoxycholate/10 mM Tris·HCl, pH 7.5) containing a mixture of protease inhibitors (Roche Applied Science), followed by centrifugation at 3,000 × g for 10 min at 4°C to remove debris. Immunoprecipitation (IP) was then performed as described (23) by using the clarified homogenate (10 μl for human and 2 μl for animal specimens) and OCD4-conjugated beads (10 μg of mAb/6 × 107 beads) in 1 ml of lysis buffer. After incubation with constant mixing for 3 h at room temperature, OCD4 beads were attracted to the side wall of the plastic tubes by external magnetic force, allowing easy removal of all unbound materials in the solution. After three washes in wash buffer (2% Tween 20 and 2% Nonidet P-40 in PBS, pH 7.5), OCD4 beads were collected and were heated at 95°C for 5 min in SDS sample buffer (3% SDS/2 mM EDTA/10% glycerol/50 mM Tris·HCl, pH 6.8).
Immunoblotting. Proteins were separated by SDS/PAGE (15% Tris-glycine precast gel, Bio-Rad), electrotransferred onto a polyvinylidene difluoride membrane. PrP was detected by immunoblotting using the following anti-PrP Abs: 3F4 mAb used at 1:50,000, 6H4 mAb used at 1:5,000, and anti-C antiserum used at 1:3,000. For DNA dot blot analysis, purified salmon sperm DNA (Sigma) was dissolved in distilled and deionized H2O (Millipore) and a 1-μl aliquot was dotted onto a nitrocellulose membrane. The blot was dried at 80°C for 1 h, blocked with 5% milk that was pretreated with nuclease (benzonase; 20 units/ml for 1 h at room temperature), followed by incubation with horseradish peroxidase-conjugated OCD4 (1:2,000 dilution) at room temperature for 1 h. Immunoreactivity was visualized on Kodak x-ray film by enhanced chemiluminescence with the ECL plus kit (Amersham Biosciences).
PK Digestion. Samples were incubated in lysis buffer with PK at 50 μg/ml at 37°C for 1 h. The digestion was terminated by addition of 5 mM phenylmethylsulfonyl fluoride. The digested samples were mixed with an equal volume of 2× SDS sample buffer before SDS/PAGE and immunoblotting with an anti-PrP Ab.
Conformation-Dependent Immunoassay. The conformation-dependent stability of PrPSc was analyzed by using a modified procedure based on published methods (24, 25). BH (10%, wt/vol) from a CJD subject was treated with 0–3 M guanidine hydrochloride (GdnHCl) in lysis buffer at room temperature for 1 h. GdnHCl was subsequently removed by precipitation with 5-fold volumes of prechilled methanol at –20°C for 2 h, followed by centrifugation at 16,000 × g for 20 min at 4°C. The pellets were resuspended in lysis buffer. PrPSc in the GdnHCl-treated samples were evaluated either by PK digestion (50 μg/ml PK for 1 h at 37°C) or by immunocapture with OCD4, followed by immunoblotting with 3F4 as described above.
Results
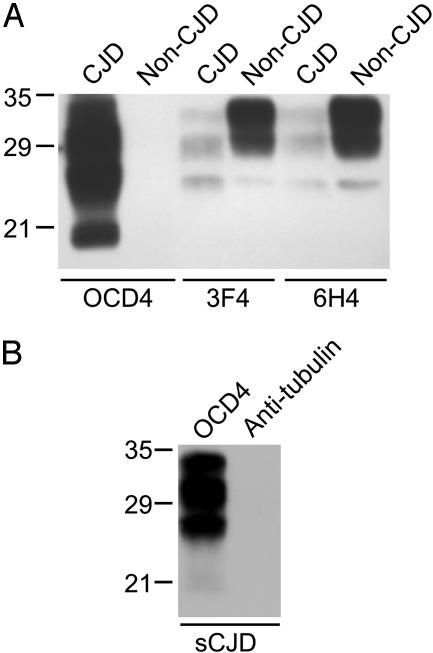
An Anti-DNA Ab Specifically Captures PrPSc but Not PrPC. We performed the immunocapture assay of PrP by using the anti-DNA Ab-conjugated magnetic beads, followed by immunoblotting with an anti-PrP Ab. Immunocapture by the mAb to DNA OCD4 yielded abundant PrP from BH of CJD subjects, whereas no PrP was recovered from control brains free of neurological disease, as well as brains with other neurodegenerative diseases, such as Alzheimer's disease (Fig. 1A and data not shown). None of the other mAbs recognizing PrP sequences demonstrated such high specificity. Under the same experimental conditions, two of these Abs to PrP widely used in current research on prion diseases, mAbs 3F4 (17) and 6H4 (16), immunocaptured PrP from both diseased and normal brains. The mAbs 3F4 and 6H4 actually showed higher affinity for PrPC than for the PrP associated with CJD brains (Fig. 1 A). To exclude the possibility that binding of PrPSc to OCD4-conjugated beads is nonspecific, we conjugated an Ab against tubulin to the beads and assayed for PrP in CJD BH after immunocapture. No PrP was captured by this irrelevant Ab, whereas abundant amounts of PrP were recovered by OCD4 from the same preparation (Fig. 1B).
Fig. 1.
Immunocapture of disease-associated PrP by anti-DNA mAb OCD4. (A) PrP captured from BHs by various mAbs. Ten microliters of 10% (wt/vol) BH from a subject with sporadic CJD or a non-CJD control was incubated in 1 ml of lysis buffer for 3 h at room temperature with OCD4, 3F4, and 6H4 that were conjugated to magnetic beads (23). After washing with a solution of 2% Tween 20 and 2% Nonidet P-40 in PBS, the samples recovered by the Ab-conjugated beads were subjected to SDS/PAGE. PrP was detected by immunoblotting with 3F4. (B) Immunocapture of PrP from a subject with sCJD by antitubulin Ab and OCD4. Experimental procedures were the same as those described for A. Molecular mass markers (in kDa) are indicated on the left.
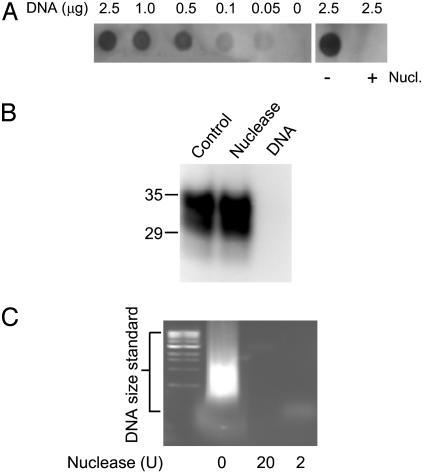
OCD4 Immunoreacts with DNA, and OCD4 Detection of PrP Is Competed by Input DNA. To determine whether DNA is indeed the antigen recognized by OCD4, we performed the DNA dot blot assay. We found that OCD4 immunoreacts with purified DNA preparations and that the reaction is sensitive to nuclease (Fig. 2A). We also preabsorbed the OCD4-conjugated beads with purified DNA before performing the immunocapture assay. Alternatively, we treated the BHs with nuclease (benzonase) followed by immunocapture with OCD4. No PrP was immunocaptured by OCD4 in the presence of the added DNA. However, pretreatment of the homogenate with nuclease had no effect on PrP immunocapture by OCD4, even although, in a control experiment, the amount of nuclease used was sufficient to digest the DNA added to the preparation (Fig. 2 B and C). Taken together, these findings argue that the OCD4 antigen is DNA, or a DNA-related molecule that is part of a molecular complex with disease-associated PrPSc. The ineffectiveness of the nuclease digestion suggests that the DNA present in the DNA–PrPSc complex may be protected from, or inaccessible to, nuclease digestion while maintaining the capacity to bind OCD4. Nucleic acid specifically isolated from CJD brain has been reported to resist nuclease treatment (26). Furthermore, we also observed that disease-associated PrPSc could be specifically isolated with other mAbs raised with different DNA preparations (data not shown). Alternatively, OCD4 and other mAbs to DNA as well, might recognize a conformation shared by PrPSc and DNA.
Fig. 2.
Immunoreactivity of OCD4 toward DNA and effect of DNA and nuclease on PrP capture by OCD4. (A) Dot blot of DNA probed with OCD4. (Left) Dot blot containing indicated amounts of purified salmon DNA was incubated with horseradish peroxidase-conjugated OCD4. (Right) Salmon sperm DNA was treated with or without nuclease (Nucl., benzonase) at 20 units/ml at 37°C for 1 h before dot blot with horseradish peroxidase-conjugated OCD4. Immunoreactivity was visualized by enhanced chemiluminescence. (B) Immunocapture of PrP by OCD4 after incubation with nuclease and salmon DNA. Scrapie-infected hamster BH (2 μl each) was either untreated (control) or treated with benzonase (nuclease) at 100 units/ml, followed by incubation with OCD4-conjugated beads. In a separate sample, purified salmon DNA (5 μg/ml) was preincubated with the OCD4-conjugated beads at 37°C for 1 h before the immunocapture assay (DNA). PrP was detected by immunoblotting with 3F4. Molecular mass is indicated on the left (in kDa). (C) Enzymatic digestion of DNA by nuclease. Salmon DNA (5 μg/ml) was treated with benzonase at 20 units/ml or 2 units/ml at 37°C for 1 h, or was left untreated. The samples were subjected to 1.0% agarose gel electrophoresis, with a 1-kb DNA ladder (Sigma) run in parallel as DNA size standard.
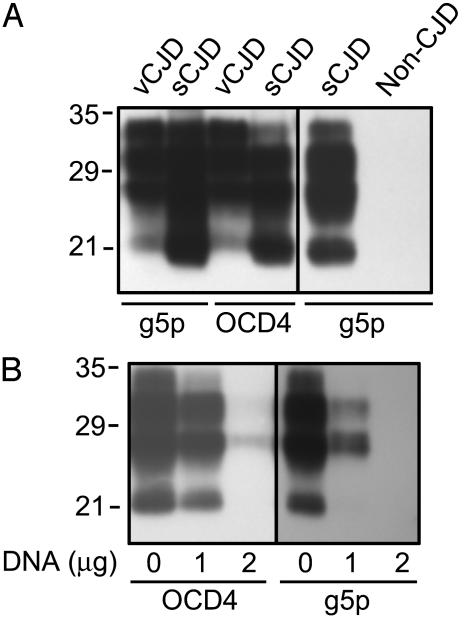
The DNA-Binding Protein g5p also Captures PrPSc in a DNA-Dependent Manner. The DNA-binding protein g5p, encoded by the filamentous bacteriophage Ff and purified from E. coli (21, 22), was conjugated to magnetic beads. The g5p-conjugated beads were then incubated with BHs from vCJD, sCJD, or normal controls. Like OCD4, g5p precipitated PrP from vCJD and sCJD brains, but not from normal control brains (Fig. 3A). Moreover, the amount of the PrP captured by both g5p and OCD4 decreased in a concentration-dependent manner after the addition of DNA (Fig. 3B), further arguing that the capture of PrP by OCD4 and g5p is mediated by, or can be competed with, nucleic acid.
Fig. 3.
Capture of PrP from sCJD, vCJD, and normal control brains by g5p. (A) BH from subjects affected by vCJD and sCJD and from a control subject was incubated with g5p- or OCD4-conjugated beads, followed by immunoblotting with 3F4. (B) The g5p- or OCD4-conjugated beads were preincubated with indicated amounts of salmon DNA, followed by incubation with BH of sCJD and immunoblotting with 3F4.
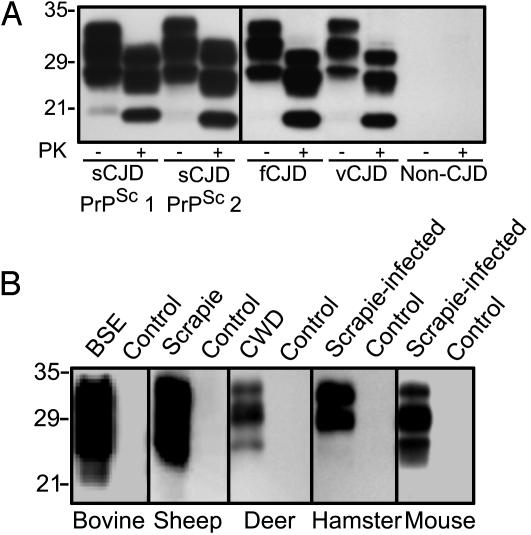
OCD4-Captured PrPSc Is PK-Resistant, and OCD4 Detects PrPSc of Different Prion Strains in Both Humans and Animals. A major portion of the PrP captured by OCD4 is resistant to PK digestion, which is the biochemical hallmark of PrPSc (Fig. 4A). Furthermore, whereas the currently available mAbs to PrP are species-sensitive because their specificity is derived from a particular PrP sequence (14, 16, 27), OCD4 captured all disease-associated PrPSc of different prion strains, which were obtained from various forms of human prion diseases, including sporadic and fCJD, GSS, and vCJD, as well as naturally occurring and experimentally transmitted animal diseases (Fig. 4). These findings further argue that under nondenaturing conditions, OCD4 captures disease-associated PrPSc but not PrPC. Furthermore, OCD4 must immunoreact with an antigen that is bound to, but distinct from, PrP, because the OCD4 capture of PrP is independent of PrP amino acid sequence.
Fig. 4.
OCD4 captures disease-associated PrPSc from various prion-infected human and animal brains. (A) Immunocapture of PrP by OCD4 from various human prion diseases. BH (10 μl each) from subjects affected by sCJD associated with PrPSc type 1 or PrPSc type 2 (28, 29), fCJD linked to the E200K mutation (fCJD), and vCJD, as well as from a non-CJD control, was incubated with OCD4-conjugated beads. The bound materials were either untreated (PK–) or treated with PK (PK+) at 50 μg/ml at 37°C for 1 h. PrP was detected by immunoblotting with 3F4. (B) Immunocapture of PrP by OCD4 from various animal species. BH (2 μl each) from normal animals and those affected by BSE (cattle), scrapie (sheep), and CWD (deer), as well as experimental scrapie adapted in hamster (263 K prion strain) and mouse (ME7 prion strain), was subjected to immunocapture by OCD4 as described in A. Captured PrP was detected by immunoblotting with 6H4.
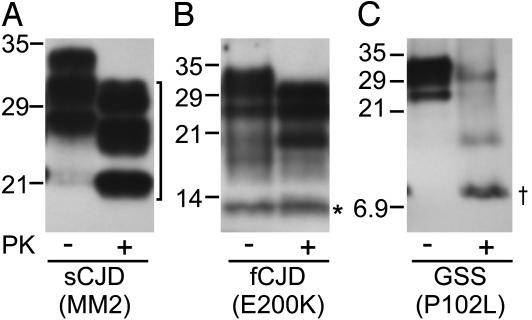
OCD4 Captures all PK-Resistant PrP Fragments Associated with Various Human Prion Diseases. To gain insight into the PrP bound to the putative DNA-containing complex, we took advantage of the different size of the PK-resistant PrPSc core fragments present in various prion diseases. Immunocapture of PrP by OCD4 was not affected by pretreatment of homogenates from diseased brains with PK (Fig. 5). Furthermore, all major PK-resistant fragments of PrPSc were recovered, including the 21- and 19-kDa core fragment in CJD (PrPSc type 1 and 2; Fig. 5A) spanning residues 82–97 to 231 (28, 29), which accounts for nearly two-thirds of the 23–231 PrP molecule; an internal 7- to 8-kDa fragment in GSS (Fig. 5C), including residues 74–90 to 146–153 (30–32), and the C-terminal 12- to 13-kDa fragment in CJD (Fig. 5B), extending from residues 154–162 to 231 (33). The cocapture by OCD4 of all major PrPSc fragments generated by PK treatment regardless of size argues that most (if not all) of the PK-resistant region of PrPSc is stably bound to the DNA-containing complex and remains with the complex after PK treatment. It also further supports the contention that OCD4 is unlikely to react directly with PrP in the complex because the 7- to 8-kDa internal fragment and the 12- to 13-kDa C-terminal fragment share no consensus in sequence, yet they are both efficiently captured by OCD4.
Fig. 5.
Immunocapture of various PK-resistant PrP fragments by OCD4. BH from subjects affected by sCJD associated with codon 129 MM and type 2 PrPSc (sCJD MM2; A), fCJD with the E200K mutation (fCJD E200K; B), and GSS with the P102L mutation (GSS P102L; C) was treated with or without PK at 50 μg/ml at 37°C for 1 h. PrP was captured by the OCD4-conjugated beads and was then detected by immunoblotting with either 3F4 recognizing PrP residues 109–112 (A and C) or the anti-C Ab against human PrP residues 220–231 (B). Three groups of PK-resistant PrP fragments were efficiently captured by OCD4 (with their position on the blots indicated), including the ≈19-kDa PrP core fragment spanning residues 97–103 to 231 (bracket; ref. 28), the internal 7- to 8-kDa fragment spanning residues 74–90 to 146–153 (dagger; refs. 30–32), and the C-terminal 12- to 13-kDa fragment spanning residues 154–162 to 231 (asterisk; ref. 33).
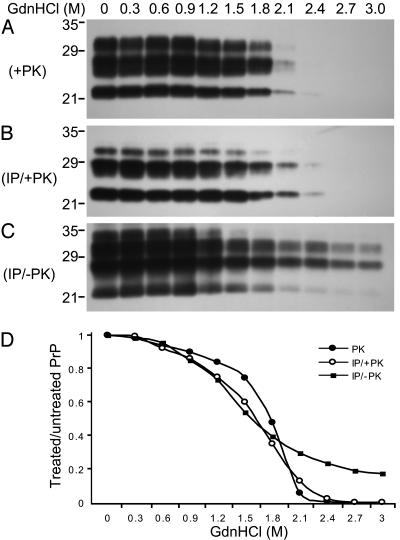
The Immunocapture of PrPSc by OCD4 Is Conformation-Dependent. Because the protease resistance and other physicochemical features of PrPSc are thought to depend on protein conformation, distinct conformers of PrPSc have been characterized based on the extent of the loss in protease resistance as a function of increased exposure to denaturants (24). We applied such a conformation-dependent immunoassay to compare the conformational characteristics of the PrP captured by OCD4 with those of native PrPSc present in the homogenate from the same CJD brain. The decrease of PK resistance with rising concentrations of GdnHCl took place at a comparable rate for native PrPSc- and OCD4-captured PrP (Fig. 6 A and B), suggesting that PK-resistant forms of native PrPSc- and OCD4-captured PrP have comparable conformational properties. We then examined the affinity of OCD4 for PrP as a function of GdnHCl treatment. Decreasing amounts of PrP were recovered by immunocapture with OCD4 after protein denaturation by increasing concentrations of GdnHCl (Fig. 6C), arguing that the affinity of OCD4 for PrP is also conformation-dependent. Whether and how the conformation of the complex and that of PrP are related remain to be clarified. The report that recombinant PrP carrying a largely α-helical conformation was converted to an isoform rich in β-structure after exposure to a DNA sequence is pertinent to these questions (15). Furthermore, by comparing B and C of Fig. 6, it is obvious that the total PrP captured by OCD4 exceeds that in the PK-resistant fraction. Densitometry showed that >20% of PrP was still detectable after treatment with 2.4 M GdnHCl that renders virtually all PrPSc sensitive to PK (Fig. 6C), suggesting that at least 20% of PrP recovered by OCD4 is PK-sensitive. Therefore, PrP captured by OCD4 comprises both PK-resistant and PK-sensitive PrP species as has been reported for PrPSc after a conformation-dependent assay and other analyses (25, 34). Taken together, the similarity in gel migration pattern and in the conformation dependence of the PK resistance, as well as the presence of both PK-sensitive and -resistant components, argue that PrP captured by OCD4 and native PrPSc are the same.
Fig. 6.
Conformation-dependent immunocapture of disease-associated PrP by OCD4. (A) PK resistance of PrPSc as a function of GdnHCl concentrations. BH from CJD was treated with various concentrations of GdnHCl. The GdnHCl-treated samples were digested with 50 μg/ml PK for 1 h at 37°C (PK+) followed by immunoblotting with 3F4. (B) PK resistance of PrP captured by OCD4 as a function of GdnHCl concentrations. The GdnHCl-treated BH of CJD was subjected to IP with OCD4, followed by digestion with PK (IP/+PK). PrP was then detected by immunoblotting with 3F4. (C) Capture of PrP by OCD4 as a function of GdnHCl concentrations. The GdnHC-treated BH of CJD was subjected to IP with OCD4 without the PK treatment (IP/–PK). PrP was detected by immunoblotting with 3F4. (D) Comparison of GdnHCl-dependent loss of PrP in BH of CJD after PK digestion (•), IP with OCD4 followed by PK digestion (○), and IP with OCD4 without PK digestion (▪). Intensity of PrP bands on immunoblots was quantified by densitometry, and the ratio of PrP in the GdnHCl-treated and untreated samples was plotted as a function of GdnHCl concentrations. Similar results were obtained in two additional experiments.
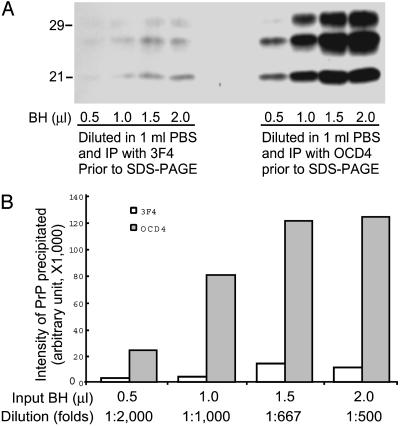
OCD4 Possesses High Affinity to PrPSc. The specificity of OCD4 to disease-associated PrP makes it potentially a suitable reagent to detect small concentrations of infectious PrP that may be present in body fluids and peripheral organs. In a crude affinity assay, small amounts of PK-treated homogenate from a CJD brain were diluted in large volume and PrP was then captured with either OCD4 or the commercially available mAb 3F4 (Fig. 7A). In four dilutions between 500- and 2,000-fold, OCD4, on average, detected the PK-resistant PrP >10-fold more efficiently than did 3F4 (11.7- ± 5.2-fold more; P = 0.004, Welch's t test; Fig. 7B). Because OCD4 was also observed to have a better capture efficiency for PrPSc than another anti-PrP Ab, 6H4 (Fig. 1), the above findings suggest that OCD4 is indeed a high-affinity PrPSc capture reagent.
Fig. 7.
Recovery of PK-resistant PrP from diluted CJD BH by OCD4 and 3F4. (A) BH (10%, wt/vol) from a CJD subject was treated with 50 μg/ml PK for 1 h at 37°C. Four aliquots of PK-treated BH ranging from 0.5 to 2.0 μl were diluted into 1 ml of PBS representing a dilution of 2,000- to 500-fold. Samples were then subjected to IP with either 3F4 or OCD4, followed by immunoblotting with 3F4. (B) Comparison of PrP recovered by IP with 3F4 and OCD4. Signal intensity (arbitrary unit) of PrP on immunoblots was quantified by densitometry. Compared with 3F4, OCD4 recovered significantly higher amounts (11.7 ± 5.2 times more) of PK-resistant PrP (P = 0.004; Welch's t test).
Discussion
In this study we observed, for the first time, to our knowledge, that the mAb to DNA OCD4 and the single-stranded DNA-binding protein g5p can capture PrPSc but not PrPC. These findings suggest that the capture of PrPSc by OCD4 and g5p depends on nucleic acid and that PrPSc forms a complex with nucleic acid. The sequence of the nucleic acid associated with PrPSc, and whether the PrPSc–nucleic acid complexes are present in vivo as previously suggested (26, 35–38), or form only in the homogenized tissue, remain to be determined.
Several recent studies (39, 40) have reported the generation of conformation-dependent mAbs that react with disease-related amyloid proteins. The Abs described by O'Nuallain and Wetzel (39) react with a variety of amyloid fibrils made of different proteins but not with their respective precursor proteins. Kayed et al. (40) also observed a polyclonal Ab that recognizes soluble amyloid oligomers of the amyloid β peptides associated with Alzheimer's disease (but not amyloid fibrils) as well as with soluble oligomeric aggregates formed by other disease-related proteins (40). It has been proposed that these Abs recognize a conformation shared by amyloid fibrils or soluble amyloid oligomers regardless of their primary structure (39, 40). These reports raise an alternative possibility that OCD4 is also a conformational Ab that recognizes a conformation shared by PrPSc and DNA. Both DNA and amyloids contain repeat structures that might result in a conformation that binds OCD4 despite the different nature of the core molecules (39). Future tests are warranted to resolve these possibilities. There is also a recent paper from Paramithiotis et al. (41) that polyclonal Abs and mAbs directed against the PrP repeat motif Tyr-Tyr-Arg specifically react with PrPSc apparently in a non-DNA-dependent fashion.
The mAb OCD4 is remarkable in several respects. First, it captures the abnormal PrP associated with prion diseases with a high level of specificity and affinity while not recognizing PrPC at all. Second, it detects both protease-sensitive and protease-resistant species of prion disease-associated PrP. Third, it suggests that both species of abnormal PrP form a complex that includes DNA or DNA-related molecules. Fourth, because several other Abs to DNA preparations also capture the disease-associated PrP and they are easy to produce, OCD4 is likely to be the first of many novel prion-specific reagents. Abs to DNA may help develop new strategies for the study, detection, and treatment of prion diseases.
Acknowledgments
We thank Jim Goodarzi for comments and statistical analysis; Jue Yuan for technical assistance; Jin-Der Wen (University of Texas at Dallas) for the purification of g5p; Cathy Burzik and Charles Tackney (Ortho-Clinical Diagnostics) for encouragement; and Katherine O'Rourke (U.S. Department of Agriculture Agricultural Research Service, Pullman, WA), Roy Jackman (Veterinary Laboratories Agency, Weybridge, U.K.), Robert Will, James Ironside, and Mark Head (U.K. Creutzfeldt–Jakob Disease Surveillance Unit) for access to clinical materials and facilities. This work was supported in part by the Britton Fund (P.G.), Robert A. Welch Foundation Grant AT-503 (to D.M.G.), National Institutes of Health Grant AG14359 (to P.G. and S.G.C.), Centers for Disease Control and Prevention Contract CCU515004 (to P.G.), Department of Agriculture Grant 2002-35201-12608 (to S.G.C.), and Department of Defense National Prion Research Program Grant DAMD17-03-1-0283 (to S.G.C.).
Abbreviations: PrP, prion protein; PrPC, cellular PrP; PrPSc, scrapie isoform of PrP; CJD, Creutzfeldt-Jakob disease; vCJD, variant CJD; sCJD, sporadic CJD; fCJD, familial CJD; GSS, Gerstmann–Sträussler–Scheinker disease; IP, immunoprecipitation; BH, brain homogenate; PK, proteinase K.
References
- 1.Gambetti, P., Kong, Q., Zou, W. Q., Parchi, P. & Chen, S. G. (2003) Br. Med. Bull. 66, 213–239. [DOI] [PubMed] [Google Scholar]
- 2.Will, R. G., Ironside, J. W., Zeidler, M., Cousens, S. N., Estibeiro, K., Alperovitch, A., Poser, S., Pocchiari, M., Hofman, A. & Smith, P. G. (1996) Lancet 347, 921–925. [DOI] [PubMed] [Google Scholar]
- 3.Bruce, M. E., Will, R. G., Ironside, J. W., McConnell, I., Drummond, D., Suttie, A., McCardle, L., Chree, A., Hope, J., Birkett, C., et al. (1997) Nature 389, 498–501. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caughey, B. W., Dong, A., Bhat, K. S., Ernst, D., Hayes, S. F. & Caughey, W. S. (1991) Biochemistry 30, 7672–7682. [DOI] [PubMed] [Google Scholar]
- 6.Pan, K. M., Baldwin, M., Nguyen, J., Gasset, M., Serban, A., Groth, D., Mehlhorn, I., Huang, Z., Fletterick, R. J., Cohen, F. E., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Safar, J., Roller, P. P., Gajdusek, D. C. & Gibbs, C. J., Jr. (1993) J. Biol. Chem. 268, 20276–20284. [PubMed] [Google Scholar]
- 8.Cohen, F. E., Pan, K. M., Huang, Z., Baldwin, M., Fletterick, R. J & Prusiner, S. B. (1994) Science 264, 530–531. [DOI] [PubMed] [Google Scholar]
- 9.Telling, G. C., Telling, G. C., Parchi, P., DeArmond, S. J., Cortelli, P., Montagna, P., Gabizon, R., Mastrianni, J., Lugaresi, E., Gambetti, P., et al. (1996) Science 274, 2079–2082. [DOI] [PubMed] [Google Scholar]
- 10.Gajdusek, D. C. (1977) Science 197, 943–960. [DOI] [PubMed] [Google Scholar]
- 11.Bruce, M. E. & Dickinson, A. G. (1987) J. Gen. Virol. 68, 79–89. [DOI] [PubMed] [Google Scholar]
- 12.Prusiner, S. B. (1997) Science 278, 245–251. [DOI] [PubMed] [Google Scholar]
- 13.Momcilovic, D. & Rasooly, A. (2000) J. Food Prot. 63, 1602–1609. [DOI] [PubMed] [Google Scholar]
- 14.Korth, C., Stierli, B., Streit, P., Moser, M., Schaller, O., Fischer, R., Schulz-Schaeffer, W., Kretzschmar, H., Raeber, A., Braun, U., et al. (1997) Nature 390, 74–77. [DOI] [PubMed] [Google Scholar]
- 15.Cordeiro, Y., Machado, F., Juliano, L., Juliano, M. A., Brentani, R. R., Foguel, D. & Silva, J. L. (2001) J. Biol. Chem. 276, 49400–49409. [DOI] [PubMed] [Google Scholar]
- 16.Korth, C., Streit, P. & Oesch, B. (1999) Methods Enzymol. 309, 106–122. [DOI] [PubMed] [Google Scholar]
- 17.Kascsak, R. J., Rubenstein, R., Merz, P. A., Tonna-DeMasi, M., Fersko, R., Carp, R. I., Wisniewski, H. M. & Diringer, H. (1987) J. Virol. 61, 3688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, S. G., Teplow, D. B., Parchi, P., Teller, J. K., Gambetti, P. & Autilio-Gambetti, L. (1995) J. Biol. Chem. 270, 19173–19180. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) in Molecular Cloning: A Laboratory Manual, eds. Sambrook, J., Fritsch, E. F. & Maniatis, T. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 9.14–9.23.
- 20.Yokoyama, W. M. (2001) in Current Protocols in Cell Biology, eds. Bonifacino, J. S., Dasso, M., Harford, J. B., Lippincott-Schwartz, J. & Yamada, K. M. (Wiley, New York), pp. 16.1.1–16.1.17.
- 21.Thompson, T. M., Mark, B. L., Gray, C. W., Terwilliger, T. C., Sreerama, N., Woody, R. W. & Gray, D. M. (1998) Biochemistry 37, 7463–7477. [DOI] [PubMed] [Google Scholar]
- 22.Mou, T.-C., Gray, C. W. & Gray, D. M. (1999) Biophys. J. 76, 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou, W. Q. & Cashman, N. R. (2002) J. Biol. Chem. 277, 43942–43947. [DOI] [PubMed] [Google Scholar]
- 24.Peretz, D., Scott, M. R., Groth, D., Williamson, R. A., Burton, D. R., Cohen, F. E. & Prusiner, S. B. (2001) Protein Sci. 10, 854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safar, J., Wille, H., Itri, V., Groth, D., Serban, H., Torchia, M., Cohen, F. E. & Prusiner, S. B. (1998) Nat. Med. 4, 1157–1165. [DOI] [PubMed] [Google Scholar]
- 26.Sklaviadis, T., Akowitz, A., Manuelidis, E. E. & Manuelidis, L. (1990) Arch. Virol. 112, 215–228. [DOI] [PubMed] [Google Scholar]
- 27.Peretz, D., Williamson, R. A., Matsunaga, Y., Serban, H., Pinilla, C., Bastidas, R. B., Rozenshteyn, R., James, T. L., Houghten, R. A., Cohen, F. E., et al. (1997) J. Mol. Biol. 273, 614–622. [DOI] [PubMed] [Google Scholar]
- 28.Parchi, P., Zou, W., Wang, W., Brown, P., Capellari, S., Ghetti, B., Kopp, N., Schulz-Schaeffer, W. J., Kretzschmar, H. A., Head, M. W., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 10168–10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou, W., Colucci, M., Gambetti, P. & Chen, S. G. (2002) in Methods in Molecular Biology-Neurogenetics: Methods and Protocols, ed. Potter, N. T. (Humana, Totowa, NJ), pp. 305–314.
- 30.Tagliavini, F., Prelli, F., Ghiso, J., Bugiani, O., Serban, D., Prusiner, S. B., Farlow, M. R., Ghetti, B. & Frangione, B. (1991) EMBO. J. 10, 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tagliavini, F., Prelli, F., Porro, M., Rossi, G., Giaccone, G., Farlow, M. R., Dlouhy, S. R., Ghetti, B., Bugiani, O. & Frangione, B. (1994) Cell 79, 695–703. [DOI] [PubMed] [Google Scholar]
- 32.Parchi, P., Chen, S. G., Brown, P., Zou, W., Capellari, S., Budka, H., Hainfellner, J., Reyes, P. F., Golden, G. T., Hauw, J. J., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 8322–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou, W. Q., Capellari, S., Parchi, P., Sy, M. S., Gambetti, P. & Chen, S. G. (2003) J. Biol. Chem. 278, 40429–40436. [DOI] [PubMed] [Google Scholar]
- 34.Caughey, B., Raymond, G. J., Callahan, M. A., Wong, C., Baron, G. S. & Xiong, L. W. (2001) Adv. Protein Chem. 57, 139–169. [DOI] [PubMed] [Google Scholar]
- 35.Marsh, R. F., Malone, T. G., Semancik, J. S., Lancaster, W. D. & Hanson, R. P. (1978) Nature 275, 146–147. [DOI] [PubMed] [Google Scholar]
- 36.Aiken, J. M., Williamson, J. L., Borchardt, L. M. & Marsh, R. F. (1990) J. Virol. 64, 3265–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akowitz, A., Sklaviadis, T. & Manuelidis, L. (1994) Nucleic Acids Res. 22, 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narang, H. K., Asher, D. M. & Gajdusek, D. C. (1988) Proc. Natl. Acad. Sci. USA 85, 3575–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Nuallain, B. & Wetzel, R. (2002) Proc. Natl. Acad. Sci. USA 99, 1485–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W. & Glabe, C. G. (2003) Science 300, 486–489. [DOI] [PubMed] [Google Scholar]
- 41.Paramithiotis, E., Pinard, M., Lawton, T., LaBoissiere, S., Leathers, V. L., Zou, W. Q., Estey, L. A., Lamontagne, J., Lehto, M. T., Kondejewski, L. H., et al. (2003) Nat. Med. 9, 893–899. [DOI] [PubMed] [Google Scholar]