Abstract
Agr is an autoinducing, quorum-sensing system that functions in many Gram-positive species and is best characterized in the pathogen Staphylococcus aureus, in which it is a global regulator of virulence gene expression. Allelic variations in the agr genes have resulted in the emergence of four quorum-sensing specificity groups in S. aureus, which correlate with different strain pathotypes. The basis for these predilections is unclear but is hypothesized to involve the phenomenon of quorum-sensing interference between strains of different agr groups, which may drive S. aureus strain isolation and divergence. Whether properties intrinsic to each agr allele directly influence virulence phenotypes within S. aureus is unknown. In this study, we examined group-specific differences in agr autoinduction and virulence gene regulation by utilizing congenic strains, each harboring a unique S. aureus agr allele, enabling a dissection of agr locus-dependent versus genotype-dependent effects on quorum-sensing dynamics and virulence factor production. Employing a reporter fusion to the principal agr promoter, P3, we observed allele-dependent differences in the timing and magnitude of agr activation. These differences were mediated by polymorphisms within the agrBDCA genes and translated to significant variations in the expression of a key transcriptional regulator, Rot, and of several important exoproteins and surface factors involved in pathogenesis. This work uncovers the contribution of divergent quorum-sensing alleles to variant expression of virulence determinants within a bacterial species.
INTRODUCTION
Bacterial pathogenicity results from a complex interplay of virulence factors with regulatory systems that respond to multiple external signals from the host environment and the bacterial population. Polymorphisms within critical regulatory genes are increasingly recognized within members of the same species and are thought to contribute to strain-specific differences in disease outcomes (13, 57). In the major human pathogen Staphylococcus aureus, virulence gene expression is controlled by several interacting regulatory systems, including two-component modules, alternative sigma factors, and transcription factors (53). Many of these regulatory genes are observed to vary across strains, often associated with profound effects on the virulon (2, 8, 12, 40).
A key multiallelic regulatory system in S. aureus that influences the overall regulatory network and the virulon is the accessory gene regulator (agr) quorum-sensing system. agr is an autoinducing, two-component system that reciprocally controls surface factors and secreted enzymes and cytotoxins in coordination with changes in the concentration of a secreted pheromone (54) and is critical for pathogenesis in many contexts (1, 17, 25, 75). agr encodes the autoinducing peptide (AIP) pheromone (known as AgrD in its proform), its processor, AgrB, and its detector, the AgrC receptor histidine kinase. At a critical AIP concentration, which may mirror a threshold population density, or quorum, AgrC activation leads to phosphorylation of its response regulator, AgrA, and autoinduction of the agr promoters P2 and P3. P3 activation results in expression of the regulatory RNA, RNAIII, which affects the expression of a plethora of virulence genes both directly, by binding to their transcripts (31, 56), and indirectly, by acting on intermediary transcription factors such as Rot (10, 22, 65). Additional genes implicated in virulence are regulated by agr independently of RNAIII in an AgrA-dependent manner (62). Expression of agr is influenced by other regulatory factors (8, 28, 46–48, 68, 80), indicating that quorum-sensing inputs are integrated with additional cellular signals in the control of the bacterial virulon.
The agr locus has diverged widely among strains of S. aureus, with polymorphisms centering on agrB, agrD, and agrC (see Fig. S1 in the supplemental material), genes whose products interact molecularly. This diversity has resulted in four S. aureus agr pherotypes or specificity groups, which are referred to as groups I through IV (33, 35) (the corresponding agr alleles, which here denote the entire group of genes comprising the agr locus, are, respectively, referred to as agr-I through -IV). The specificity groups have the interesting functional property of heterologous mutual inhibition; that is, the unique AIP of a given group will generally cross-inhibit the agr signal in strains belonging to a heterologous group (35). Diversity in the agr locus extends far beyond S. aureus: all other staphylococcal species examined to date possess a unique agr locus with one or more allelic variants (20, 35, 37, 59), several of which are known to mediate heterologous interference with S. aureus agr (24, 37, 58; J. S. Wright and R. P. Novick, unpublished data), and agr homologs have been identified in many other Gram-positive bacterial genera (4, 19, 51, 54, 67, 78). agr divergence and cross-interference may therefore be a wide-ranging phenomenon among the staphylococci and related organisms.
Interestingly, S. aureus agr specificity groups appear to correlate with different infection types. For example, strains belonging to agr group III are the primary cause of menstrual toxic shock syndrome (TSS) (34, 35) and are responsible for a large subset of Panton-Valentine leukocidin (PVL)-associated necrotizing pneumonia cases (26, 70), while most strains producing the toxin responsible for staphylococcal scalded skin syndrome, exfoliatin, belong to agr group IV (33, 50). These associations appear to involve the acquisition by strains in each group of particular mobile genetic elements (MGEs) carrying toxins (77); however, other transmissible elements and virulence genes correlate poorly with agr alleles (34, 77). The molecular basis underlying the predilections of agr pherotypes for S. aureus pathotypes is currently unknown.
At least two not mutually exclusive hypotheses have been proposed to explain the broader biological significance of agr diversity. The first hypothesis is that agr interference may serve to isolate bacterial populations through niche competition, limiting genetic exchange and promoting evolutionary diversification. In support of this prediction, each S. aureus agr allele is observed to be associated with a specific set of overall genotypes (34, 77), pointing to agr group differentiation as a primary evolutionary event that preceded genotypic divergence; however, as mentioned above, examples of virulence determinants that strongly correlate with agr alleles are limited (34, 77). The second hypothesis is that agr diversity results in quorum-sensing systems with variant signaling properties, ultimately affecting the overall virulence gene regulatory network. In support of this hypothesis are previous observations that relate to other groups; induction of RNAIII occurs early (in early exponential phase) in agr-IV strains (33) and later (in late exponential phase) in agr-III strains (69; our unpublished data). These timing differences are associated with expression of virulence genes at different phases of cell growth (33, 69; A. M. Figueiredo, H. F. Ross, and R. P. Novick, unpublished data). It is unknown to what extent the temporal differences in agr activation are due to intrinsic properties particular to each quorum-sensing allele as opposed to extrinsic influences of the overall bacterial regulatory network (which are particular to each strain) on the agr promoters and gene products. The direct impact of intrinsically different agr signaling dynamics on the expression of the virulon, and by extension on potential strain pathotypes, is similarly unknown.
In this study, we have addressed these questions by analyzing the four divergent S. aureus agr alleles in identical genomic contexts. This approach has allowed us to dissect the contribution of each allele, independent of the background genotype, to signal transduction and accessory gene production and begin to test the hypothesis that concerted allelic variants in an autoinducing regulatory system provide the framework for adaptive virulence phenotypes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. aureus strains used in this study are listed in Table S1 in the supplemental material. RN6734 is a derivative of 8325-4 (52) and is our standard agr group I laboratory strain. RN7206 is a derivative of 8325-4, in which the agr locus has been replaced by tetM. Newman, an agr group I prototype and a human clinical isolate, was originally isolated from a secondarily infected tubercular osteomyelitis lesion (21). HF6122 is a Newman derivative carrying the agr∷tetM replacement. RN6607 (also known as 502A), an agr group II prototype, was originally isolated from the nasal mucosa of a healthy carrier (66). RN3984 is a prototypical agr group III strain and a menstrual toxic shock syndrome isolate (39). RN4850 is a group IV prototype isolated from a patient with staphylococcal scalded skin syndrome (33). Cloning was performed with Escherichia coli strain DH5α. All clones were first transformed into RN4220, our standard recipient for E. coli DNA, or its derivatives containing a site-specific integrase before transduction to other strains.
S. aureus cells from overnight plates containing the appropriate selective antibiotics (chloramphenicol 10 μg/ml, erythromycin 10 μg/ml, and/or cadmium 0.1 mM) were used as base inocula for all experiments. Subsequent growth in Casamino Acids-yeast extract-glycerophosphate (CYGP) broth without glucose and without antibiotics was performed at 37°C with shaking. Cell density was determined by using a ThermoMax microplate reader (Molecular Devices) to measure the optical density at 650 nm (OD650) of 100 μl samples in 96-well round-bottom assay plates.
Plasmid construction.
The plasmids used in this study (see Table S1 in the supplemental material) were constructed by cloning PCR products amplified with oligonucleotide primers obtained from Integrated DNA Technologies (Coralville, IA), as listed in Table S2 in the supplemental material. Clones were sequenced by the Skirball DNA Sequencing Core Facility or Macrogen (Rockville, MD). Plasmid pJC111, which contains a cadmium resistance cassette and the SaPI-I attS sequence (23), was used as the backbone vector for agr locus constructs. agr alleles II through IV were amplified from RN6607, RN3984, and RN4850, respectively, and introduced via blunt ligation into the polylinker of pUC18 at the HincII site. agr-I was obtained from pJC1000, which contains the agr locus cloned from RN6734 in pUC18 (23). Constructs were then subcloned into pJC1111 using restriction enzymes SphI and KpnI. Chimeric agr loci were constructed by swapping SphI-BseRI and BseRI-KpnI fragments involving the unique BseRI site in a conserved region immediately upstream of agrB. pJC1111 derivatives containing agr locus constructs or the vector alone were integrated into the S. aureus genome at the SaPI-1 attC site by electroporating into RN9011, which expresses the SaPI-1 integrase. Chromosomal agr constructs within RN9011 derivatives were amplified and resequenced to confirm the absence of adventitious mutations, and multiple independent transductants were utilized in subsequent experiments.
An agrp3 β-lactamase reporter was constructed by moving the agrp3-blaZ fragment from pJC1349 (containing agrp3 cloned from RN6734) to pEG813, a suicide vector containing a cloned ϕ11 attP cassette (amplified from pLL29 [44]). An analogous plasmid lacking a promoter was constructed by moving the promoterless blaZ gene from pCN41 to pEG813. These plasmids were individually integrated into the ϕ11 attP site by electroporating into RN11679, which contains the ϕ11 integrase encoded in pLL2787 (44). pLL29 and pLL2787 were kindly provided by Chia Lee. β-Lactamase fusions to virulence gene promoters were constructed by PCR amplification of the lukSF-PV promoter from ϕSLT, hla promoter from RN6734, and the tsst-1 promoter from RN4282 and cloned into pJC1280. A constitutive variant of the β-lactamase promoter was moved from pJC1122 to the multiple-cloning site (MCS) of pCN41.
Reporter assays.
Strains containing β-lactamase fusions were transferred from overnight plates into CYGP without glucose, washed once with fresh media to remove carry-over AIP, and then inoculated into CYGP without glucose at OD650 of 0.005. Cells were grown at 37°C with shaking to OD650 of 0.05 and were diluted with fresh media to OD650 of 0.01, representing T0 and corresponding to approximately 1 × 107 CFU/ml. This dilution step permitted further reduction of residual agr and reporter transcripts, cell density normalization, and synchronization in early exponential phase. Samples were collected at the indicated time points and stored at −80°C. Samples were thawed on ice and diluted with media to appropriate densities for turbidity measurements and enzyme assays. Assay of β-lactamase activity was performed by the nitrocefin method as described previously (36). Assay data were normalized to β-lactamase units (Vmax/OD650).
Exoprotein analysis.
For qualitative analysis of hemolysin production, cells were patched on sheep's blood agar (SBA) plates, followed by overnight growth at 37°C. For SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of exoprotein profiles, cells were washed and grown as described for reporter assays, samples were collected at the indicated time points, and cell density was measured. Culture supernatants were isolated by centrifugation, and sample volumes within each time point were normalized on the basis of culture densities. Protein was precipitated with trichloroacetic acid, resuspended in SDS loading buffer, and separated by SDS-PAGE in a 12% polyacrylamide gel, followed by visualization using Coomassie blue dye (41). Secreted β-lactamase from the same samples was analyzed by zymography with a method adapted from the work of others (49, 63). Samples were run on a 15% polyacrylamide gel, and the gel was washed in water twice for 20 min, incubated in 500 ml renaturation buffer (50 mM sodium phosphate buffer [pH 7.0], 1% Triton X-100, 0.1 mM ZnSO4) at 37°C for 4 h, and then overlaid with filter paper presoaked in 50 mM sodium phosphate buffer, pH 7.0, containing the chromogenic substrate nitrocefin at 0.25 mM. Protein identification by mass spectrometry was performed by proteolytic digestion with trypsin of excised gel slices containing the protein band of interest, followed by liquid chromatography-mass spectrometry (LC-MS) (Skirball Protein Analysis Facility).
RESULTS
Construction of congenic strains containing unique agr alleles.
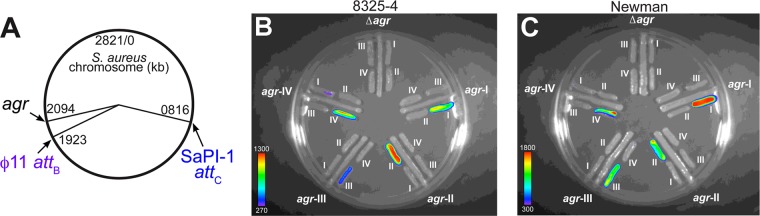
In order to construct congenic strains, each harboring a unique agr allele, we inserted the agr genes, cloned from the chromosome of prototypical S. aureus strains belonging to each agr specificity group (see Table S1 in the supplemental material), in single copy within a Δagr strain. We utilized the staphylococcal pathogenicity island (SaPI)-1 attC locus (16, 23, 24) as the insertion site, noting that this position is at a roughly similar distance from the chromosomal origin of replication relative to the native agr locus (Fig. 1A), enabling similar gene copy numbers at these locations in exponentially growing cells (14, 81). We employed two S. aureus strain backgrounds, RN7206 and HF6122, which are Δagr mutant derivatives of NCTC 8325-4 and Newman, respectively (see Table S1). These strains were selected because of their differences in expression of transcriptional regulators, including Sae (2) and σB (8), their differences in agr activity (8), their global differences in virulence factor production (29), and their wide use by several labs as prototypical wild-type (WT) strains (6, 15, 17, 61, 65). Integration of SaPI-1 into its attC site invariantly occurs in the same orientation (64), and we verified the identical orientation of each agr locus in the chromosome by PCR (see Fig. S2A in the supplemental material). The functionality of the agr system from this site and the specificity group identity for each resulting congenic strain were confirmed using the plate-based agr typing assay (77) (Fig. 1B and C).
Fig 1.
Chromosomal positions of loci under study and bioluminescent typing assay of congenic S. aureus strains. (A) Chromosomal positions in NCTC 8325 of the native agr locus and SaPI-1 and ϕ11 attachment sites. The ϕ11 attB site is identical to the integration site of phage NM1 in Newman (5), and the indicated sites are located at analogous distances in the Newman chromosome. (B, C) Bioluminescent plate assay. Congenic strains with the indicated agr allele are streaked radially. The reporter strains (RN9688, RN9689, RN9690, and RN9691 [77]), each carrying an allele-specific AgrCA two-component signaling pair plus a agrp3-lux fusion, are patched alongside the congenic strains, each of which activates one of the reporters according to the specificity of its AIP. Reporters are identified by roman numerals designating their specificity group. Bioluminescence was detected with a Hamamatsu charge-coupled-device camera and is presented as a pseudocolor image, with the color bar indicating the signal intensity in counts.
Cell growth in culture was observed to be identical for congenic strains of the Newman background (see Fig. S2C in the supplemental material) and nearly identical for those of the 8325-4 background (see Fig. S2B); the agr-III and Δagr strains in this background were associated with slightly increased culture turbidity over time, consistent with earlier observations with agr mutant cells (our unpublished data). These differences were eliminated when early exponential-phase cells were diluted to a lower cell density (see Fig. S2B, inset), presumably because of dilution of residual agr transcripts and AIP carried over during subculture.
Allelic differences in agr induction dynamics.
We have previously observed differences in the timing of RNAIII transcription, the immediate consequence of agr autoinduction, across divergent S. aureus strains belonging to different agr groups; most notably, RNAIII is induced very early during growth in group IV isolates, about 3 h earlier than in other groups during standard culture conditions (33), and group III strains have been observed to induce RNAIII transcription late compared to the other groups (69; our unpublished data). The contribution of the intrinsic properties of each agr allele in determining the timing of RNAIII expression, relative to the effects of extrinsic regulatory factors elsewhere in the genome, is not known. Similarly, a uniform analysis of group-specific differences in agr activation magnitude has not been reported. In order to explore the activation timing and maximal signal strength of each agr allele in the congenic strains, we employed a fusion of the β-lactamase reporter gene to the principal agr promoter, P3. The agrp3-blaZ construct was cloned into a suicide plasmid containing the phage ϕ11 attP sequence (42) and inserted in single copy into the corresponding chromosomal attB site (Fig. 1A). A promoterless blaZ construct at this position resulted in reporter activity barely detectable over background (see Fig. S3A in the supplemental material), indicating that gene expression from this chromosomal site is not affected by exogenous transcriptional read-through.
Using the agrp3-blaZ reporter, we next determined whether the chromosomal position of the agr locus affected its activation. We observed similar agrp3 induction kinetics and maximal signal levels during growth in culture in cells containing the agr-I locus at its native site or at the SaPI-1 attC site (see Fig. S3B in the supplemental material). We also observed similar exoprotein production levels, as measured by SDS-PAGE of culture supernatants (see Fig. S4 in the supplemental material) and SBA analysis (see Fig. 3B, top row), with agr in its native site or at the SaPI-1 site within the same genomic background. These results indicate that the SaPI-1 attC site functionally substitutes for the native agr site for these studies.
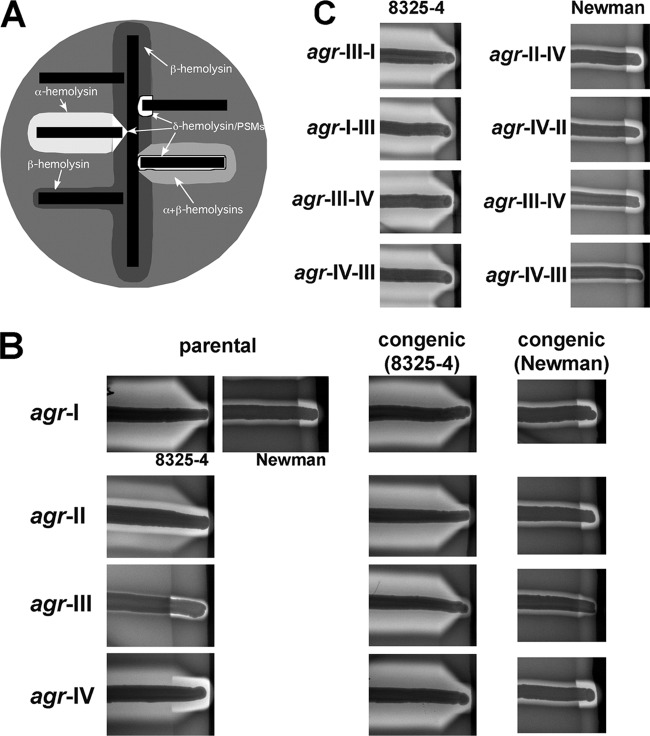
Fig 3.
Analysis of hemolytic activity of S. aureus strains on sheep's blood agar (SBA). (A) Schematic of hemolytic activities on SBA. Tested bacteria (horizontal black bars) are streaked at a right angle to RN4220 (vertical black bar), a β-hemolysin producer, and the plate is incubated overnight. β-Hemolysin forms a turbid zone of hemolysis; β is synergistic with δ-hemolysin and other PSMs, producing an amplified zone of clearing where they intersect; β inhibits α-hemolysin, producing the V-shaped zone of clearance where they intersect. (B, C) Hemolytic activities on SBA for parental and congenic strains with the indicated agr allele cross-streaked with RN4220. Images from each column are from the same plate. Parental strains: agr-II, RN6607; agr-III, RN3964; agr-IV, RN4850. The pattern seen with native agr-II is due to the production of β-hemolysin, as well as α and δ and/or other PSMs; Newman has a prophage in the β-hemolysin gene. The α-β mutual inhibition is seen only with Newman agr-III because of the low δ-hemolysin and/or other PSM production by group III. In panel C, the chimeras tested correspond to those tested for reporter activity in Fig. 2C and D.
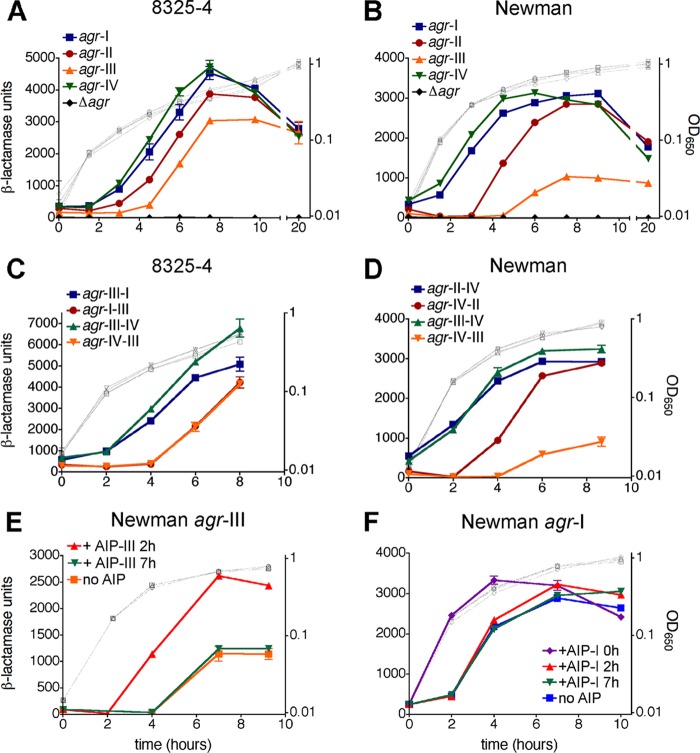
We next used the agrp3-blaZ reporter to analyze agr activation by the congenic strains containing unique agr alleles. The timing and maximal activity of reporter induction varied for both 8325-4 and Newman backgrounds and was significantly dependent on the particular agr allele (Fig. 2). With the 8325-4 strains harboring agr-IV and -I, reporter activity was induced with the earliest kinetics, with 50% maximal activities reached at approximately 4.4 and 4.7 h, respectively (Fig. 2A). This was followed by agr-II and -III cells at approximately 5.3 and 5.8 h, respectively (Fig. 2A). With the Newman strains, a similar ordering in allele-specific induction timing was observed (Fig. 2B): agr-IV cells displayed the earliest induction kinetics, followed in order by agr-I, -II, and -III (Fig. 2B; 50% maximal activities reached at approximately 2.4, 2.9, 4.6, and 5.6 h, respectively). Overall, with respect to maximal activation levels, these were higher for the 8325-4 strains compared to the Newman strains, consistent with previous reports with the wild-type strains (8, 27). With both congenic strain sets, the greatest induction levels were observed with agr-I and agr-IV and the lowest with agr-III (Fig. 2). Reporter induction by the agr-III cells was particularly low in the Newman background (Fig. 2B).
Fig 2.
Tests of P3 promoter activity in congenic strains containing unique agr alleles. S. aureus cells of the 8325-4 (A, C) or Newman (B, D) background containing the indicated agr allele (at SaPI-1 attC) and a chromosomal agrp3-blaZ transcriptional fusion (ϕ11 attB∷pEG835) were assayed for β-lactamase activity during growth in triplicate cultures. (C, D) agr-x-y refers to chimeric agr loci, with x representing the 5′ fragment containing RNAIII and the intergenic region, and y representing the 3′ fragment containing agrBDCA (see Fig. S1 in the supplemental material). (E, F) Effect of exogenous AIP on agrp3 activity. S. aureus cells derived from strain Newman containing agr-III (E) or agr-I (F) and the agrp3-blaZ reporter were assayed before or after addition at the indicated time points of AIP at 1 μM. Assay data are presented as β-lactamase units ± standard errors of the means (SEM) (closed symbols, left axis) and growth as optical density measurements (gray open symbols, right axis).
To determine whether the observed allele-dependent variations in reporter activity are due specifically to effects on the P3 promoter, as opposed to general effects of the agr alleles on cellular transcript levels, protein levels, or protein export, we tested strains expressing the β-lactamase reporter from a promoter known to be unaffected by agr. To this end, we utilized a stable constitutive variant of the blaZ promoter (blaZpc; see Fig. S3C in the supplemental material), the native version of which is known to be agr independent (55). In strains carrying either of the two agr alleles most variant in induction properties, agr-III and -IV, we observed that the reporter signal from blaZpc was largely unaffected by the agr allele present (see Fig. S3D and E). The allelic differences in agrp3 reporter activity are therefore due to specific effects on transcription from this promoter and are not due to general effects on intra- or extracellular reporter protein levels. Together these results demonstrate that the agr alleles are characterized intrinsically by different activation dynamics, with significant variations in the timing and maximal strength of the agr signal. agr-III mediates the most delayed and weakest induction signal, agr-IV the earliest, and agr-I and -IV the strongest. These differences are consistent with previous observations of RNAIII production by S. aureus isolates belonging to different agr specificity groups (33, 69; our unpublished data). These experiments also demonstrate that strain genomic background affects both the timing and magnitude of agr induction, in agreement with previous reports on the effects of σB levels on agr activity (30).
The agrBDCA region is responsible for variations in induction properties.
Intrinsic properties of the agr locus that could influence induction dynamics include differences in the rate of AIP processing and secretion, AIP-AgrC interaction kinetics, AgrC phosphorylation and phosphotransfer rates, and agr promoter activities. The vast majority of nucleotide polymorphisms among the agr alleles occurs in the hypervariable agrB-agrD-agrC region (see Fig. S1 in the supplemental material), while a much smaller subset of polymorphisms occurs in RNAIII, the agr intergenic region (containing agr promoters P2 and P3), and agrA, for which the resulting protein sequence is absolutely conserved. Single nucleotide changes in the agr intergenic region, however, can have a large impact on expression of this system (71). To determine the relative contribution of each set of polymorphisms to differences in agr activation properties, we analyzed agr locus chimeras in which locus segments were swapped between alleles at a conserved site immediately upstream of agrB (see Fig. S1). These chimeras were functional, as indicated by agrp3 reporter activity (Fig. 2C and D). In each case, the chimeras demonstrated activation signal timing and magnitude that behaved according to the group identity of the fragment containing the P2 transcript genes (agrBDCA). For example, in all backgrounds, chimeras involving this region of agr-III demonstrated activation kinetics virtually identical to that of the WT agr-III allele, and in the Newman background in which chimeras involving agr-II were tested, the construct containing the agr-II P2 transcript genes behaved similarly to WT agr-II (Fig. 2C and D). These results indicate that the P2 open reading frames (ORFs) (agrBDCA), as opposed to the P2 and P3 promoters and RNAIII, determine allelic differences in agr induction properties.
Effects on agr induction by addition of exogenous AIP.
The magnitude of induction by the agr-III allele is lower than other groups, particularly in the Newman background. In order to determine whether this weak signal was due to a decreased capacity for AgrC-III to activate in response to ligand, we tested whether the signal could be increased by addition of exogenous AIP-III at different time points during the autoinduction process. We added a saturating concentration of AIP-III to Newman cells expressing the agr-III allele at a time point before the onset of agrp3 induction (2 h) and after maximal induction levels were reached (7 h) and measured agr reporter activity. Addition of AIP-III at 2 h resulted in agrp3 activity that was earlier and of higher magnitude than that observed with no exogenous AIP (Fig. 2E), and the maximal signal level reached was roughly commensurate with that of the other agr alleles in this background. Addition of exogenous AIP-III at 7 h, however, did not result in any further increase in agr signal strength (Fig. 2E). This result can be interpreted in two ways: either all AgrC receptor molecules are already saturated with endogenous AIP at this late time point or additional AgrC activation does not lead to further transcription from agrp3 or expression of the transcript at this stage of growth, perhaps due to extrinsic, stationary phase-dependent limits on gene expression (32, 38). Activation of agr-I was also hastened by adding exogenous AIP-I at the early time point (T0), but the maximal signal level was not increased (Fig. 2F); further, addition of AIP-I to agr-I cells at later time points (2 and 7 h) did not affect induction kinetics or signal strength, consistent with receptor saturation at 2 h and saturation or growth-phase limits at 7 h. These data indicate that the delayed and weakened induction of agr-III relative to other alleles is not due to a decreased maximal signaling capacity of AgrC-III and is likely owing, at least in part, to a reduced production of AIP-III, controlled by the products of agrB or agrD.
Influence of agr allele on hemolysin production.
Production of exoproteins such as hemolysins in S. aureus is controlled by both the agr system and by regulatory factors encoded elsewhere in the genome, the expression and activity of which often vary across strains (9, 15). Given the differences in induction timing and magnitude among the agr alleles, we hypothesized that these differences would lead to variations in exoprotein production when examined in congenic backgrounds. As an initial test of this hypothesis, we analyzed hemolysin activity qualitatively on sheep's blood agar (SBA) plates. Strains were cross-streaked with RN4220, which produces only β-hemolysin, to facilitate identification of the activities of different types of hemolysins after overnight growth. An interpretation of the resulting hemolytic patterns is shown in Fig. 3A. As seen in Fig. 3B, the parental strains (the agr specificity group prototypes from which each allele was cloned) each produced a unique hemolytic pattern on SBA. The agr-III prototype, a toxic shock syndrome (TSS) isolate, produced very low hemolysin levels, characteristic of strains producing the TSST-1 toxin, which is associated with repression of exoproteins (53); in contrast, the agr-IV prototype produced large amounts of hemolysins that synergize with β. The global differences across specificity groups were no longer apparent with the congenic strains (Fig. 3B), illustrating the role of background genotype on hemolysin activity. Reproducible decreases in the production of β-synergizing hemolysins, however, were observed with the agr-III strains compared to their congenic counterparts, particularly in the Newman background; likewise, the agr-IV strains appear to produce more β-synergizing hemolysins relative to their congenic strains. β-Synergistic hemolysis on SBA has traditionally been attributed to δ-hemolysin; it is noted, however, that this toxin belongs to the family of phenol-soluble modulins (PSMs) (72), additional members of which can also contribute to the β-synergistic hemolysis phenotype (18) and are also agr regulated (72). This contribution may be more relevant in Newman versus 8325-4, because of the very low levels of many non-δ-toxin PSMs in the latter strain background (18). These experiments demonstrate that while differences in hemolysin production in strains with a functional agr system are globally determined by the background genotype, allelic variation of the agr locus has an effect on the production of at least one hemolysin type, the β-synergizing hemolysins.
SBA tests of S. aureus congenic strains containing the interallele agr chimeras tested in Fig. 2 agreed with the corresponding reporter data: chimeras involving the group III agrBDCA region generally demonstrated reduced hemolysis, predominantly due to a reduction in β-synergizing hemolysin activity, comparable to the hemolysin pattern observed with strains containing a WT agr-III allele (Fig. 3C). The delayed and weakened induction signal mediated by the group III agrBDCA region thus results in decreased hemolysin production.
Influence of agr allele on exoprotein profiles.
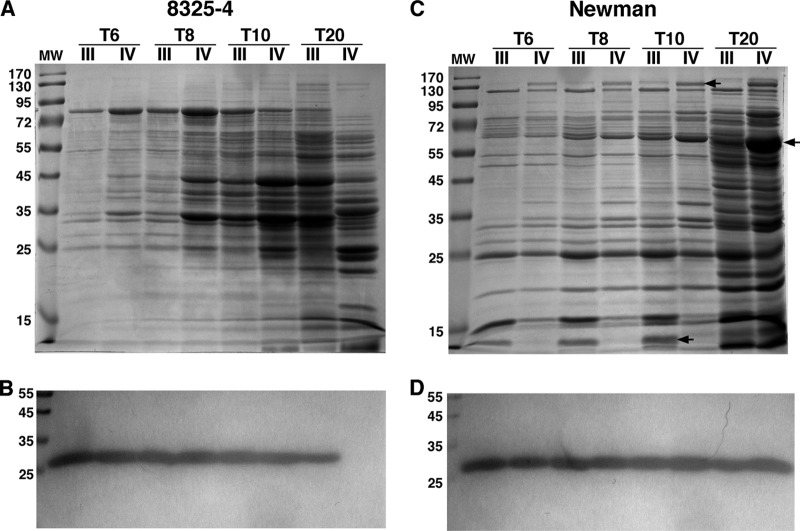
To explore the temporal effects of agr alleles on production of a range of exoproteins, we analyzed, by SDS-PAGE, exoprotein levels in culture supernatants collected at multiple time points during autoinduction. For these experiments, we focused on the two agr alleles with the most widely variant induction properties, agr-III and agr-IV, and we employed strains carrying the blaZpc-blaZ construct (see Fig. S3 in the supplemental material), which enabled detection, via zymography, of secreted β-lactamase as an agr-independent internal control. Over the time points analyzed, β-lactamase levels were similar between agr-III and agr-IV samples (Fig. 4B and D), with the exception of one sample (Fig. 4B, T20 section, lane IV), for which the corresponding cells had presumably lost the reporter plasmid after overnight growth without selection. In contrast, overall exoprotein profiles, as revealed by Coomassie staining of the same samples, were visibly different across all time points between the two alleles in both 8325-4 and Newman backgrounds (Fig. 4A and C). With respect to overall exoproteins produced, agr-III cells of the 8325-4 background appeared to lag behind the congenic agr-IV cells by about 2 h, consistent with the temporal gap in agrp3 reporter induction between these strains (Fig. 2A), resulting in differences in exoprotein patterns at each time point that were quite dramatic (Fig. 4A). The differences in exoprotein production between the two alleles was less dramatic with the Newman strains and, unlike the 8325-4 background, these differences did not appear to reflect a temporal shift between the two agr groups. Several distinct bands appeared in agr-IV supernatants that were much less abundant in that of agr-III, as well as one band present in agr-III samples that was not visible in agr-IV samples (Fig. 4C). We determined the identity of three of these bands by mass spectrometry. The band at approximately 140 kDa was identified as bifunctional autolysin precursor (Atl), the band at approximately 60 kDa was identified as catalase (KatA), and the band at approximately 12 kDa, which was less abundant in agr-IV cells, was identified as fibrinogen binding protein (Fib/Efb). Extracellular fibrinogen binding protein from strain Newman has a molecular mass of 19 kDa, so the band we analyzed may represent a degradation product, as observed in previous studies (29, 43); the differential presence of this band may thus represent differential Efb levels or differential exoproteolytic activities associated with the two strains. These results demonstrate that the agr allele can have multiple effects on exoprotein expression in S. aureus.
Fig 4.
Exoprotein analysis. Culture supernatants were collected at the indicated time points, and exoproteins were isolated and separated via SDS-PAGE. Cells contained the indicated agr allele and the blaZpc-blaZ construct (pEG832). (A, C) Coomassie staining. Bands indicated by arrows were analyzed by LC-MS. (B, D) Results of β-lactamase zymogram analysis performed after renaturing SDS-PAGE with the chromogenic substrate nitrocefin.
Influence of agr allele on regulation of virulence genes.
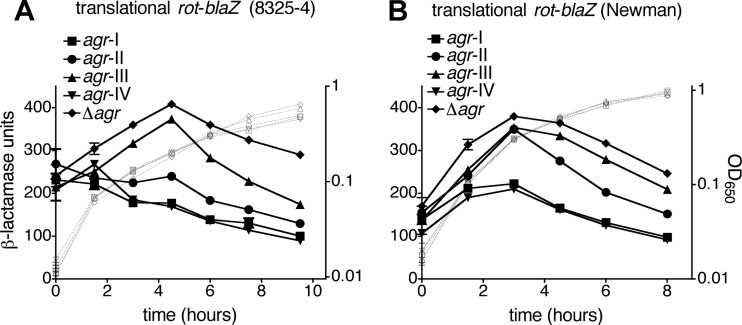
agr affects exoprotein expression to a large extent through intermediary transcription factors (53), including the global regulator Rot, the translation of which is blocked by the agr effector, RNAIII (10, 22). We reasoned that allelic variation in agr activity could result in altered regulation of Rot translation, which may contribute to the observed differences in exoprotein production. To test this prediction, we employed a translational rot-blaZ fusion. In previous studies, we observed that the activity of this reporter is decreased in agr+ relative to Δagr mutant cells, whereas the reverse is true for the corresponding transcriptional rotp-blaZ fusion, indicating translational inhibition of Rot by agr superimposed on Rot-mediated transcriptional autorepression (22). Here, in both strain backgrounds, we again observed decreased reporter activity in agr+ cells relative to Δagr mutant cells (Fig. 5). Notably, the degree of this relative decrease depends on the agr allele present and is commensurate with the timing and magnitude of agrp3 activation by each allele (see Fig. 2). For both strain backgrounds, maximal inhibition of Rot translation is observed with agr-I and -IV, the two alleles with the earliest and strongest agr activation dynamics. This is followed by agr-II, with an intermediate degree of inhibition (which is also delayed until after 3 h in the Newman strain), and agr-III, which demonstrates delayed inhibition of Rot translation in the 8325-4 and very low-level inhibition in the Newman context (Fig. 5).
Fig 5.
Effect of agr allele on inhibition of Rot translation. S. aureus cells of the 8325-4 (A) or Newman (B) background containing the indicated agr allele and a rot-blaZ translational fusion (pRN9162) were assayed for β-lactamase activity. Assay data are presented as in Fig. 2.
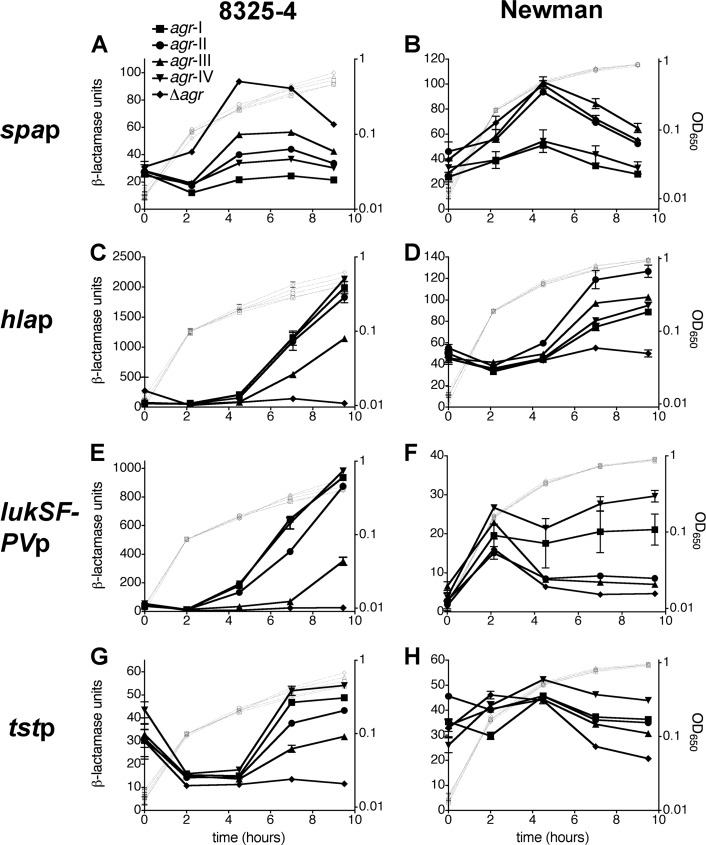
We next examined the influence of each allele on the temporal activity of several virulence gene promoters that are up- or downregulated by agr, including those for virulence genes encoded in the chromosome (spa and hla) and encoded by MGEs (lukSF-PV and tst). As shown in Fig. 6, transcription from these promoters was affected in an allele-dependent manner. In the 8325-4 background, promoter regulation followed the trend previously observed with agrp3 activity and Rot inhibition: relative to Δagr mutant cells, cells expressing agr-I and -IV demonstrated the greatest temporal change in promoter activity, followed in order by agr-II and agr-III (Fig. 6A, C, E, and G). In the Newman background, this trend generally applied for spap and tstp reporter constructs (Fig. 6B and H), although tstp was only minimally induced in agr+ strains relative to the Δagr mutant (Fig. 6H). As expected, the hla promoter was much more weakly active in the Newman background compared to 8325-4, and interestingly, the activities with each agr allele (Fig. 6D) did not follow the trend observed with reporters for agrp3, Rot, and other virulence factor promoters. Finally, although the PVL promoter was very strongly activated by agr in 8325-4 strains, its activity was weak in Newman derivatives and either decreased or plateaued after a short period of induction (Fig. 6F); nonetheless, this range of activity depended on the specific agr allele, in agreement with the pattern of expression associated with the agrp3 and Rot reporters (Fig. 2B and 5B). These results are consistent with previous findings that PVL expression is dependent on agr and is affected by the host background (12, 74). Together these data demonstrate that allelic differences in quorum-sensing signal timing and intensity result in significant variations in the regulation of agr-dependent virulence genes.
Fig 6.
Effects of agr allele on virulence gene promoters. S. aureus cells of the 8325-4 (A, C, E, G) or Newman (B, D, F, H) background containing the indicated agr allele and transcriptional fusion of the spa (A, B), hla (C, D), lukSF-PV (E, F), or tst (G, H) promoters to the blaZ reporter were tested for β-lactamase activity. Assay data are presented as in Fig. 2.
DISCUSSION
In this study, we have probed the differences in quorum-sensing dynamics by the four S. aureus agr alleles expressed by congenic strains. We found significant intrinsic variation in the timing and magnitude of the signal generated from the principal agr promoter, P3, indicating that each of the divergent quorum-sensing alleles responds with a unique cell density set point. These signaling variations had consequences for the overall regulatory network and the virulon in S. aureus, as they resulted in altered expression of the agr-targeted global regulator, rot, and several important virulence factors. A hierarchical order in induction timing and strength can be established for the four S. aureus agr alleles based on these results, with agr-IV and -I the earliest and strongest, followed in order by agr-II and agr-III. Knowledge of these signaling differences will aid comparative analyses of virulence gene regulation between diverse S. aureus isolates.
The differences in agr signal generation among the alleles were not due to polymorphisms in the agr promoters themselves or RNAIII and instead relied on the agrBDCA genes, which contain the region of hypervariability. Aside from the four agr ORFs in this segment, we note that an uncharacterized weak promoter, P1, is also present, located between the 5′-most RsaI site in agrC and the PvuII site within agrA, based on a previous gene fusion study (60); this promoter is believed to function as a constitutive, basal source of agrA transcription. A computationally predicted promoter within a conserved region of agrC (see Fig. S1 in the supplemental material) may represent the P1 promoter, although this has yet to be tested experimentally. No polymorphism specific to agr-II or agr-III exists within this predicted promoter segment, so it is unlikely that this DNA region is responsible for the delayed kinetics observed with these two alleles. Because the agrA polymorphisms across the four alleles do not result in changes at the amino acid level (the AgrA polypeptide sequence is 100% conserved), the most likely conclusion is that differences in the activities and/or interactions among the hypervariable proteins AgrB, AgrD, and AgrC form the molecular basis of the observed allelic differences in agr signaling. Because addition of AIP at an early time point can bypass the delayed and weakened induction characteristic of agr-III cells (Fig. 2E), a logical conclusion is that the agr-III phenotype is owing at least in part to a reduced production of AIP (controlled presumably by allelic variations in the agrB or agrD genes) or possibly to its accelerated degradation. Biochemical quantitation of secreted AIP levels determined by each allele over time would address this prediction. If differences in secreted AIP levels are not apparent, a remaining potential source of altered induction properties among the alleles would be variability in cognate AIP-AgrC interaction kinetics. Such differences may be difficult to reveal, however, with the indirect reporter methods currently used (23, 24, 45, 76). Future studies employing purified and reconstituted AgrC will enable quantitative determination of allelic differences in ligand binding and receptor phosphorylation kinetics among cognate ligand-receptor pairs.
In general, different quorum-sensing dynamics resulted in analogous differences in the timing and level of virulence gene production, with interesting features dependent on the host background. With the 8325-4 strain containing the agr-III allele, overall exoproteins followed a temporal delay in expression compared to the congenic agr-IV strain, and the activities of the four analyzed virulence gene promoters varied according to the activity of the agr allele in each strain. In the Newman background, differences in overall exoproteins did not appear to reflect a temporal shift between the alleles analyzed, and allelic differences in transcription from the hla promoter did not correlate with agr activities. A caveat of the latter experiment is that the maximal agr+/Δagr mutant difference in hlap activity was much lower for the Newman background (approximately 3-fold) compared to the 8325-4 background (approximately 50-fold). In the case of the spa and lukSF-PV promoters in the Newman background, activities with the agr alleles displaying the most delayed kinetics, agr-III and agr-II, were virtually indistinguishable from that of the Δagr strain, while these two alleles behaved quite differently from the Δagr mutant in the 8325-4 background. The impact of agr allele-dependent differences in virulence factor expression thus depends significantly on the background genotype, which may affect expression of agr, virulence genes, or both. The 8325-4 and Newman strain chromosomes are known to differ in at least three areas that influence the virulon and/or pathogenesis: expression of σB (8), expression of Sae (2), and presence of prophages (5). Our findings that agr and certain of its upregulated targets are expressed at lower levels in Newman compared to 8325-4 are consistent with previous studies on σB, an agr downregulator whose activity is greatly reduced in 8325-4 (8, 30). The precise extent to which other strain-specific factors contribute to variations in agr expression remains to be determined. Nonetheless, these observations underscore the complex interactions of quorum sensing with the overall strain regulatory network.
We examined the variable expression of chromosomal and MGE-encoded virulence genes with the hypothesis that, if expression of horizontally acquired, agr-regulated virulence genes varies with the activity of the resident agr allele, certain alleles may determine an adaptive virulence phenotype due to a favorable combination of virulence and regulatory elements. With promoters of the lukSF-PV and tst genes, encoded by phage and SaPI, respectively, we indeed observed a range of activities that depended on the agr allele, and the activities and allele-dependent patterns of variability differed significantly between the two host backgrounds. In both backgrounds, expression was the weakest with agr-III, the allele that is most commonly represented in strains containing either the phage-encoded PVL toxin or the SaPI-encoded TSST-1 toxin. It is possible that temporal differences in expression of these toxins are beneficial to the organism in particular contexts, although this remains speculative. We note that our experiments involved reporter fusions to the toxin gene promoters in the absence of their native MGE, which may have affected the degree to which their expression varies with each agr allele.
While the data presented here demonstrate that divergent quorum-sensing alleles translate to variant regulation of virulence genes, a key area of future experimentation will involve examining the effects of the agr alleles on pathogenesis using animal models of infection. In addition, because the current model for S. aureus biofilm development involves localized, agr-mediated cellular dispersal from biofilm surfaces (11, 79), temporal differences in quorum-sensing activation may have profound effects on biofilm dynamics. Examining the congenic strains in host infection and biofilm models will enable further tests of the hypothesis that variant quorum-sensing dynamics may constitute a determinant of adaptive virulence phenotypes. Previous observations indicate that the 8325-4 and Newman strain backgrounds differ in their virulence phenotypes in the mouse sepsis model (29) and the mouse subcutaneous abscess model (J. Liese and R. P. Novick, unpublished data), as well as in biofilm formation (7, 29), so these strain backgrounds would again provide two distinct platforms with which to study differences in virulence and biofilm development associated with the agr alleles. Finally, although our focus was on virulence factors regulated primarily through the RNAIII-rot axis, an additional area of future experimentation will be to examine the effects of agr alleles on the subset of agr-dependent genes recently identified to be controlled independently of RNAIII, including the PSM family cytolysins (62).
This study indicates that altered virulon regulation can arise from concerted natural allelic variants in multiple genes, adding to previously established mechanisms of allelic variations in S. aureus regulators, including deletion mutants that result in premature protein termination (such as that in rsbU [8] and arlR [40]), and single-nucleotide polymorphisms resulting in missense substitutions (such as that in saeS [2]). Concerted allelic differences in the agr loci of other staphylococci and Gram-positive bacteria (20, 78), and in other polymorphic microbial autocrine systems (3, 73), may similarly provide the basis for altered regulatory properties that can affect pathogenicity, in addition to their widely accepted function as determinants of pherotype specificity. As this study demonstrates, the use of congenic strains combined with reporter gene fusions is a sensitive approach to quantitate variations in signaling dynamics among homologous autoinduction systems.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01-AI30138 (to R.P.N.).
We are grateful to Tom Muir and Elizabeth George Cisar for providing synthetic AIP, and we thank members of the Novick lab, especially Hope Ross, for helpful discussions.
Footnotes
Published ahead of print 30 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. 1993. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61:3879–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adhikari RP, Novick RP. 2008. Regulatory organization of the staphylococcal sae locus. Microbiology 154:949–959 [DOI] [PubMed] [Google Scholar]
- 3. Ansaldi M, Dubnau D. 2004. Diversifying selection at the Bacillus quorum-sensing locus and determinants of modification specificity during synthesis of the ComX pheromone. J. Bacteriol. 186:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Autret N, Raynaud C, Dubail I, Berche P, Charbit A. 2003. Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect. Immun. 71:4463–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bae T, Baba T, Hiramatsu K, Schneewind O. 2006. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 62:1035–1047 [DOI] [PubMed] [Google Scholar]
- 6. Bae T, et al. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. U. S. A. 101:12312–12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beenken KE, Blevins JS, Smeltzer MS. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bischoff M, Entenza JM, Giachino P. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blevins JS, Beenken KE, Elasri MO, Hurlburt BK, Smeltzer MS. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boisset S, et al. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21:1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bronner S, Stoessel P, Gravet A, Monteil H, Prevost G. 2000. Variable expressions of Staphylococcus aureus bicomponent leucotoxins semiquantified by competitive reverse transcription-PCR. Appl. Environ. Microbiol. 66:3931–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carroll RK, et al. 2011. Naturally occurring single amino acid replacements in a regulatory protein alter streptococcal gene expression and virulence in mice. J. Clin. Invest. 121:1956–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chai Y, Norman T, Kolter R, Losick R. 2011. Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis. EMBO J. 30:1402–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan PF, Foster SJ. 1998. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 144:2469–2479 [DOI] [PubMed] [Google Scholar]
- 16. Charpentier E, et al. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 70:6076–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheung AL, et al. 1994. Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Invest. 94:1815–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheung GY, Duong AC, Otto M. 2011. Direct and synergistic hemolysis caused by Staphylococcus phenol-soluble modulins: implications for diagnosis and pathogenesis. Microbes Infect. 14:380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cooksley CM, et al. 2010. Regulation of neurotoxin production and sporulation by a putative agrBD signaling system in proteolytic Clostridium botulinum. Appl. Environ. Microbiol. 76:4448–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dufour P, et al. 2002. High genetic variability of the agr locus in Staphylococcus species. J. Bacteriol. 184:1180–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duthie ES, Lorenz LL. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95–107 [DOI] [PubMed] [Google Scholar]
- 22. Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. 2006. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 61:1038–1048 [DOI] [PubMed] [Google Scholar]
- 23. Geisinger E, George EA, Muir TW, Novick RP. 2008. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J. Biol. Chem. 283:8930–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Geisinger E, Muir TW, Novick RP. 2009. agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc. Natl. Acad. Sci. U. S. A. 106:1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gillaspy AF, et al. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gillet Y, et al. 2002. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753–759 [DOI] [PubMed] [Google Scholar]
- 27. Goerke C, Fluckiger U, Steinhuber A, Zimmerli W, Wolz C. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439–1447 [DOI] [PubMed] [Google Scholar]
- 28. Heinrichs JH, Bayer MG, Cheung AL. 1996. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 178:418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herbert S, et al. 2010. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 78:2877–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horsburgh MJ, et al. 2002. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huntzinger E, et al. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24:824–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ishihama A. 1997. Adaptation of gene expression in stationary phase bacteria. Curr. Opin. Genet. Dev. 7:582–588 [DOI] [PubMed] [Google Scholar]
- 33. Jarraud S, et al. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517–6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jarraud S, et al. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ji G, Beavis R, Novick RP. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030 [DOI] [PubMed] [Google Scholar]
- 36. Ji G, Beavis RC, Novick RP. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. U. S. A. 92:12055–12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ji G, et al. 2005. Staphylococcus intermedius produces a functional agr autoinducing peptide containing a cyclic lactone. J. Bacteriol. 187:3139–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kolter R, Siegele DA, Tormo A. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855–874 [DOI] [PubMed] [Google Scholar]
- 39. Kreiswirth BN, Novick RP, Schlievert PM, Bergdoll M. 1982. Genetic studies on Staphylococcal strains from patients with toxic shock syndrome. Ann. Intern. Med. 96:974–977 [DOI] [PubMed] [Google Scholar]
- 40. Kuroda M, et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240 [DOI] [PubMed] [Google Scholar]
- 41. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 42. Lee CY, Iandolo JJ. 1988. Structural analysis of staphylococcal bacteriophage phi 11 attachment sites. J. Bacteriol. 170:2409–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee LY, et al. 2004. Inhibition of complement activation by a secreted Staphylococcus aureus protein. J. Infect. Dis. 190:571–579 [DOI] [PubMed] [Google Scholar]
- 44. Luong TT, Lee CY. 2007. Improved single-copy integration vectors for Staphylococcus aureus. J. Microbiol. Methods 70:186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lyon GJ, Wright JS, Christopoulos A, Novick RP, Muir TW. 2002. Reversible and specific extracellular antagonism of receptor-histidine kinase signaling. J. Biol. Chem. 277:6247–6253 [DOI] [PubMed] [Google Scholar]
- 46. Majerczyk CD, et al. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190:2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Manna AC, Cheung AL. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Manna AC, Cheung AL. 2006. Transcriptional regulation of the agr locus and the identification of DNA binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol. Microbiol. 60:1289–1301 [DOI] [PubMed] [Google Scholar]
- 49. Massidda O, Rossolini GM, Satta G. 1991. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-beta-lactamases. J. Bacteriol. 173:4611–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McDowell P, et al. 2001. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol. Microbiol. 41:503–512 [DOI] [PubMed] [Google Scholar]
- 51. Nakayama J, et al. 2006. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal agrd. J. Bacteriol. 188:8321–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Novick R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155–166 [DOI] [PubMed] [Google Scholar]
- 53. Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449 [DOI] [PubMed] [Google Scholar]
- 54. Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541–564 [DOI] [PubMed] [Google Scholar]
- 55. Novick RP, Kornblum J, Kreiswirth B, Projan S, Ross H. 1989. agr: a complex locus regulating post-exponential phase exoprotein synthesis in Staphylococcus aureus, p 495–510 In Butler LO, Moseley BEB. (ed), Genetic transformation and expression. Intercept, Ltd., Andover, United Kingdom [Google Scholar]
- 56. Novick RP, et al. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Olsen RJ, et al. 2010. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc. Natl. Acad. Sci. U. S. A. 107:888–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Otto M, Echner H, Voelter W, Gotz F. 2001. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:1957–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Otto M, Sussmuth R, Vuong C, Jung G, Gotz F. 1999. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 450:257–262 [DOI] [PubMed] [Google Scholar]
- 60. Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pishchany G, et al. 2010. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe 8:544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Queck SY, et al. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rossolini GM, et al. 1999. Cloning of a Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaACME) encoding an extended-spectrum class A β-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER β-lactamases. Antimicrob. Agents Chemother. 43:2193–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ruzin A, Lindsay J, Novick RP. 2001. Molecular genetics of SaPI1—a mobile pathogenicity island in Staphylococcus aureus. Mol. Microbiol. 41:365–377 [DOI] [PubMed] [Google Scholar]
- 65. Saïd-Salim B, et al. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shinefield HR, Ribble JC, Boris M, Eichenwald HF. 1963. Bacterial interference: its effect on nursery-acquired infection with Staphylococcus aureus. I. Preliminary observations on artificial colonization of newborns. Am. J. Dis. Child 105:646–654 [PubMed] [Google Scholar]
- 67. Sturme MH, et al. 2005. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 187:5224–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tamber S, et al. 2010. The staphylococcus-specific gene rsr represses agr and virulence in Staphylococcus aureus. Infect. Immun. 78:4384–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Traber KE, et al. 2008. agr function in clinical Staphylococcus aureus isolates. Microbiology 154:2265–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tristan A, et al. 2007. Global distribution of Panton-Valentine leukocidin–positive methicillin-resistant Staphylococcus aureus, 2006. Emerg. Infect. Dis. 13:594–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Villaruz AE, et al. 2009. A point mutation in the agr locus rather than expression of the Panton-Valentine leukocidin caused previously reported phenotypes in Staphylococcus aureus pneumonia and gene regulation. J. Infect. Dis. 200:724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang R, et al. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13:1510–1514 [DOI] [PubMed] [Google Scholar]
- 73. Whatmore AM, Barcus VA, Dowson CG. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181:3144–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wirtz C, Witte W, Wolz C, Goerke C. 2009. Transcription of the phage-encoded Panton-Valentine leukocidin of Staphylococcus aureus is dependent on the phage life-cycle and on the host background. Microbiology 155:3491–3499 [DOI] [PubMed] [Google Scholar]
- 75. Wright JS, III, Jin R, Novick RP. 2005. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. U. S. A. 102:1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wright JS, III, Lyon GJ, George EA, Muir TW, Novick RP. 2004. Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 101:16168–16173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wright JS, III, et al. 2005. The agr radiation: an early event in the evolution of staphylococci. J. Bacteriol. 187:5585–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wuster A, Babu MM. 2008. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J. Bacteriol. 190:743–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yarwood JM, McCormick JK, Schlievert PM. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yoshikawa H, Sueoka N. 1963. Sequential replication of Bacillus subtilis chromosome. I. Comparison of marker frequencies in exponential and stationary growth phases. Proc. Natl. Acad. Sci. U. S. A. 49:559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.