Abstract
MtfA of Escherichia coli (formerly YeeI) was previously identified as a regulator of the phosphoenolpyruvate (PEP)-dependent:glucose phosphotransferase system. MtfA homolog proteins are highly conserved, especially among beta- and gammaproteobacteria. We determined the crystal structures of the full-length MtfA apoenzyme from Klebsiella pneumoniae and its complex with zinc (holoenzyme) at 2.2 and 1.95 Å, respectively. MtfA contains a conserved H149E150XXH153+E212+Y205 metallopeptidase motif. The presence of zinc in the active site induces significant conformational changes in the region around Tyr205 compared to the conformation of the apoenzyme. Additionally, the zinc-bound MtfA structure is in a self-inhibitory conformation where a region that was disordered in the unliganded structure is now observed in the active site and a nonproductive state of the enzyme is formed. MtfA is related to the catalytic domain of the anthrax lethal factor and the Mop protein involved in the virulence of Vibrio cholerae, with conservation in both overall structure and in the residues around the active site. These results clearly provide support for MtfA as a prototypical zinc metallopeptidase (gluzincin clan).
INTRODUCTION
The phosphoenolpyruvate (PEP):carbohydrate phosphotransferase system (PTS) is utilized by many bacteria for the transport and concomitant phosphorylation of carbohydrates from the environment (43). In general, such a system consists of three different components: the two general cytoplasmic enzymes, enzyme I (EI) and histidine-phosphotransfer protein (HPr), and one of at least 20 different carbohydrate-specific, membrane-bound, transport protein enzymes II (EII), which usually include three functional subunits corresponding to individual domains or proteins—EIIA, EIIB, and the membrane-spanning domain, EIIC. In the case of the glucose-specific PTS, the phosphorylation cascade starts with autophosphorylation of the protein kinase EI using PEP as a phosphoryl group donor. The phosphoryl group is then shuttled from EI to HPr, subsequently to EIIAGlc, and finally, to EIICBGlc, which mediates the uptake and phosphorylation of glucose (see Fig. S1 in the supplemental material).
In nutrient-rich environments, bacteria generally have a preferred carbon source, such as d-glucose. Two complex regulatory phenomena, known as carbon catabolite repression (CCR) and inducer exclusion, enable enteric bacteria to take advantage of a preferred carbon source by suppressing the expression and activity of the catabolic systems that facilitate metabolism of the secondary carbon sources (13, 14, 21). Both regulatory mechanisms are controlled by the glucose PTS via several complex reactions, including the control of the global transcription repressor Mlc (makes large colonies) (33). In Escherichia coli, Mlc represses the expression of several genes for carbohydrate transport and utilization, including the ptsG gene that encodes EIICBGlc. In contrast to the activities of other repressors, Mlc's activity is not modulated by direct binding of glucose or any other small-molecule inducer. Instead, in a glucose-rich environment, EIIBGlc becomes dephosphorylated by transferring the phosphoryl group to the incoming glucose. Mlc interacts with the membrane-bound, dephosphorylated EIIBGlc and becomes sequestered from the promoters (inactivation), allowing increased expression of glucose-utilizing genes (50, 54). Recently, a new mechanism of Mlc inactivation was identified during a search for new factors that influence ptsG expression (4). A YeeI mutant was shown to exhibit slower growth on glucose and decreased expression of ptsG. The corresponding E. coli protein YeeI (subsequently renamed MtfA, for Mlc titration factor A) activates ptsG gene expression through an interaction with Mlc (4). The mechanism of MtfA-dependent Mlc inactivation is not currently well understood.
The mtfA gene in E. coli and Klebsiella pneumoniae is located between two tRNA genes (serU and asnT) in their respective genomes. The 3′ end of asnT is a conserved integration site for a well-characterized high-pathogenicity island (HPI) (49), which was initially described in pathogenic Yersinia (6) and later identified in other Enterobacteriaceae, such as E. coli (48) and K. pneumoniae (31). However, MtfA is probably not part of this HPI as it is also present in the genomes of nonpathogenic strains where this HPI is absent. MtfA is widespread among various bacterial species, forming a protein family of more than 600 unique family members (Pfam PF06167). MtfA-like proteins are distributed predominately in proteobacteria (∼89%), especially in the beta and gamma classes. They are also present in bacteroidetes, cyanobacteria, and planctomycetes (∼11%). One remote eukaryotic homolog is present in Nematostella vectensis (starlet sea anemone), a primitive animal. Interestingly, MtfA contains an HEXXH zinc metallopeptidase sequence motif that is highly conserved across the MtfA protein family.
Here, we report the structural studies of an MtfA ortholog from Klebsiella pneumoniae, a human pathogen involved in urinary tract infections, nosocomial pneumonia, and intra-abdominal infections. MtfA of K. pneumoniae shares 76% sequence identity with E. coli MtfA and encodes a protein of 266 residues with a calculated molecular weight of 30.3 kDa and an isoelectric point of 4.8. We determined the crystal structure of MtfA as the apoenzyme and in complex with zinc (holoenzyme). We show here that MtfA is a zinc-dependent metallopeptidase. Despite limited overall sequence similarity, its structure displays significant similarity to the catalytic domain of the anthrax lethal factor (LF) both in its overall folding and the arrangement of the active site and surrounding residues. The structure also helps to establish an evolutionary link between LF and the Mop protein that modulates the pathogenesis and reactogenicity of epidemic Vibrio cholerae (64), thus providing new insights into the evolution of bacterial toxins.
MATERIALS AND METHODS
Cloning and protein purification.
Clones were generated using the polymerase incomplete primer extension (PIPE) cloning method (30). The gene encoding MtfA (gi:152970975, Uniprot identifier MTFA_KLEP7) was amplified by PCR from K. pneumoniae MGH 78578 genomic DNA using PfuTurbo DNA polymerase (Stratagene) and I-PIPE (insert) primers that included sequences for the predicted 5′ and 3′ ends. The expression vector, pSpeedET, which encodes an amino-terminal tobacco etch virus (TEV) protease-cleavable expression and purification tag (MGSDKIHHHHHHENLYFQG), was PCR amplified with V-PIPE (vector) primers. V-PIPE and I-PIPE PCR products were mixed to anneal the amplified DNA fragments together. Escherichia coli GeneHogs (Invitrogen) competent cells were transformed with the V-PIPE–I-PIPE mixture and dispensed onto selective LB agar plates. The cloning junctions were confirmed by DNA sequencing. Expression was performed in a selenomethionine-containing medium, and selenomethionine was incorporated via inhibition of methionine biosynthesis (59), which does not require a methionine auxotrophic strain. At the end of fermentation, lysozyme was added to the culture to a final concentration of 250 μg/ml, and the cells were harvested and frozen. After one freeze/thaw cycle, the cells were homogenized in lysis buffer [50 mM HEPES, pH 8.0, 50 mM NaCl, 10 mM imidazole, 1 mM Tris(2-carboxyethyl)phosphine-HCl (TCEP)] and the lysate was clarified by centrifugation at 32,500 × g for 30 min. The soluble fraction was passed over nickel-chelating resin (GE Healthcare) preequilibrated with lysis buffer, the resin washed with wash buffer (50 mM HEPES, pH 8.0, 300 mM NaCl, 40 mM imidazole, 10% [vol/vol] glycerol, 1 mM TCEP), and the protein eluted with elution buffer (20 mM HEPES, pH 8.0, 300 mM imidazole, 10% [vol/vol] glycerol, 1 mM TCEP). The eluate was buffer exchanged with HEPES crystallization buffer (20 mM HEPES, pH 8.0, 200 mM NaCl, 40 mM imidazole, 1 mM TCEP) using a PD-10 column (GE Healthcare) and incubated with 1 mg of TEV protease per 15 mg of eluted protein. The protease-treated eluate was passed over nickel-chelating resin (GE Healthcare) preequilibrated with HEPES crystallization buffer, and the resin was washed with the same buffer. The flowthrough and wash fractions were combined and concentrated for crystallization trials to 18 mg/ml by centrifugal ultrafiltration (Millipore). The molecular weight and oligomeric state of MtfA were determined using a 0.8- by 30-cm Shodex protein KW-803 column (Thomson Instruments) precalibrated with gel filtration standards (Bio-Rad).
Crystallization and diffraction screening of MtfA.
MtfA from K. pneumoniae was crystallized using the nanodroplet vapor diffusion method (45) with standard Joint Center for Structural Genomics (JCSG) crystallization protocols (15, 34). The crystallization reagent that produced the MtfA crystal at 4°C that was used for structure determination contained 1.0 M (NH4)2HPO4, 0.2 M NaCl, and 0.1 M imidazole, pH 8.0. Glycerol was added to the crystal as a cryoprotectant to a final concentration of 20% (vol/vol). The crystallization reagent that produced the MtfA-Zn crystal at 20°C for structure determination consisted of 10.0% polyethylene glycol 6000, 1.0 M lithium chloride, 0.1 M bicine, pH 9.0, 1 mM Leu-CMK (chloromethyl ketone), and 1 mM ZnCl2. The cryoprotectant was 15% (vol/vol) ethylene glycol.
MtfA could easily be crystallized, and crystals were harvested from ∼50 conditions. However, most crystals did not diffract very well. To identify the crystals with the best possible diffraction, we screened all harvestable crystals using the Stanford Automated Mounting system (SAM) (7) at the Stanford Synchrotron Radiation Lightsource (SSRL, Menlo Park, CA). A total of 230 crystals were screened. The space group for both crystals used for data collection was P41212 with similar cell dimensions (Table 1).
Table 1.
Data collection, phasing, and refinement statistics for MtfA and MtfA-Zn
| Parametera | Value(s) for indicated data set |
|||
|---|---|---|---|---|
| MtfA Se peak | MtfA-Zn |
|||
| Se peak | Zn remote | Zn peak | ||
| Beamline | SSRL 11-1 | SSRL 9-2 | SSRL 9-2 | SSRL 9-2 |
| Wavelength (Å) | 0.97874 | 0.97951 | 1.28744 | 1.27946 |
| Space group | P41212 | P41212 | ||
| Unit cell parameters | a = 131.1 Å, c = 37.1 Å | a = 131.2 Å, c = 37.1 Å | ||
| Resolution range (Å) | 29.3–2.20 | 46.4–1.95 | 36.4–2.19 | 43.7–2.18 |
| No. of observations | 121,682 | 172,542 | 78,650 | 79,786 |
| No. of unique reflections | 17,061 | 24,283 | 17,228 | 17,455 |
| Completeness (%) | 99.9 (99.9)b | 99.9 (99.8) | 99.8 (99.3) | 99.7 (99.5) |
| Mean I/σ (I) | 16.9 (3.5)b | 14.5 (2.2) | 16.0 (2.5) | 16.9 (2.8) |
| Rmerge on I (%) | 8.1 (54.9)b | 7.1 (93.1) | 6.1 (58.6) | 6.0 (52.0) |
| Rmeas on I (%) | 8.8 (59.1)b | 7.7 (100.0) | 6.9 (67.7) | 6.7 (60.0) |
| Rpim on I (%) | 3.3 (21.8)b | 2.9 (37.1) | 3.2 (33.2) | 3.1 (29.3) |
| Highest resolution shell (Å) | 2.32–2.20 | 2.06–1.95 | 2.31–2.19 | 2.3–2.18 |
| Phasing | ||||
| Resolution range (Å) | 29.3–2.20 | 46.4–1.95 | ||
| No. of heavy atom sites | 4 | 5 | ||
| Figure of merit/phasing power | 0.28/1.1 | 0.33/1.5 | ||
| Refinement | ||||
| PDB code | 3dl1 | 3khi | ||
| Resolution range (Å) | 29.3-2.20 | 46.4-1.95 | ||
| Cutoff criteria | |F| > 0 | |F| > 0 | ||
| No. of reflections (total) | 16,997 | 24,240 | ||
| No. of reflections (test) | 861 | 1,244 | ||
| Completeness (% total) | 99.7 | 99.9 | ||
| Rcryst/Rfree (%) | 17.1/20.2 | 18.3/21.2 | ||
| Restraints (RMSD observed) | ||||
| Bond angle (°) | 1.38 | 1.28 | ||
| Bond length (Å) | 0.016 | 0.014 | ||
| Average isotropic B value (Å2) | 48.1 (43.8)c | 42.5 (36.6) | ||
| Molprobity | ||||
| Ramachandran favored (outliers, %) | 98.5 (0.0) | 98.6 (0.0) | ||
| All-atom clashscore | 3.1 | 5.8 | ||
| Poor rotamers (%) | 0.6 | 2.3 | ||
| ESU based on Rfree (Å) | 0.15 | 0.12 | ||
| Protein residues/atoms | 210/1,680 | 223/1,785 | ||
| Water molecules/ions | 97/1 | 123/3 | ||
Rmerge is ΣhklΣi|Ii(hkl) − <I(hkl)>|/ΣhklΣiIi(hkl); Rmeas (redundancy-independent Rmerge) is Σhkl[Nhkl/(Nhkl − 1)]1/2Σi|Ii(hkl) − <I(hkl)>|/ΣhklΣiIi(hkl); Rpim (precision-indicating Rmerge) is Σhkl[1/(Nhkl − 1)]1/2Σi|Ii(hkl) − <I(hkl)>|/ΣhklΣiIi(hkl); ESU is estimated overall coordinate error based on Rfree; Rcryst is Σ‖Fobs|-|Fcalc‖/Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively; Rfree is as for Rcryst but for 5.0% of the total reflections chosen at random and omitted from refinement.
Highest resolution shell. The high-resolution cutoff was chosen such that the mean I/σ(I) in the highest resolution shell is around 2. These statistics were calculated assuming the equivalence of Friedel pairs.
Wilson B value.
Data collection, structure solution, and refinement.
Single-wavelength anomalous diffraction (SAD) data were collected at the SSRL beamlines 11-1 and 9-2 for the apoenzyme and the holoenzyme, respectively. The data were collected at wavelengths corresponding to the peak energy of a selenium multiwavelength anomalous diffraction (MAD) experiment. The data sets were collected at 100 K using a MarCCD 325 detector (Rayonix, United States). The data processing and structure solution were carried out using an automatic structure solution pipeline developed at the JCSG (58). The MtfA data were integrated using MOSFLM (35) and scaled with SCALA (17). The holoenzyme data were integrated using XDS and then scaled with XSCALE (26). The structures of the apoenzyme and the holoenzyme were solved independently using the selenium SAD method. Selenium sites were located with SHELXD (51). Phase refinement and automatic model building were performed using autoSHARP (61) and wARP (8) or RESOLVE (55). Model completion and refinement were performed with COOT (16) and REFMAC (62). The protein molecule was refined as a single TLS group. Additional CCP4 programs were used for data conversion and other calculations (10). Data reduction and refinement statistics are summarized in Table 1.
Identification of zinc in the active site by fluorescence microscopy and anomalous difference Fourier analysis.
To confirm the identity of the metal in the active site as zinc, Zn-MAD data were collected at SSRL beamline 9-2. XAFS measurements were carried out around the zinc K absorption edge using the same holoenzyme crystal as used for X-ray data collection in a cold nitrogen gas stream at 100 K. Subsequently, two data sets were collected above (λ = 1.2795 Å, “peak”) and below (λ = 1.2874 Å, “low energy remote”) the zinc absorption edge. Data processing was performed in a manner similar to the method described above for the holoenzyme (Table 1). Anomalous difference maps were calculated using the anomalous difference amplitudes and Se-SAD experimental phases (after density modification) from the same crystal.
Refinement of the holoenzyme.
It was clear from the experimental map that zinc is coordinated by four residues; the first three (His149, His153, and Glu212) are from the conserved zinc binding motif (HEXXH+E) as expected, and the fourth ligand is located on a helix in a disordered region, with a reasonable quality of electron density only for the main chain. Mass spectrometry and SDS-PAGE analyses confirmed that the protein used for crystallization is the full-length MtfA construct and is not contaminated by other proteins; thus, this helical fragment belongs to MtfA. Its spatial location suggests that the helix most likely corresponds to the disordered regions between residues 99 and 123 (99IVDDEWEDDIGLVHNQRVVQSGQSW123). Since both ends of this helix and most of its side chains are disordered, it is difficult to unambiguously assign the identity of this helical fragment. However, based on electron density and the interaction environment, we were able to derive a possible motif for side chains of residues near the zinc. This motif (E/Q/H)-X3-(V/T/I) (starting from the first residue that coordinates with the zinc, where X indicates nonglycine/nontryptophan residues) can correspond only with 112HNQRV116 within the 99 to 123 region. The helical fragment can be uniquely assigned to the closest MtfA molecule in the crystal lattice based on end-to-end distance analysis (i.e., the distance between two open ends must be within the reach of the disordered peptide region). The unmodeled density near Gly109 and His112 could not be unambiguously assigned.
The atomic coordinates of apoenzyme (MtfA) and holoenzyme (MtfA-Zn) have been deposited in the RCSB Protein Data Bank (http://www.rcsb.org/) with the entry codes 3dl1 and 3khi, respectively. All molecular graphics were prepared with PyMOL (Schrödinger; http://www.pymol.org). Figure 4A was generated with WEBLOGO (11) using a sequence alignment of the top 100 BLAST hits (2). Sequence alignments were calculated with CLUSTAL W (32). Electrostatic potentials were calculated using APBS (3). The secondary structure assignment was made with DSSP (27). The secondary structure prediction was performed using the JNET server (9).
Fig 4.
Active site and self-inhibitory complex of MtfA. (A) Consensus sequences of three highly conserved regions near the active site (β2-to-β3 region, α5 and α7) in a sequence logo representation, where the overall height of the stack indicates the sequence conservation and the height of the symbols indicates the relative frequency of each amino acid at that position. The multiple sequence alignment was generated using the top 100 unique hits of the E. coli MtfA of a BLAST search. (B) Mapping of sequence conservation onto the MtfA structure. The most conserved residues are shown in purple, the least conserved residues in cyan. A β-strand corresponding to the edge strand (yellow) was transferred from the anthrax lethal factor (LF) structure (PDB ID 1j7n) to show a potential conformation for part of the disordered 99 to 123 segment. (C) Electrostatic potentials of MtfA. The color is scaled from −10 to 10 kT/e (blue, positive electrostatic potential; red, negative electrostatic potential). (D) Interaction between the self-inhibitory peptide (residues 109 to 118) and MtfA-Zn. The modeled edge strand in panel B is shown as a stick/sphere representation.
Peptidase assay.
For peptidase assays, MtfA was cloned and purified based on a protocol described previously (20) that is independent from the protocol used for the crystallographic studies. Purified MtfA-His5 and derivatives were expressed from the plasmid pTM30MtfAkpnHis. They were generated by amplifying the gene or mutants encoding mtfA of the K. pneumoniae strain KAY 2026 (the MtfA gene product is identical to that of the K. pneumoniae strain MGH 78578 used in the crystallographic studies). PCR was carried out using TaKaRa Taq DNA polymerase (Stratagene) and primers MtfAkpnEcoRV+ and MtfAkpnKpnI. The PCR product was ligated into the expression vector pTM30 (40) using restriction enzymes EcoRV and KpnI (New England BioLabs). The expression vector was confirmed by DNA sequencing. Mutations of the mtfA gene were generated using the vector described above as a template for the QuikChange II site-directed mutagenesis kit (Stratagene). The oligonucleotides are listed in Table S1 in the supplemental material.
For purification of MtfA-His5 and derivatives thereof, E. coli cells harboring pTM30MtfAkpnHis or derivatives with relevant mutations were grown in LB medium with ampicillin and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). When cultures reached an optical density of 3 at 650 nm, cells were harvested by centrifugation. Cells were washed and resuspended in buffer A (300 mM NaCl, 2 mM β-mercaptoethanol, 100 mM KPi, and 10% glycerol, pH 8.0) and disrupted by sonication using a Branson W-250D sonifier (Branson, Danbury, CT). Cell debris was removed by low-speed centrifugation (14,000 rpm for 20 min at 4°C). Affinity chromatography was done with an ÄKTA-FPLC (GE Healthcare, Munich, Germany) using a His-Trap FF column (GE Healthcare). Elution was performed with buffer A supplemented with 300 mM imidazole. Subsequently, the sample obtained was dialyzed against 20 mM Tris buffer, pH 8.0. The concentrations of purified proteins were determined by the Bradford method, and samples were stored at 4°C. Purified samples were analyzed by mass spectrometry, which indicated that MtfA was the sole protein in the preparations.
The aminopeptidase activity of MtfA was determined by measuring the hydrolysis of different l-amino 4-nitroanilide substrates. l-Alanine 4-nitroanilide, l-arginine 4-nitroanilide, l-glutamic acid 4-nitroanilide, l-proline 4-nitroanilide, l-valine 4-nitroanilide, or l-alanine–l-alanine–l-alanine 4-nitroanilide (Sigma, Taufkirchen, Germany) were tested as substrates. Measurement of the chromophoric end product 4-nitroanilide was performed by spectral photometric determination of the absorbance at 405 nm (Shimadzu UV mini 1240 photometer). The assay was carried out in semimicrocuvettes (Sarstedt, Nümbrecht, Germany). To 500 μl of 10 mM l-amino acid 4-nitroanilide, 10 to 30 μg of purified protein was added. The sample was supplemented with 20 mM Tris buffer (pH 8) to a final volume of 1 ml and incubated at 37°C for 4 h. No zinc was added in the enzymatic assays as it precipitated the protein, probably due to differences in buffer conditions or other factors compared to the protein used in crystallization trials. The absorbance was measured against a blank sample without protein (500 μl l-amino 4-nitroanilide and 500 μl Tris buffer) at 405 nm. The extinction difference (ΔE) was determined. A similar assay testing carboxypeptidase activity was also performed. The release of hippuric acid from hippuryl–l-phenylalanine and hippuryl–l-arginine by MtfA was monitored by measuring the optical density at 254 nm (OD254).
The following protease inhibitors were subsequently tested in the assay: 1,10-phenanthroline monohydrate, EDTA, E64, pepstatin A, and iodoacetamide (Sigma-Aldrich, Taufkirchen, Germany) and AEBSF [4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride; AppliChem, Darmstadt, Germany]. Additionally, a protease assay (protease assay kit; Calbiochem, Merck, Darmstadt, Germany) with fluorescein thiocarbamoyl (FTC)-casein as a cleavable substrate was performed against the MtfA wild-type protein following the manufacturer's instructions.
Gene cooccurrence analysis.
Amino acid sequences from 951 bacteria with completed genomes were downloaded from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria). BLAST (1) was run to search for homologs of MtfA (265 amino acids [aa]), Mlc (406 aa), and EIICBGlc (477 aa) of E. coli in each bacterium. The resulting log files were parsed using BioPERL (http://www.bioperl.org). A significant homolog was considered present in the genome if it met all of the following criteria: (i) E <1.0e−6 and (ii) sequence identity >25% with alignment length greater than 150 aa (MtfA), 250 aa (Mlc), and 300 aa (EIICBGlc). For Mlc, a relaxed condition of E <0.01 and alignment length greater than 150 aa with sequence identity >20% was also used.
RESULTS
Structure determination.
The crystal structures of the MtfA apoenzyme and the holoenzyme with zinc (MtfA-Zn) were determined using the semiautomated, high-throughput pipeline of the Joint Center for Structural Genomics (JCSG; http://www.jcsg.org) (15, 34) as part of the National Institute of General Medical Sciences' Protein Structure Initiative (PSI; http://www.nigms.nih.gov/Research/FeaturedPrograms/PSI/). The selenomethionine derivative of the full-length MtfA from K. pneumoniae was expressed in E. coli with an N-terminal TEV protease-cleavable His tag and purified by metal affinity chromatography (see Materials and Methods). The crystal structure of the MtfA apoenzyme was determined in space group P41212 at 2.2-Å resolution using the SAD method. The final model of MtfA contains one monomer (residues 17 to 253), 97 water molecules, and 1 chloride ion (PDB ID code 3dl1). Residues 1 to 16, 99 to 123, 159 to 160, and 254 to 266 of the apoenzyme were omitted due to the lack of interpretable electron density. The structure was refined to an Rcryst of 17.1% and Rfree of 20.2%, and the model displays good geometry with an all-atom clash score of 3.06, 98.5% residues in favorable regions of the Ramachandran plot (no outliers), and 99.4% favorable side-chain rotamers according to MolProbity (12).
The holoenzyme crystal was obtained with different crystallization reagents but resulted in the same space group and cell dimensions as the apo crystal. The structure was determined and refined independently at 1.95-Å resolution using the selenium SAD data (Rcryst/Rfree = 18.3%/21.2%). The final model of the holoenzyme contains residues 16 to 253 (except for two gaps at residues 99 to 108 and 119 to 123), one zinc ion, two chloride ions, five ethylene glycols, and 123 water molecules (PDB ID code 3khi). The model quality is similar to that of the MtfA apo structure. Data collection, phasing, refinement, and model statistics for both structures are summarized in Table 1.
MtfA binds zinc.
Although our initial MtfA apoenzyme structure did not contain zinc, it confirmed the existence of a conserved zinc binding motif, 149HEXXH153, that is common in metallopeptidases, indicating that MtfA would very likely bind zinc. As a result, we cocrystallized MtfA, as indicated below, in the presence of 1 mM ZnCl2 and obtained the structure of the MtfA holoenzyme.
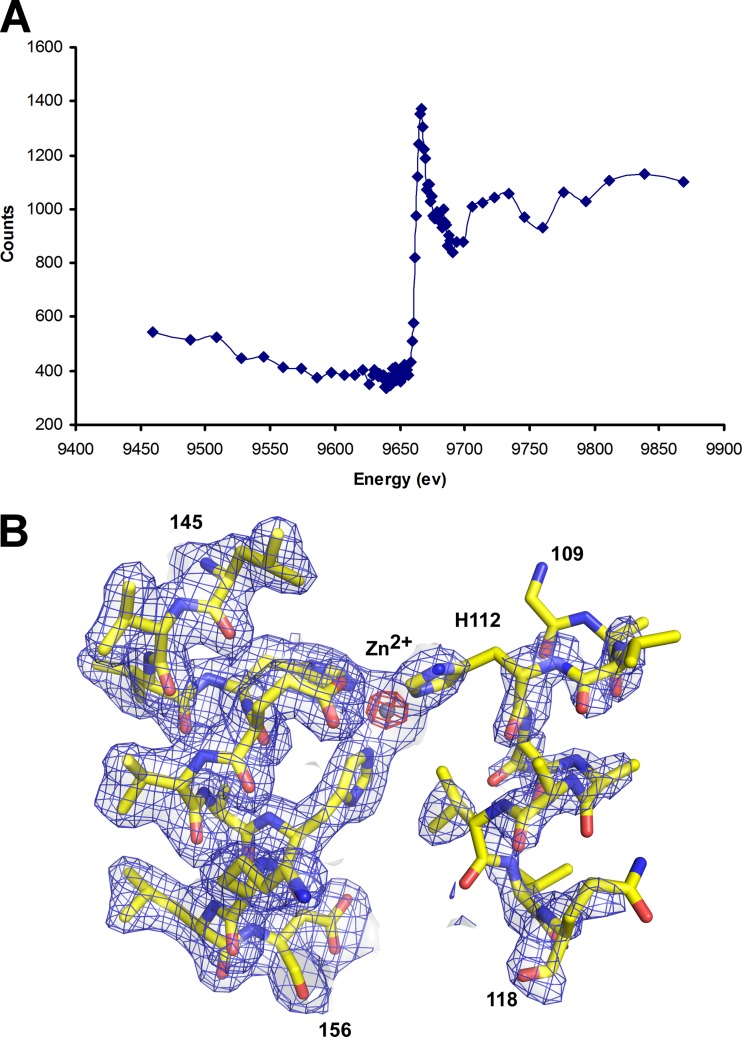
A fluorescence excitation scan (not shown) indicated that zinc was present in the holoenzyme crystal, which was then confirmed by X-ray fluorescence measurements (Fig. 1A). Small traces of other metals (Cu and Ni) detected in the excitation scan were probably due to contaminants from the Cu crystal mounting pin (Hampton) and the Ni affinity column used for protein purification. To further confirm that the metal observed in the active site was a zinc ion (Fig. 1B), we collected a MAD data set at wavelengths corresponding to the low-energy remote (9,630 eV) and peak (9,690 eV) of the zinc K absorption edge using the same crystal used for structure determination (Table 1). The anomalous difference map of the peak data (f″ 160, ∼3.4) indicated that the metal at the active site was the only significant peak, with a height of 30 σ, compared to Se peaks (∼5 σ; theoretical f″ 160, ∼0.8). The peak heights for the same metal from the low-energy remote (f″ 160, ∼0.8) data were 4.8 σ, similar to those of Se (∼5.0 σ; theoretical f″ 160, ∼0.8). These results support the presence of zinc in the crystal structure, thus confirming that MtfA is capable of chelating zinc in the active site.
Fig 1.
MtfA binds zinc. (A) X-ray energy scan of the fluorescence emitted by the sample near the zinc absorption K edge (9,690 eV). (B) Electron density map near the zinc. The experimental map (after density modification) is shown in blue (1.0 σ), and the anomalous difference map calculated above the zinc edge is in red (20.0 σ). The refined model is shown as sticks, and the zinc as a sphere.
Overall structural description.
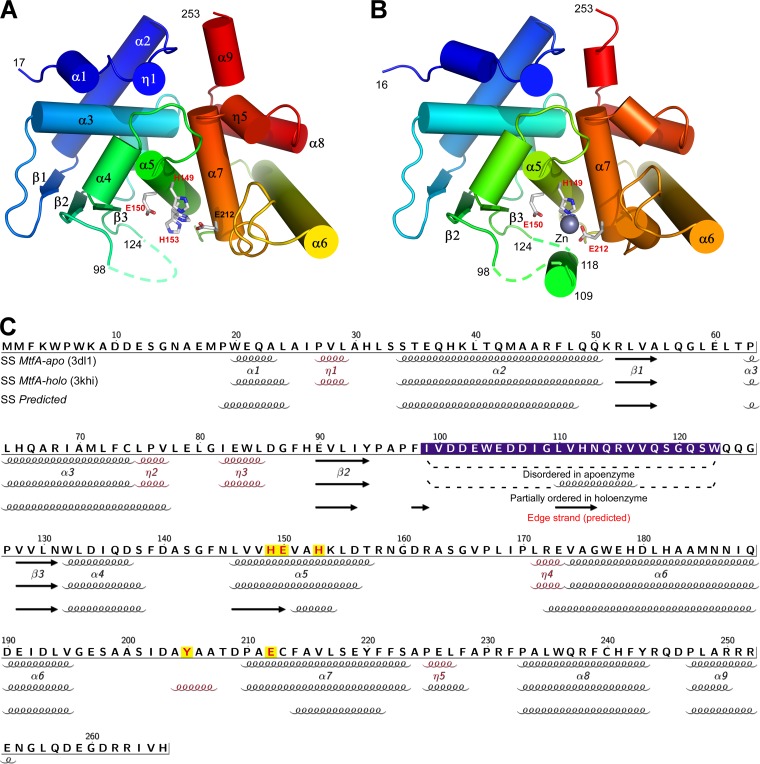
The crystal packing suggests that MtfA is likely to exist as a monomer in solution, in agreement with the results of size exclusion chromatography (data not shown). MtfA is a bilobal, kidney-shaped molecule with dimensions of 52.6 Å by 40.5 Å by 35.9 Å (Fig. 2). The N-terminal lobe (residues 17 to 144) adopts a mixed α+β structure, with four α-helices (α1 to α4), three 310-helices (η1 to η3), and three parallel β-strands (β1 to β3). Residues 99 to 123 in this lobe are disordered in the apo structure (Fig. 2A), which results in a solvent-exposed hydrophobic patch across β2 and β3 (Leu92, Tyr94, Phe98, Pro127, Val129, and Leu146). The C-terminal lobe (residues 159 to 253) is an all-helical structure (α6 to α9). Overall, MtfA consists of three β-strands (β1 to β3), nine α-helices (α1 to α9), and five 310-helices (η1 to η5). The two lobes of MtfA are connected through a central helix, α5 (residues 145 to 158), where the 149HEXXH153 sequence motif is located. Helices α5 and α7 pack at an angle of 129° with an interhelix distance of 8.1 Å, similar to the equivalent helices in thermolysin (134.5° and 7.0 Å, PDB ID 1zdp) (44).
Fig 2.
Crystal structures of MtfA apoenzyme and holoenzyme from K. pneumoniae. (A) Ribbon diagram of the MtfA apoenzyme color coded from N terminus (blue) to C terminus (red). Helices α1-to-α9, β-strands β1-to-β3, and residue numbers for the boundaries of the gaps (i.e., disordered regions) in the model are indicated with a dashed trace. The conserved residues for the HEXXH zinc peptidase motif are shown as sticks. (B) Ribbon diagram of the MtfA holoenzyme with the Zn atom shown as a gray sphere. (C) Amino acid sequence of MtfA annotated with the secondary structure elements based on the apo structure (1st row), the holo structure (2nd row), and the secondary structure prediction (3rd row). The secondary structure elements are labeled based on the apo structure (310 helices are shown in red). The catalytic residues are highlighted in red/yellow.
A search for proteins structurally similar to MtfA using the DALI server (22) identified similarity to the catalytic domain of anthrax lethal factor (LF, PDB ID 1j7n, Z = 8.8, root mean square difference [RMSD] 3.3 Å for 145 aligned Cα atoms, 12% sequence identity) (42) and many other thermolysin-like zinc peptidases (∼125 to 140 equivalent Cα atoms and <15% sequence identity). All of these proteins have a similar catalytic core, first characterized in thermolysin (39), that consists of a topologically similar fold with four β-strands and two α-helices, with one of the helices bearing the HEXXH zinc-binding motif (36). Thus, the overall structural organization of MtfA is typical of thermolysin-like metallopeptidases, with the catalytic core matching its β-sheet and helices (α5 and α7) (Fig. 2A).
However, neither MtfA structure contains the strictly conserved “edge strand,” an antiparallel strand extending the N-terminal subdomain β-sheet (β1 to β3) through the open edge of the β3 strand. Structural comparisons with other related proteins, such as LF, indicate that the edge strand would be formed by a portion of the region between strands β2 and β3. However, the region that should include the edge strand is disordered in the apoenzyme (Fig. 2A) and only partially ordered in the holoenzyme (Fig. 2B).
Zinc induces structural changes.
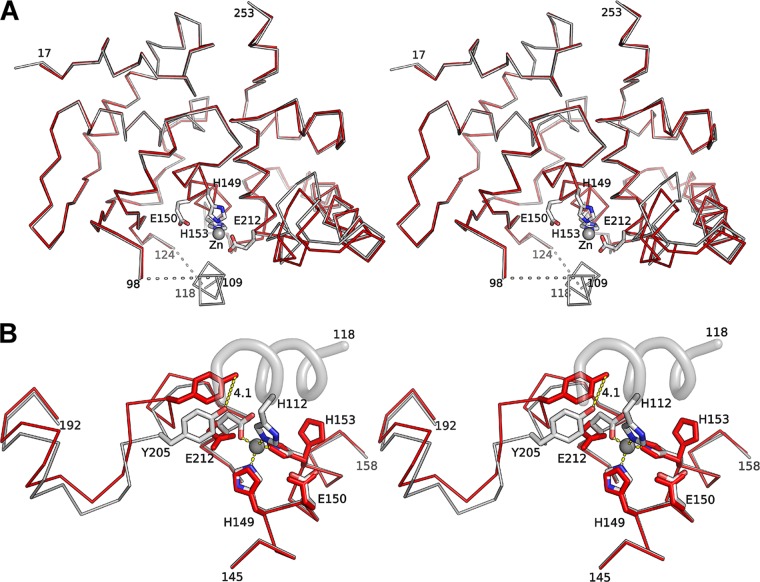
The overall structure of the holoenzyme (Fig. 2B) is very similar to that of the MtfA apoenzyme, with an RMSD of 0.7 Å for 213 Cα atoms (Fig. 3A). The most significant structural differences occur in the vicinity of the zinc binding site, where zinc binding partially orders a region (residues 99 to 123) that is disordered in the apo structure. In the holoenzyme structure, a helical segment within this region (residues 109 to 118) interacts directly with the zinc (Fig. 1B; also see Fig. S2 in the supplemental material). Furthermore, a segment of the protein in the C-terminal lobe (residues 195 to 214) shifts toward the zinc and involves side-chain rearrangements that optimize interaction with the zinc (RMSD 1.24 Å for 20 Cα atoms) (Fig. 3B), pushing Tyr205 closer to the zinc binding site. The zinc ion is coordinated by four residues (His112, His149, His153, and Glu212). In the absence of zinc, a water molecule is located in a position similar to that of the zinc, and side chains of the metal-chelating residues are more flexible, indicated by higher B values and different side chain rotamers. For example, the His153 side chain adopts two conformations in the apoenzyme but only one in the holoenzyme. Additionally, the side chain rotamers for Glu212 are different in the apo and holo structures.
Fig 3.
Zinc induces structural changes at the active site. (A) Stereo view of the structural superimposition of the MtfA apoenzyme (red) and holoenzyme (gray). (B) Close-up stereo view of the zinc binding site. The most significant changes in the holoenzyme (gray) compared to the zinc-free structure (red) occur near the active site and include the appearance of a partially ordered helix on the holoenzyme. The essential catalytic residues are shown as sticks, and the self-inhibitory helical peptide as a helical ribbon. The displacement of Tyr205 from the apo to the holo structure is marked by a dashed line (distance, 4.1 Å).
Active site.
Zinc metallopeptidases (zincins) are generally classified based on the spatial distribution of conserved sequence motifs that are essential for catalysis (23). The HEXXH motif in MtfA is located in the middle of helix α5 and has a consensus sequence of 145NhhhHEhhHKhD156 (“h” indicates a hydrophobic residue) (Fig. 4A). This motif plays an important role in zinc coordination and catalysis in zinc-dependent metallopeptidases (36). In the holoenzyme, the conformation of the strictly conserved residues (His149, Glu150, and His153) in this motif are essentially identical to those of the thermolysin-like proteins, such as LF (42) and thermolysin (44). The two histidines are required for zinc binding, while Glu150 serves as the prospective general base for catalysis. For gluzincins, an additional glutamate is required for zinc coordination (HEXXH+E) (36). This residue corresponds to the strictly conserved Glu212 of MtfA, which is located on helix α7 (consensus sequence 210PXEXFA215) (Fig. 4A). Another strictly conserved residue, Tyr205, equivalent to Tyr728 of anthrax LF (57), probably plays an important role in stabilizing the transition state (see below). The position of this tyrosine residue and the associated loop are affected by zinc binding. Thus, MtfA contains all essential catalytic residues for a gluzincin (HEXXH+E+Y). In addition to the strictly conserved zinc metallopeptidase reaction center, several other highly conserved residues that are specific to MtfA-like proteins are found on helix α5 (e.g., Asn145, Lys154, and Asp156). These residues are probably important for the structural integrity of the protein. Asn145 stabilizes α5 by forming an N-cap, while Lys154 and Asp156 stabilize the structural elements (β3 and the α5-to-η4 loop) at one end of the active-site groove.
The active-site groove is located at the top of α5, flanked at each side by the two subdomains. The wall of the groove formed by the C-terminal helical lobe has a well-defined structure that includes the α6-to-α7 loop (Asp203 and Tyr205) and helix α7 (Glu212, Val216, Ala215, and Glu219). The opposite side of the groove is not well defined due to disorder in residues 99 to 123. The active-site groove of MtfA is lined with highly conserved residues and is mostly acidic (Fig. 4B and C). Most notably, a few highly conserved residues, His149, Asp203, Tyr205, Val216, and Glu219, form a pocket that probably corresponds to the S1′ site, suggesting that this site is probably important for substrate specificity.
A self-inhibited conformation in the presence of zinc.
The fourth ligand for the bound zinc is normally a water molecule in zinc metallopeptidases and is important for attack on the scissile peptide bond during catalysis (38). In the holoenzyme, a helical peptide segment, corresponding to a disordered region in apo MtfA (residues 109 to 118) from its own subunit, was identified in the vicinity of the zinc (see “Refinement of the holoenzyme” in Materials and Methods). The imidazole of a highly conserved histidine (His112) completes the coordination sphere of the zinc (distance, 1.91 Å) (Fig. 3B). This “peptide” docks to the S subsites (Schechter and Berger nomenclature [47]) of the active site (Fig. 4D). Four residues from this helix (His112, Asn113, Val116, and Val117) contact the protein (His149, Glu150, His153, Glu212, Ala163, Thr208, Asp209, and Tyr205). The Val116 side chain fits into a small pocket contributed by His153, Ala211, and Ala163. This “peptide”-protein interface buries 578 Å2 of surface area and blocks access to the zinc and the active site. As a result, the holoenzyme conformation represents an interesting nonproductive, self-inhibitory form of the enzyme. A nonproductive peptide complex was also observed in the anthrax LF structure, but in the absence of zinc (42). However, the directionality of both of these peptides is opposite to that of canonical thermolysin and anthrax LF (46) substrates. Another example of a zinc-coordinated self-inhibitory complex can be found in a member of the M48 family, peptidase Q74D82 (PDB ID 3c37, unpublished), where the fourth coordination to the zinc is provided by the Nδ1 atom of a histidine (His206) at the start of a C-terminal disordered loop.
The observed helical self-inhibitory peptide segment in MtfA has significantly weaker density than the rest of the protein (Fig. 1B), suggesting that it is highly flexible and is probably only partially occupied (estimated occupancy, 0.85). Correspondingly, the overall average B value for this region (69.4 Å2) is also much higher than the average B value of the rest of the protein (41.6 Å2).
An evolutionary link between MtfA and LF.
Sequence analysis indicated that MtfA is evolutionarily related to anthrax LF. Using K. pneumoniae MtfA as a probe against the nr database at NCBI, PSI-BLAST (2), LF was identified as a significant hit (E = 0.005 after 3 iterations), sharing significant homology (24% sequence identity) in an 82-residue region around the active site. Other profile-based methods, such as FFAS (24) and HHpred (53), which were designed to detect remote homologs, provided even clearer evidence. The evolutionary relationship predicted by sequence analysis is further confirmed by structural comparison results. LF is the top hit when searching the Dali database using the MtfA structure. A search using the LF structure as a probe also showed MtfA as the second most significant hit (Z = 8.5), with the protective antigen-binding domain (PABD) of the anthrax EF (52) ranking first (Z = 10.7).
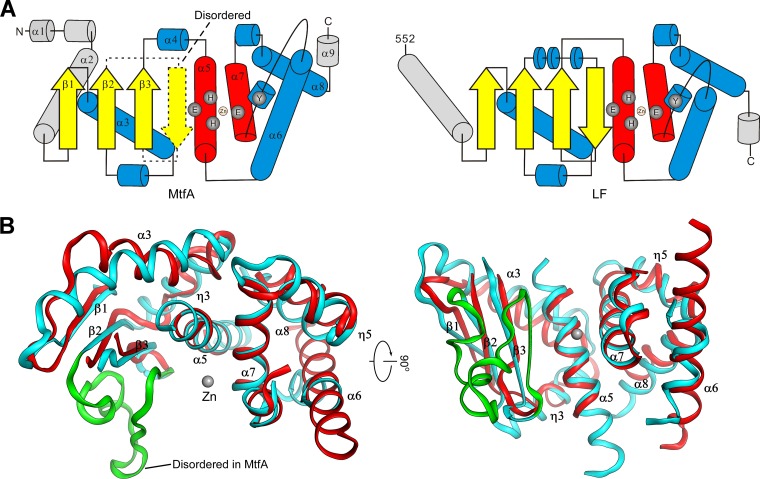
Despite low similarity at the sequence level, MtfA and the catalytic domain of LF share significant structural similarity both at the global fold level and in identical arrangements of catalytic residues. The topologies of the two domains are essentially identical (Fig. 5A). The structurally equivalent secondary elements include β-strands (β1, β2, and β3), as well as helices α3 and α5 of the N-terminal subdomain and helices α6, α7, α8, and η5 of the C-terminal helical subdomain (Fig. 5B). The conserved core of the two proteins can be aligned with an RMSD of 2.7 Å for 119 Cα atoms.
Fig 5.
Structural comparison between MtfA and anthrax lethal factor (LF). (A) Topology diagrams of MtfA and the catalytic domain of LF. Nonequivalent secondary structures are shown in gray. The disordered region of MtfA (residues 99 to 124) is shown as dashed lines. (B) Two orthogonal views of the common core region of MtfA (red) and the catalytic domain of LF (cyan, PDB ID 1j7n) consisting of equivalent secondary structures of both structures. The disordered region of MtfA (residues 99 to 123) is shown on the LF structure in green.
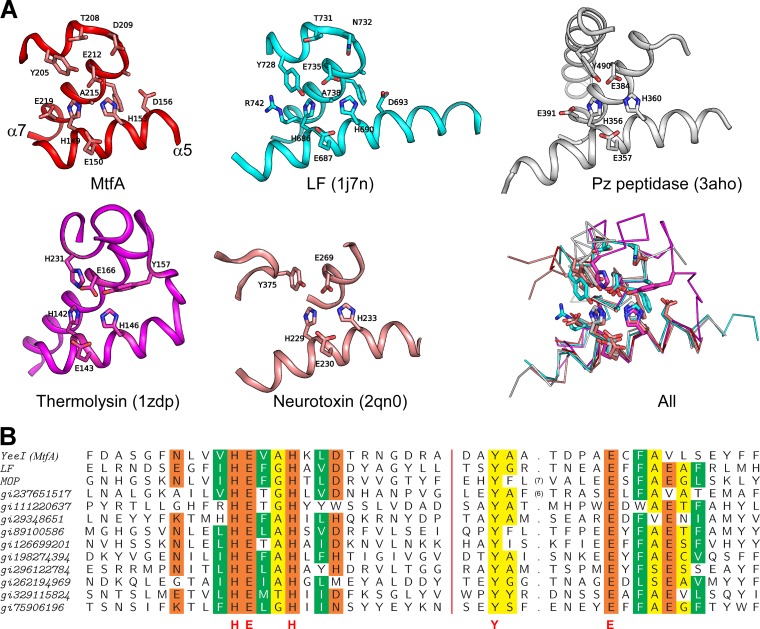
As expected, the most structurally and sequence-conserved regions are located near the zinc binding site and residues that are important for catalysis (α5 and α7 of MtfA), i.e., in all HEXXH+E+Y regions (Fig. 6). This region (residues 145 to 158 and 203 to 223) of MtfA can be aligned to the equivalent region of the LF catalytic domain with an RMSD of 0.85 Å for 35 aligned Cα (sequence identity, 31%). This substructure is also highly conserved in the thermolysin structure (PDB ID 1zdp, RMSD 1.47 Å for 30 aligned Cα), but with lower sequence identity (sequence identity, 14%). Tyr728 of LF is essential for its enzymatic activity (57). Interestingly, the locations of the tyrosines of MtfA (Tyr205) and LF (Tyr728) correspond to that of His231 in thermolysin and not the expected, conserved Tyr157 (Fig. 6A). An essential catalytic tyrosine from a different part of the molecule is also present at a similar position in botulinum neurotoxin (25) (Fig. 6A). The placement of this functionally important residue may suggest differences in the mode of transition state stabilization in MtfA, LF, and botulinum neurotoxin compared to that of thermolysin. The HEXXH+E+Y motif is also present in the oligopeptidase F family of proteins (Fig. 6A) (19, 29).
Fig 6.
Structural and sequence comparisons. (A) Structural comparisons of the active sites of MtfA (red), LF (cyan), Pz peptidase (white), thermolysin (magenta), and botulinum neurotoxin (salmon). Cα superposition of all five active sites with important residues represented as sticks is shown at the bottom right (All). (B) Sequence alignment of the active-site regions of representative MtfA homologs (see the text).
The most significant structural difference between MtfA and the catalytic domain of LF is in the N-terminal regions. This helical region of MtfA (residues 17 to 52), which packs up against α3 and reaches over to the C terminus, is important for maintaining the overall structural integrity by capping the top of the protein and linking the N and C lobes (Fig. 2A and B). The corresponding region in LF is a long helix that interacts with β1 and attaches catalytic domain IV to the rest of LF. In addition, the central helix of LF is longer than the corresponding regions in MtfA and thermolysin and results in a more extensive binding site. Moreover, MtfA has a deeper groove on the N-terminal side of α5 than LF due to a different conformation of the β3-to-α5 region in MtfA (residues 131 to 146). These differences probably indicate a difference in the substrate specificities of MtfA and LF.
Other remotely related HEXXH+E+Y peptidases.
Structural comparison between MtfA and LF clearly indicates a highly conserved core harboring all of the important catalytic residues (HEXXH+E+Y). This core corresponds to a helix containing the HEXXH motif, which is common to all zincins, followed by a 310-helix-turn-helix containing a YX6E motif. Inspection of PSI-BLAST search results revealed the presence of this conserved core in more than 850 unique proteins. A multiple sequence alignment of the core region of representative proteins is shown in Fig. 6B.
Most of these remotely related homologs are functionally uncharacterized. However, a distant MtfA homolog (Mop or VC0823) is a part of the pathogenicity island (VPI) associated with epidemic and pandemic strains of V. cholerae (28). Mop is involved in the modulation of pathogenesis, since the mop deletion mutant is hypervirulent (63, 64). Previous studies suggested that Mop probably functions as an extracellular zinc metallopeptidase (63). Mop shares sequence similarity with both LF (HHpred probability = 97.3, E = 7.8E−05) and MtfA (probability = 96.3, E = 0.002). More importantly, the catalytic residues HEXXH+E+Y are also conserved in Mop (Fig. 6B). Moreover, this catalytic core is also embedded in other larger proteins, such as glycoside hydrolases (e.g., gi:198274394), phage virion-encapsulated RNA polymerases (e.g., gi:237651517), membrane-bound proteases (e.g., gi:75906196), and multiple-peptidase fusion proteins (e.g., gi:296122784).
Peptidase assays of MtfA wild type and mutants.
We performed several nonspecific protease activity assays for the K. pneumoniae MtfA, based on those recently reported for the E. coli ortholog (20). The activity of the K. pneumoniae MtfA is similar to that of the E. coli MtfA. Only very weak aminopeptidase activity was detected using smaller substrates containing a single amino acid fused to 4-nitroanilide. The highest activity and specificity were obtained for l-alanine fused to 4-nitroanilide (ΔOD405 of 0.27 with 30 μg protein and 5 mM substrate after 4 h of incubation), which is about 40 times lower than for LF with the optimized substrate LFPS-2, consisting of an N-acetylated 14-mer 4-nitroanilide (5) (ΔOD405 of ∼0.3 with 50 μg protein and 0.14 mM substrate after 3 h of incubation). Metal chelators partially inhibit the aminopeptidase activity (see Fig. S3 in the supplemental material). However, significant residual activity (∼40% to 60%) was detected for two mutants that involve important catalytic residues (E212A and Y205A), based on assays using l-alanine 4-nitroanilide (data not shown).
The identification of the peptidase activity in MtfA is complicated by several factors. First, the substrate specificity of MtfA is currently unknown. Second, the crystal structures reveal that the active site is partially disordered. The MtfA holoenzyme adopts a self-inhibited conformation and is most likely not productive. Furthermore, the above-described aminopeptidase activity is very weak. As a result, it is difficult to interpret the aminopeptidase assay results unambiguously at present. On the other hand, lack of (or poor) peptidase activity in MtfA is consistent with the crystal structures, which implies that additional regulatory factor(s) are needed to activate the peptidase (see below).
DISCUSSION
LF is an extraordinary peptidase (MEROPS family M34), primarily because of its limited distribution. The modular design of anthrax toxins, such as LF and edema factor (EF), clearly suggests that these proteins have evolved from gene fusion (42, 52). Besides having similar overall folds, MtfA and LF also share an HEXXH+E+Y motif, which is distinct from the HEXXH+E+H+Y motif of thermolysin. These results strongly suggest that MtfA, as well as the MtfA-like domains involved in bacterial pathogenesis, such as in anthrax toxins and Mop, have evolved from a more recent common ancestor. The arrangement of catalytic residues in the MtfA-like domain is more similar to that of botulinum neurotoxin despite different evolutionary origins (56). The overall sequence similarity between MtfA and the LF family or other known metallopeptidase families is very low, and as a result, MtfA cannot be assigned to any known metallopeptidase families. Based on the evidence presented here, we conclude that MtfA and its homologs define a novel large family of gluzincins in the class of metallopeptidases (metallopeptidases/zincins/gluzincins/MtfA-like).
Currently, the substrate specificity and physiological substrates of MtfA are unknown. Based on the deduced configuration of the putative active site, we cannot distinguish between an exo- or endopeptidase activity. As shown above, the conserved residues within this family of metallopeptidases are concentrated near the catalytic center and are probably important in substrate recognition and specificity, as for example, the S′ site (probably S1′) of MtfA. Moreover, the self-inhibitory peptide identified in the crystal structure suggests that there might be an S site (probably S2) that recognizes an amino acid residue with a small side chain. Sites that are further removed from the catalytic center are more divergent across bacterial species. However, the properties of the S2 and S1′ binding sites of MtfA resemble those of LF, whose specificity is dictated by the P2, P1′, and P4-to-P7 positions (60), with the P2 and P1′ positions being occupied by two hydrophobic residues and the P4-to-P7 positions by one or more basic residues. Interestingly, the active-site groove of MtfA is less extended than that of LF, which may indicate a shorter recognition sequence.
The edge strand of zincins in general plays a critical role in substrate binding through the formation of a ladder of main-chain hydrogen bonds with the substrate (37). The edge strand is the only antiparallel strand of the N-terminal β-sheet. However, the edge strand was not observed in the MtfA structures since the region (residues 99 to 123) harboring the edge strand is mostly disordered. Even in the presence of zinc, the disordered region only becomes partially ordered to form the self-inhibitory α-helix, but not as a β-strand (Fig. 4D). More intriguingly, this same partially ordered region is predicted to form the edge strand based on secondary structure prediction algorithms whose accuracy for other regions of the protein correlates well with the crystal structure (Fig. 2C). Thus, the conformational flexibility of the region containing the edge strand (Fig. 4A), which includes several highly conserved residues, such as Asp106, Gly109, and His112, is probably important for the function of MtfA.
The self-inhibitory conformation of the MtfA holoenzyme is reminiscent of the conformation of inhibitory propeptides in other zinc peptidases, such as MMPs, ADAMs, and astacins. His112 occupies the same position as the catalytically active water in thermolysin-like enzymes. In order to probe the role of His112, we mutated this histidine into an alanine, which would be expected to disrupt its interaction with zinc, thus potentially releasing the helical fragment from blocking the active site. However, the peptidase activity of the mutant was only slightly reduced (by 14% based on an assay using l-alanine 4-nitroanilide). Since the disordered region (99 to 123) near the catalytic center may interfere with ligand binding, a properly folded edge strand may also be required for full peptidase activity. Thus, a single mutation, H112A, may not significantly alter the overall unstructured nature of the edge strand region, resulting in a similar weak activity compared to that of the wild-type protein. Thus, the disorder of the edge strand region and the presence of a α-helix covering the active site in the MtfA holoenzyme suggest that the enzyme is initially in an inactive state, which may serve to regulate the function for this enzyme. The physiological relevance of the flexible conformation of the edge strand region remains to be explored, but one cannot completely exclude the possibility that the disorder may be a crystallization artifact. To our knowledge, no other similar example of structural plasticity in the active site has been observed in thermolysin-like enzymes. In order to activate the peptidase activity, the edge strand region is expected to fold as in other zincins. The proper folding of this region could be influenced by the presence of other factors, such as interaction with another, yet-unidentified protein partner, binding of a small-molecule regulator or the natural substrate.
In vitro studies previously showed that E. coli MtfA interacts with the C-terminal amphipathic helix region of the glucose repressor Mlc located at the interface between the DNA binding domain (DBD) and the middle domain (4). Interestingly, a similar region of Mlc is also involved in interactions with EIIBGlc (41). The overall structures of MtfA and EIIBGlc are different. However, the N-terminal subdomain of MtfA displays some resemblance to EIIBGlc, both containing an exposed β-sheet, which is involved in intermolecular interactions in the EIIBGlc-Mlc complex (see Fig. S4 in the supplemental material). The active-site region of MtfA seems to be important for the interaction with Mlc (20). However, Mlc is not a target for the peptidase activity of MtfA. In fact, the addition of Mlc stimulated MtfA activity in the cleavage assay with l-alanine 4-nitroanilide (18, 20), which seems to further support our structural data on the existence of an inactive state that could be converted to the active form upon the binding of one or several interaction partners. Thus, we speculate that association between Mlc and MtfA probably induces structural changes that activate the peptidase activity of MtfA while inactivating the DNA-binding ability of Mlc. A feedback loop may exist involving a yet-unknown molecule that disassociates the MtfA-Mlc complex by binding to one of the proteins (see Fig. S5 in the supplemental material). Further experiments are clearly needed to explore the regulatory role of Mlc on MtfA.
Gene cooccurrence analysis of MtfA, Mlc, and EIICBGlc in various bacterial genomes suggests that MtfA homologs, at least in some bacteria, may have physiological roles that are independent of Mlc. Two hundred six of 951 sequenced bacterial genomes contain at least one MtfA ortholog that bears significant similarity to MtfA of E. coli. Eighty-five (41%) of these genomes do not contain significant Mlc or EIICBGlc homologs, and another 38 (18%) have EIICBGlc and MtfA but not Mlc. Even if we assume that Mlc is more diverse in sequence, there are still a significant number of bacteria (53 out of 206, 26%) where MtfA or both MtfA and EIICBGlc homologs are present in the genome while Mlc is not, such as Pseudomonas aeruginosa, Bordetella pertussis, Idiomarina loihiensis, Psychrobacter arcticus, Xanthomonas campestris, and Nitrosomonas europaea.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the JCSG high-throughput structural biology pipeline for their contribution to this work. Genomic DNA from K. pneumoniae MGH 78578 (ATCC 700721D) was obtained from the American Type Culture Collection (ATCC). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL). The SSRL is a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University.
The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (grant P41RR001209), and the National Institute of General Medical Sciences. This work was supported by the NIH, National Institute of General Medical Sciences, Protein Structure Initiative (grants U54 GM094586 and GM074898), the German Federal Ministry of Education and Research (grant FKZ 0315285C), and the German Research Foundation (grant SFB431).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Published ahead of print 30 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Becker AK, et al. 2006. YeeI, a novel protein involved in modulation of the activity of the glucose-phosphotransferase system in Escherichia coli K-12. J. Bacteriol. 188:5439–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao S, et al. 2010. Residue histidine 669 is essential for the catalytic activity of Bacillus anthracis lethal factor. J. Bacteriol. 192:5799–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carniel E, Guilvout I, Prentice M. 1996. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 178:6743–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. 2002. An automated system to mount cryo-cooled protein crystals on a synchrotron beamline, using compact samples cassettes and a small-scale robot. J. Appl. Cryst. 35:720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen SX, et al. 2004. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr. D Biol. Crystallogr. 60:2222–2229 [DOI] [PubMed] [Google Scholar]
- 9. Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collaborative Computational Project Number 4 1994. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [Google Scholar]
- 11. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis IW, Murray LW, Richardson JS, Richardson DC. 2004. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32:W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87–93 [DOI] [PubMed] [Google Scholar]
- 14. Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elsliger M-A, et al. 2010. The JCSG high-throughput structural biology pipeline. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 66:1137–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 17. Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72–82 [DOI] [PubMed] [Google Scholar]
- 18. Gabor E, et al. 2011. The phosphoenolpyruvate-dependent glucose-phosphotransferase system from Escherichia coli K-12 as the center of a network regulating carbohydrate flux in the cell. Eur. J. Cell Biol. 90:711–720 [DOI] [PubMed] [Google Scholar]
- 19. Gerdts CJ, et al. 2006. Time-controlled microfluidic seeding in nL-volume droplets to separate nucleation and growth stages of protein crystallization. Angew. Chem. Intl. Ed. Engl. 45:8156–8160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Göhler AK, et al. 2012. Characterization of MtfA, a novel regulatory output signal protein of the glucose-phosphotransferase system in Escherichia coli K-12. J. Bacteriol. 194:1024–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624 [DOI] [PubMed] [Google Scholar]
- 22. Holm L, Sander C. 1995. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20:478–480 [DOI] [PubMed] [Google Scholar]
- 23. Hooper NM. 1994. Families of zinc metalloproteases. FEBS Lett. 354:1–6 [DOI] [PubMed] [Google Scholar]
- 24. Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. 2005. FFAS03: a server for profile-profile sequence alignments. Nucleic Acids Res. 33:W284–W288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin R, et al. 2007. Structural and biochemical studies of botulinum neurotoxin serotype C1 light chain protease: implications for dual substrate specificity. Biochemistry 46:10685–10693 [DOI] [PubMed] [Google Scholar]
- 26. Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsch W, Sander C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637 [DOI] [PubMed] [Google Scholar]
- 28. Karaolis DK, et al. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. U. S. A. 95:3134–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kawasaki A, et al. 2010. The exquisite structure and reaction mechanism of bacterial Pz-peptidase A toward collagenous peptides: X-ray crystallographic structure analysis of PZ-peptidase A reveals differences from mammalian thimet oligopeptidase. J. Biol. Chem. 285:34972–34980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klock HE, Koesema EJ, Knuth MW, Lesley SA. 2008. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71:982–994 [DOI] [PubMed] [Google Scholar]
- 31. Koczura R, Kaznowski A. 2003. Occurrence of the Yersinia high-pathogenicity island and iron uptake systems in clinical isolates of Klebsiella pneumoniae. Microb. Pathog. 35:197–202 [DOI] [PubMed] [Google Scholar]
- 32. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 33. Lengeler JW, Jahreis K. 2009. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib. Microbiol. 16:65–87 [DOI] [PubMed] [Google Scholar]
- 34. Lesley SA, et al. 2002. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. U. S. A. 99:11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leslie AGW. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 ESF EAMCB Newsl. Protein Crystallogr. 26 [Google Scholar]
- 36. Lipscomb WN, Strater N. 1996. Recent advances in zinc enzymology. Chem. Rev. 96:2375–2434 [DOI] [PubMed] [Google Scholar]
- 37. Madala PK, Tyndall JD, Nall T, Fairlie DP. 2010. Update 1 of: Proteases universally recognize beta strands in their active sites. Chem. Rev. 110:PR1–PR31 [DOI] [PubMed] [Google Scholar]
- 38. Matthews BW. 1988. Structural basis of the action of thermolysin and related zinc peptidases. Acc. Chem. Res. 21:333–340 [Google Scholar]
- 39. Matthews BW, Jansonius JN, Colman PM, Schoenborn BP, Dupourque D. 1972. Three-dimensional structure of thermolysin. Nat. New Biol. 238:37–41 [DOI] [PubMed] [Google Scholar]
- 40. Morrison TB, Parkinson JS. 1994. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 91:5485–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nam TW, et al. 2008. Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration. Proc. Natl. Acad. Sci. U. S. A. 105:3751–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pannifer AD, et al. 2001. Crystal structure of the anthrax lethal factor. Nature 414:229–233 [DOI] [PubMed] [Google Scholar]
- 43. Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roderick SL, Fournie-Zaluski MC, Roques BP, Matthews BW. 1989. Thiorphan and retro-thiorphan display equivalent interactions when bound to crystalline thermolysin. Biochemistry 28:1493–1497 [DOI] [PubMed] [Google Scholar]
- 45. Santarsiero BD, et al. 2002. An approach to rapid protein crystallization using nanodroplets. J. Appl. Crystallogr. 35:278–281 [Google Scholar]
- 46. Santelli E, Bankston LA, Leppla SH, Liddington RC. 2004. Crystal structure of a complex between anthrax toxin and its host cell receptor. Nature 430:905–908 [DOI] [PubMed] [Google Scholar]
- 47. Schechter I, Berger A. 1967. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27:157–162 [DOI] [PubMed] [Google Scholar]
- 48. Schubert S, Rakin A, Fischer D, Sorsa J, Heesemann J. 1999. Characterization of the integration site of Yersinia high-pathogenicity island in Escherichia coli. FEMS Microbiol. Lett. 179:409–414 [DOI] [PubMed] [Google Scholar]
- 49. Schubert S, Rakin A, Heesemann J. 2004. The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int. J. Med. Microbiol. 294:83–94 [DOI] [PubMed] [Google Scholar]
- 50. Seitz S, Lee SJ, Pennetier C, Boos W, Plumbridge J. 2003. Analysis of the interaction between the global regulator Mlc and EIIBGlc of the glucose-specific phosphotransferase system in Escherichia coli. J. Biol. Chem. 278:10744–10751 [DOI] [PubMed] [Google Scholar]
- 51. Sheldrick GM. 2008. A short history of SHELX. Acta Crystallogr. A 64(Pt 1):112–122 [DOI] [PubMed] [Google Scholar]
- 52. Shen Y, Zhukovskaya NL, Guo Q, Florian J, Tang WJ. 2005. Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor. EMBO J. 24:929–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tanaka Y, Kimata K, Aiba H. 2000. A novel regulatory role of glucose transporter of Escherichia coli: membrane sequestration of a global repressor Mlc. EMBO J. 19:5344–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Terwilliger TC, Berendzen J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55:849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tonello F, Montecucco C. 2009. The anthrax lethal factor and its MAPK kinase-specific metalloprotease activity. Mol. Aspects Med. 30:431–438 [DOI] [PubMed] [Google Scholar]
- 57. Tonello F, Naletto L, Romanello V, Dal Molin F, Montecucco C. 2004. Tyrosine-728 and glutamic acid-735 are essential for the metalloproteolytic activity of the lethal factor of Bacillus anthracis. Biochem. Biophys. Res. Commun. 313:496–502 [DOI] [PubMed] [Google Scholar]
- 58. van den Bedem H, Wolf G, Xu Q, Deacon AM. 2011. Distributed structure determination at the JCSG. Acta Crystallogr. D Biol. Crystallogr. 67:368–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. 1993. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229:105–124 [DOI] [PubMed] [Google Scholar]
- 60. Vitale G, Bernardi L, Napolitani G, Mock M, Montecucco C. 2000. Susceptibility of mitogen-activated protein kinase kinase family members to proteolysis by anthrax lethal factor. Biochem. J. 352:739–745 [PMC free article] [PubMed] [Google Scholar]
- 61. Vonrhein C, Blanc E, Roversi P, Bricogne G. 2007. Automated structure solution with autoSHARP. Methods Mol. Biol. 364:215–230 [DOI] [PubMed] [Google Scholar]
- 62. Winn MD, Murshudov GN, Papiz MZ. 2003. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374:300–321 [DOI] [PubMed] [Google Scholar]
- 63. Zhang D, Rajanna C, Sun W, Karaolis DK. 2003. Analysis of the Vibrio pathogenicity island-encoded Mop protein suggests a pleiotropic role in the virulence of epidemic Vibrio cholerae. FEMS Microbiol. Lett. 225:311–318 [DOI] [PubMed] [Google Scholar]
- 64. Zhang D, Xu Z, Sun W, Karaolis DK. 2003. The vibrio pathogenicity island-encoded mop protein modulates the pathogenesis and reactogenicity of epidemic vibrio cholerae. Infect. Immun. 71:510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.