Abstract
Heterocyst differentiation is orchestrated by the N control transcriptional regulator NtcA and the differentiation-specific factor HetR. In Anabaena sp. strain PCC 7120, the devBCA operon is expressed from two different promoters activated upon N stepdown. The distal devB promoter (transcription start point [TSP] located at position −704) represents a canonical class II NtcA-activated promoter, including a consensus NtcA-binding site centered 39.5 nucleotides upstream from the TSP. Transcription activation from a second TSP (−454) requires NtcA and is impaired in hetR mutants. In a wild-type background, three different DNA fragments, including both or each individual promoter, directed gfp expression localized mainly to proheterocysts and heterocysts. Expression was undetectable in an ntcA background and, for the fragment including the proximal promoter alone, also in a hetR background. In spite of the absence of consensus NtcA-binding sequences between the two TSPs, NtcA was shown to interact with this DNA region, and NtcA and its effector, 2-oxoglutarate, were necessary and sufficient for in vitro transcription from the −454 TSP. No HetR binding to the DNA or in vitro transcription from the proximal devB TSP promoted by HetR alone were detected. However, a moderate positive effect of HetR on NtcA binding to the DNA between the two devB TSPs was observed. The proximal devB promoter appears to represent a suboptimal NtcA-activated promoter for which HetR may act as a coactivator, with the physiological effect of restricting gene activation to conditions of prevalence of high NtcA and HetR levels, such as those taking place during heterocyst differentiation.
INTRODUCTION
Cyanobacteria form a group of microorganisms characterized by a photoautotrophic mode of life relying on oxygenic photosynthesis, a process likely developed by ancestors of extant cyanobacteria. Nowadays, cyanobacteria are important primary producers in the oceans and thus play a crucial role in C and N cycling in our planet (31). Some cyanobacteria are able to undergo complex developmental processes to adapt to changes in their environment (25). The differentiation of cells, called heterocysts, that are specialized in the fixation of atmospheric nitrogen that takes place in some filamentous cyanobacteria in response to N deficiency merits special attention (1, 15, 40). In Anabaena and Nostoc strains, heterocysts differentiate at semiregular intervals along the filament, forming a truly multicellular bacterium with different cell types specialized in different nutritional tasks that exchange nutrients and regulators and contribute to the growth of the filament as the organismic unit (15).
In cyanobacteria, N control is exerted by NtcA, a member of the cyclic AMP receptor protein (CRP) family of bacterial regulators that activates and in some cases represses genes in response to nitrogen limitation (17). NtcA binds to specific DNA sites with the sequence signature GTAN8TAC that are present in the regulated promoters (24, 36). NtcA binding to DNA is increased by interaction with 2-oxoglutarate (2-OG), a metabolic signal of the N status of the cells (30, 37, 41). A conserved structure, matching that of the bacterial class II activator-dependent promoters (6), has been found upstream of many NtcA-dependent genes in different types of cyanobacteria. These canonical NtcA-activated promoters include an NtcA-binding site centered about −41 nucleotides (nt) from the transcription start point of the regulated gene and a −10 box with the consensus sequence TAN3T (17). In these promoters, NtcA and 2-OG have a positive influence on RNA polymerase (RNAP) recruitment, and both are required for the production of transcriptionally active open promoter complexes with RNAP and for in vitro transcript production (34).
The differentiation of a vegetative cell into a heterocyst involves large morphological and metabolic changes which are sustained by a specific program of gene expression. A crucial component of this program is a temporal sequence of activation events that leads to expression of the many structural and regulatory heterocyst-specific genes (1, 15, 18). Two regulators have key roles in heterocyst differentiation: the global transcription factor NtcA and the differentiation-specific factor HetR, both of which are required for initiation of heterocyst differentiation (5, 16, 39). Indeed, heterocyst differentiation genes so far studied are not activated in mutant strains lacking a functional ntcA gene or, in many cases, a functional hetR gene. Upon perception of N deficiency, the ntcA and hetR genes increase their expression in a mutually dependent manner (28) and their expression is localized mostly to certain spatially distributed cells that eventually become heterocysts (4, 29).
A remarkable feature of many characterized genes involved in heterocyst differentiation and function is that they bear multiple promoters (15). These include canonical NtcA-activated promoters (which do not require HetR), σ70 consensus-type promoters, and a special type of promoters that, depending in vivo on both NtcA and HetR, do not show any recognizable promoter structure except, in some cases, a putative −10 promoter box (e.g., see references 27, 30, and 35). Expression of these HetR-dependent promoters likely takes place in the differentiating cells, having a role in cell-specific gene activation during heterocyst differentiation. HetR has been reported to bind DNA upstream of some regulated genes in vitro (20, 33), and in the case of the hetP gene promoter, an inverted repeat DNA sequence has been identified as the DNA-binding site for HetR (19). However, this sequence is not found upstream from other HetR-dependent genes that have been analyzed (19). Recently, the crystal structure of HetR from Fischerella sp. strain MV11 has been solved, showing the protein as a dimer with a central DNA-binding unit made of the N-terminal regions of the two subunits, which provide two helix-turn-helix motifs (22). However, the precise mechanism by which HetR influences gene expression remains unknown. Also, the mechanism of the dependence on NtcA of promoters that do not exhibit recognizable NtcA-binding sites remains unknown.
In an attempt to gain insight into the mechanism of cell-specific transcription activation during the differentiation of the cyanobacterial heterocyst, we have analyzed, both in vivo and in vitro, the effects of NtcA and HetR on the complex promoter region of the Anabaena sp. strain PCC 7120 devBCA operon encoding an ABC-type exporter involved in the deposition of the glycolipid layer of the heterocyst envelope (13).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Anabaena sp. (also known as Nostoc sp.) strain PCC 7120 was grown in BG11 medium modified to contain ferric citrate instead of ferric ammonium citrate (containing NaNO3) (32), BG110 medium (BG11 further modified by omission of NaNO3), or BG110-ammonium medium {BG110 containing 4 mM NH4Cl and 8 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]-NaOH buffer, pH 7.5} at 30°C under continuous illumination (25 μE m−2 s−1) in shaken (80 to 90 rpm) liquid cultures or in medium solidified with 1% Difco agar. Alternatively, cultures (referred to as bubbled cultures) were supplemented with 10 mM NaHCO3, bubbled with a mixture of CO2 and air (1%, vol/vol), and illuminated (75 μE m−2 s−1). In this case, the ammonium-containing medium included 6 mM NH4Cl and 12 mM TES-NaOH buffer (pH 7.5). The ntcA mutant, strain CSE2 (16), and the hetR mutant strains DR884a (4) and CSSC2 (this work) were grown in BG110-ammonium medium supplemented with neomycin (Nm) in the case of strain DR884a and with streptomycin (Sm) and spectinomycin (Sp) in the case of strain CSE2. The gfp-mut2 reporter strains CSSC6 through CSSC16 were grown in BG110-ammonium medium supplemented with Nm, with Sm and Sp also added in the cases of CSSC9 to CSSC11. Antibiotics were used at the following concentrations: Sm, 2 to 5 μg ml−1; Sp, 2 to 5 μg ml−1; and Nm, 10 to 50 μg ml−1.
Escherichia coli DH5α and XL1-Blue were used for plasmid constructions. They and strains HB101 and ED8654, used for conjugations with Anabaena spp., were grown in LB medium (2) supplemented, when appropriate, with antibiotics at standard concentrations.
Strain construction.
To construct a hetR deletion mutant, two DNA fragments, one encompassing nucleotide positions −558 to +200 with regard to the hetR open reading frame (ORF) start and the other encompassing positions +714 to +1460, including 184 bp of the 3′ end of hetR and 562 bp downstream of it, were amplified by PCR using DNA from strain PCC 7120 as the template and oligodeoxynucleotide pairs alr2339-19/alr2339-20 and alr2339-22/alr2339-21, respectively (all oligodeoxynucleotide primers are listed in Table 1). The two DNA fragments were digested with NheI and ligated. The resulting fragment was amplified by PCR using primers alr2939-19 and alr2939-21 and, after digestion with BamHI and XhoI, cloned into plasmid pRL278, which carries an Nm resistance (Nmr) determinant and the sacB gene for positive selection (4, 11), producing plasmid pCSSC7, which was transferred by triparental mating to Anabaena sp. strain PCC 7120 (12). Clones exhibiting resistance to sucrose and sensitivity to Nm were selected. One such clone, which according to PCR and restriction analyses bore the deleted hetR gene and no wild-type chromosomes, was selected and named strain CSSC2.
Table 1.
Oligodeoxynucleotides used in this studya
| Oligonucleotide | Sequence (5′–3′) | Positionb |
|---|---|---|
| alr3710-1 | CCCCCTACTCCCTTTCC | −825 to −809 |
| alr3710-2 | GTGCGATAACATAACATTTCCC | −571 to −592 |
| al3710-15 | CCACAATGTACTCGTTTCTG | −209 to −228 |
| alr3710-16 | TCTCAGTAGCGTTTTTAAATAG | −292 to −313 |
| alr3710-17 | CCTGAAACAGATGAATGTA | −731 to −713 |
| alr3710-18 | GGTACAACCCCTAGCCACGG | −652 to −634 |
| alr3710-19 | GATAGATTTTCATTATGGC | −452 to −469 |
| alr3710-26 | GAGTATCTCGAGAGATTTCATTATGGCA | −443 to −470 |
| alr3710-30 | CCATCGATTAGAAAAAGCTTTTGGTGTG | −1009 to −985 |
| alr3710-31 | CCGCTCGAGGTATAACTCCTGAATTAGTC | −437 to −418 |
| alr3710-32 | CCGCTCGAGCTAAAATTGGTTATTTACTC | −687 to −668 |
| alr3710-33 | CCATCGATGCATTTGATGGAGTAAAT | −697 to −680 |
| alr3710-34 | AAAATCGATCTGGACAACACAATTACTG | −1067 to −1040 |
| alr3710-35 | ACTCTCGAGTTAGTCAGCCTGAGTATGC | −451 to −423 |
| alr2339-19 | CAATGCGATCCGTGAGTTTGGCGTAGT | −558 to −530 |
| alr2339-20 | CAAATGCTAGCTCATCCGGAGGTTTTG | +200 to +174 |
| alr2339-21 | CAACATCTCGAGAACGTCGTGATGCCTTTATT | +1460 to +1429 |
| alr2339-22 | AAAGAGCTAGCAGAAAAGCGGCCAAACG | +714 to +471 |
| Ck3-1 | CAGAGCAGCCGATTGTCTGTTG | +96 to +75 |
| CK3-2 | CTGATAAGTGAGCTATTCAC | −145 to −126 |
| gfp-5 | GTATGTTGCATCACCTTCAC | +116 to +97 |
| pDU-1 | GTGAGGGATGAGCGATGAC |
Restriction endonuclease sites incorporated are underlined.
Relative to the translation start site of the corresponding gene.
To construct strains bearing PdevB-gfp-mut2 transcriptional fusions, three DNA fragments from the devB promoter region were amplified by PCR using primer pairs alr3710-30/alr3710-35, alr3710-30/alr3710-32, and alr3710-33/alr3710-31, all pairs including ClaI and XhoI sites, and plasmid pCSAM155 (which contains a DNA fragment corresponding to positions −825 to +17 with respect to the translational start point of devB [27]) as the template and cloned in front of the gfp-mut2 gene, preceded by its own Shine-Dalgarno sequence, present in vector pCSAM211 (21), producing plasmids pCSSC18, pCSSC19, and pCSSC20, respectively. pCSAM211 is based on Nmr-encoding pDUCA7, which includes the pDU1 replicon (5), and as such, all these plasmids can be mobilized to and replicate in Anabaena. Plasmids pCSSC18, pCSSC19, and pCSSC20, as well as pCSAM211, used as a control of basal fluorescence, were transferred to Anabaena sp. strain PCC 7120, to strain CSE2 (ntcA), and to strain CSSC2 (hetR) by triparental mating selecting for Nmr. The genetic structure of selected clones from each mating was tested by PCR with total DNA from the clone and the primer pair pDU-1/gfp-5 to check the presence of the transferred plasmid and with primer pairs pDU-1/alr3710-16, pDU-1/alr3710-15, and alr3710-34/gfp-5 to check whether integration of the plasmid into the chromosomal devB region had occurred. Strains CSSC6 and CSSC12 bore the alr3710-30/alr3710-35-gfp construct (expressing gfp-mut2 from the dual devB promoter), integrated mainly in the devB chromosomal region (not shown), in the wild-type and hetR backgrounds, respectively. Strain CSSC9 bore the alr3710-30/alr3710-35-gfp construct in the transferred plasmid in the ntcA background (not shown). Strains CSSC7, CSSC10, and CSSC13 bore the alr3710-30/alr3710-32-gfp construct (expressing gfp-mut2 from the distal devB promoter) in the transferred plasmid in the wild-type, ntcA, and hetR backgrounds, respectively (not shown). Strains CSSC8, CSSC11, and CSSC14 bore the alr3710-33/alr3710-31-gfp construct (expressing gfp-mut2 from the proximal devB promoter) in the transferred plasmid in the wild-type, ntcA, and hetR backgrounds, respectively (not shown). Strains CSSC15 and CSSC16 bore control plasmid pCSAM211 in the wild-type and hetR backgrounds, respectively. Green fluorescent protein (GFP) fluorescence was analyzed by confocal microscopy using a Leica HCX PLAN-APO 63× 1.4 NA oil immersion objective attached to a Leica TCS SP2 confocal laser-scanning microscope. GFP was excited using 488-nm irradiation from an argon ion laser, and fluorescence emission was monitored from 510 to 540 nm.
DNA and RNA isolation, manipulation, and analysis.
Isolation of genomic DNA (7) and of total RNA (2, 26) from Anabaena sp. was done as described previously. The double-stranded DNA fragments used in electrophoretic mobility shift assays (EMSA) and in vitro transcription experiments were obtained by PCR amplification followed by purification of the corresponding products with the GFX gel band purification kit (Pharmacia Biotech). The double-stranded DNA fragments used in DNase I footprinting analysis were obtained by PCR using one of the primers labeled with T4 polynucleotide kinase (Roche) and [γ-32P]dATP and were extracted with phenol and chloroform and precipitated with absolute ethanol, ammonium acetate, and glycogen (Roche). The different devB promoter fragments were amplified using plasmid pCSAM155 (27) as the template and the following oligodeoxynucleotide pairs: alr3710-1/alr3710-2 (positions −825 to −571 with regard to the ORF start), alr3710-17/alr3710-19 (positions −731 to −452), alr3710-17/alr3710-26 (positions −731 to −443), alr3710-1/alr3710-16 (positions −825 to −292), alr3710-17/alr3710-16 (positions −731 to −292), and alr3710-18/alr3710-16 (positions −652 to −292). The fragment of the Amaranthus hybridus promoter used in Fig. 4 was amplified using plasmid pRL278 (4) as the template and primers CK3-1 and CK3-2.
Fig 4.
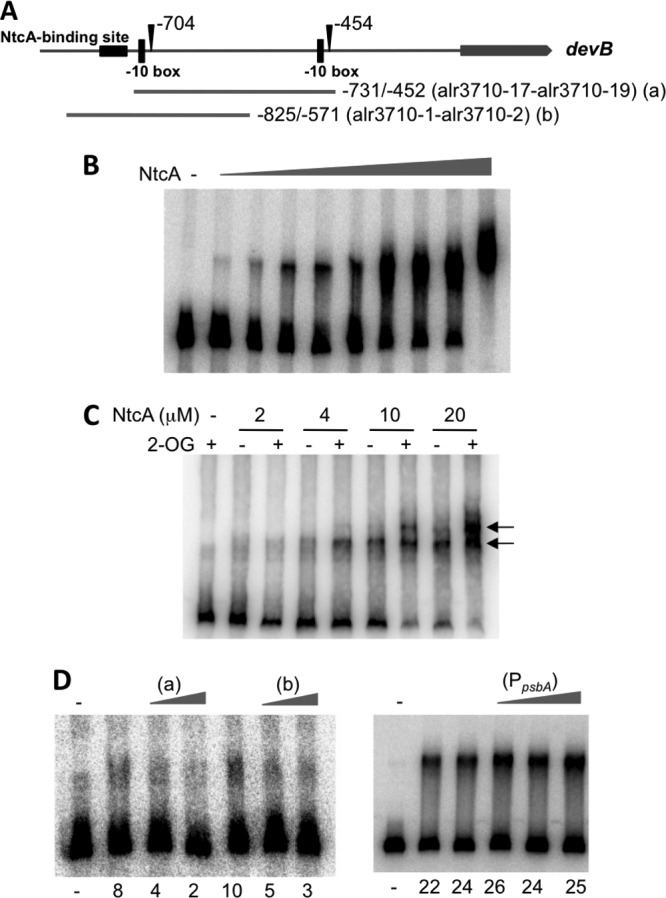
EMSA of the binding of Anabaena NtcA to DNA sequences between the two TSPs of the devB gene. The DNA fragment of the devB promoter region represented in panel A (fragment a) was incubated, as indicated, with purified NtcA (plus 0.6 mM 2-OG, except when indicated in panel C) before loading onto PAGE gels. NtcA concentrations in panel B are as follows: 0.1, 0.5, 1.0, 1.5, 2.0, 2.5 3.0, 4.0, and 10.0 μM. NtcA concentrations in panel D are 0.6 μM. −, no NtcA added. Arrows in panel C point to the positions of NtcA-DNA complexes. (D) 100× or 200× excess of unlabeled DNA fragment (a and b are as represented in panel A) or 100×, 200×, or 300× excess of a fragment of the promoter of the Amaranthus hybridus psbA gene (PpsbA), added as indicated. Numbers under the pictures in panel D indicate percentages of retarded fragments in the corresponding lane (see Materials and Methods for details).
Primer extension analyses.
Primer extension was performed with primer alr3710-16 labeled with T4 polynucleotide kinase (Roche) and [γ-32P]dATP and mixed with 20 μg RNA in the presence of 10 mM Tris-HCl (pH 8.0)-150 mM KCl-1 mM EDTA. The mixtures were incubated at 85°C for denaturation of RNA and then at 50°C for 1 h for annealing. The extension reactions were carried out at 47°C for 1 h in a final volume of 45 μl containing the whole annealing mixture, 0.25 mM concentrations of each deoxynucleoside triphosphate, 200 U of reverse transcriptase (Invitrogen), and the buffer provided by the transcriptase supplier. Reaction mixtures were then treated with RNase A (DNase free; Boehringer) and extracted with phenol. The extended fragments were precipitated with sodium acetate and ethanol, resuspended in formamide loading dye, and loaded onto 6% polyacrylamide-8 M urea sequencing gels next to the corresponding sequencing ladder. Images of radioactive gels were obtained with a Cyclone storage phosphor system (Packard).
EMSA.
EMSA were carried out as described previously (24) using 5 to 10 fmol of the specific DNA fragment end-labeled with T4 polynucleotide kinase (Roche) and [γ-32P]ATP and 0.07 mg/ml poly(dI-dC) as nonspecific competitor DNA in a final volume of 15 μl. The reaction mixtures with the corresponding promoter fragment were incubated with purified NtcA for 30 min at 30°C. The protein-DNA complexes were separated on native 5% polyacrylamide gels. Radioactive areas of the gels were visualized and quantified with a Cyclone storage phosphor system (Packard).
DNase I footprinting assays.
Protein-DNA complexes were formed in a final volume of 13.5 μl of buffer [23 mM Tris-HCl (pH 8.0), 80 mM KCl, 10 mM MgCl2, 5 mM CaCl2, 2 mM dithiothreitol (DTT), 0.07 mg/ml bovine serum albumin (BSA), 0.07 mg/ml poly(dI-dC)] with 10 fmol of the 32P-end-labeled (with T4 polynucleotide kinase and [γ-32P]ATP) DNA fragment and purified NtcA and HetR proteins as indicated. The reaction mixtures were incubated for 30 min at 30°C. Then, DNase I (1 U; Roche) was added in 1.5 μl of a buffer containing 100 mM CaCl2 and 100 mM MgCl2, and the reaction was immediately stopped with 4 μl of stop solution (100 mM EDTA, 1.5 mM sodium acetate [pH 5.2], 1 mg/ml salmon sperm DNA, 2.5 mg/ml glycogen). The products were resolved in 6% polyacrylamide-8 M urea gels next to the corresponding sequencing ladder. Images of radioactive gels were obtained as described above.
In vitro transcription assays.
The runoff transcription assays were performed in a total volume of 15 μl of buffer (40 mM Tris-HCl [pH 8.0], 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.5 mg/ml BSA, 5% glycerol) supplemented with 25 to 30 fmol of DNA fragment, 2-OG, and NtcA and HetR as indicated. Complexes were formed during a 5-min incubation at 37°C. Transcription was started by the addition of 1.5 μl of a substrate solution of 0.15 mM (each) ATP, GTP, and UTP, 20 μM CTP, and 3 μCi [α-32P]CTP. After 30 min of incubation at 37°C, the reaction was terminated by the addition of 6 μl phenol, 10 mM EDTA, and 1.5 μl of glycogen prepared at 200 ng/μl in 3 M sodium acetate (pH 5.2). The products were precipitated with ethanol and fractionated by electrophoresis on 6% polyacrylamide-8 M urea gels. Images of radioactive gels were obtained as described above.
Purification of recombinant Anabaena NtcA, HetR, and RNAP proteins.
NtcA was purified from crude extracts of E. coli containing plasmid pCSAM61 (27) through a 1-ml HiTrap Heparin HP column (Pharmacia Biotech) as described previously to be used in in vitro transcription assays (34). NtcA was purified from extracts of E. coli containing plasmid pCSAM70 (which bears a 6His-tagged ntcA gene cloned in plasmid vector pQE9) as described previously to be used in EMSA and DNase I footprinting assays (14). HetR was purified from crude extracts of E. coli containing plasmid pCSAM105 (which bears a 6His-tagged hetR gene cloned in plasmid vector pTrc99a). Extracts were passed through a 1-ml Ni2+-charged His-Select cartridge (Sigma), and HetR-containing fractions were further passed through a 1-ml HiTrap Heparin HP column (Pharmacia Biotech). HetR protein was eluted with a linear gradient of NaCl (0.15 to 1.5 M) in 20 mM sodium phosphate buffer (pH 7.0) supplemented with 20% glycerol. Recombinant RNAP from Anabaena sp. strain PCC 7120 was reconstituted from the purified cloned subunits as described previously (34).
RESULTS
Two differentially regulated transcription start sites of the devB operon.
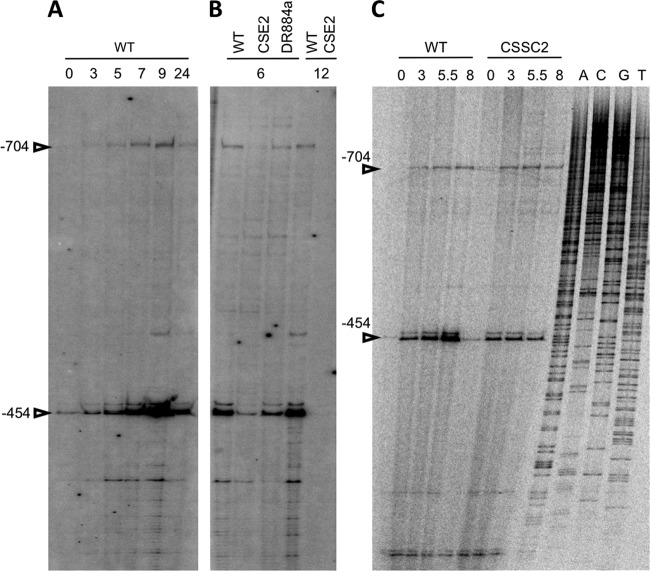
In Anabaena sp. strain PCC 7120, expression of the devBCA operon at the whole-filament level is activated upon combined N deprivation dependent on the global transcription factor NtcA and the differentiation-specific factor HetR (8, 14). Expression of the devBCA operon takes place from two different transcription start points (TSPs): a distal site located at −704 nucleotides with regard to the translation initiation site of the devB gene, which requires NtcA but not HetR, and a proximal site located at −454 that requires NtcA and is expressed at a low level in a hetR mutant (27). To get a more detailed picture about the use of the two devB TSPs during heterocyst differentiation, we performed primer extension analysis to trace transcripts with each of those 5′ RNA ends at different times after combined nitrogen stepdown in the wild-type strain, in an ntcA mutant (strain CSE2) (16), and in two different hetR mutants, strain DR884a, an insertional mutant in the hetR locus (4), and strain CSSC2, with the hetR gene deleted (see Materials and Methods). In the wild-type strain, transcription from both the −704 and −454 TSPs followed a similar time course of activation upon ammonium withdrawal, increasing until at least 9 h and decreasing thereafter (Fig. 1A). The abundance of transcripts with any of the two ends observed at 6 h and especially at 12 h was greatly impaired in the ntcA mutant (Fig. 1B). On the other hand, mutation of hetR appeared to impair the maximum accumulation of RNA with the 5′ end at −454 (see Fig. 1B for DR884a and 1C for CSSC2).
Fig 1.
Primer extension analysis of the devB gene promoter region. Primer extension was carried out with primer alr3710-16 and RNA isolated from Anabaena sp. strain PCC 7120, strain CSE2 (ntcA), and strains DR884a and CSSC2 (hetR), grown in bubbled cultures with ammonium (0) and incubated in the absence of combined nitrogen for the indicated times (in hours). The positions of the −704 and −454 RNA 5′ ends are indicated. A sequence ladder of the same DNA region is shown at the right in panel C. WT, wild type.
In vivo GFP report from the different devB promoters.
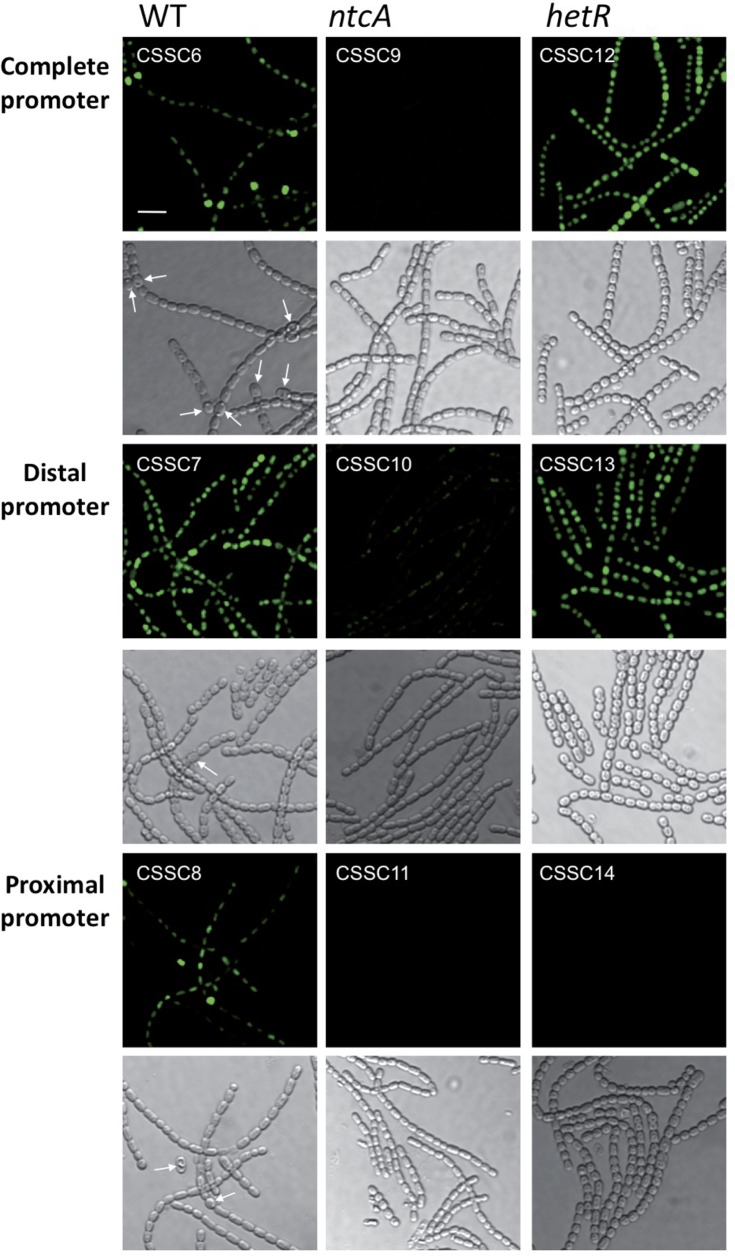
To study the cellular localization of the expression from each devB promoter, as well as from the whole promoter region, and the influence of NtcA and HetR, GFP fluorescence was monitored from strains bearing transcriptional fusions of different DNA fragments including the distal promoter, the proximal promoter, or the two devB promoters to the gfp-mut2 gene in an otherwise wild-type, ntcA, or hetR background. Incubation in the absence of combined nitrogen of strains CSSC6, CSSC7, and CSSC8 (which reported GFP from both, only the distal, and only the proximal devB promoters, respectively, in the wild-type background) produced high GFP fluorescence localized mainly to cells that, morphologically and by the loss of cyanobacterial autofluorescence, could be identified as proheterocysts or heterocysts (Fig. 2). From the three promoter fragments, fluorescence was similar to basal levels in an ntcA mutant background (strains CSSC9, CSSC10, and CSSC11). In a hetR background, expression was undetectable from the proximal promoter (strain CSSC14), whereas the strains reporting the activity of the distal promoter (strain CSSC13) or of the two promoters (strain CSSC12) exhibited GFP fluorescence levels higher than that of the background in all cells of the filament (Fig. 2). Thus, expression of the distal promoter may exhibit some activity in all cells of the filament with a strict dependence on NtcA, increasing in proheterocysts depending on HetR. Activation of the proximal promoter seems to be restricted to the differentiating cells with a requirement for both NtcA and HetR. Additionally, HetR may have a repressor effect in vegetative cells (observed with the dual promoter), as has been described for other Anabaena N-regulated genes (e.g., see reference 23).
Fig 2.
Expression of gfp-mut2 from the two promoters of the devB gene in wild-type, ntcA, and hetR backgrounds after N stepdown. GFP fluorescence (top) and bright-field (bottom) micrographs of filaments of strains CSSC6 (dual devB promoter directing expression of GFP in the wild-type background), CSSC7 (distal promoter in the wild-type background), CSSC8 (proximal promoter in the wild-type background), CSSC9 (dual promoter in the ntcA background), CSSC10 (distal promoter in the ntcA background), CSSC11 (proximal promoter in the ntcA background), CSSC12 (dual promoter in the hetR background), CSSC13 (distal promoter in the hetR background), and CSSC14 (proximal promoter in the hetR background), grown in shaken cultures with ammonium and incubated in the absence of combined nitrogen for 40 h. Settings to capture GFP fluorescence were identical only for the three strains reporting GFP from the same promoter fragment. Arrows point to some (pro)heterocysts. Size marker, 10 μm.
Effect of NtcA and HetR on in vitro transcription from the two devB promoters.
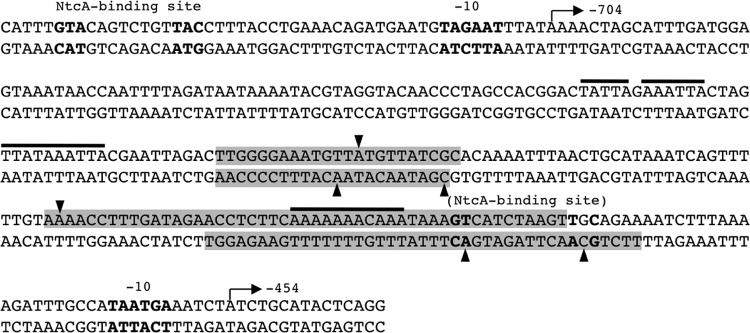
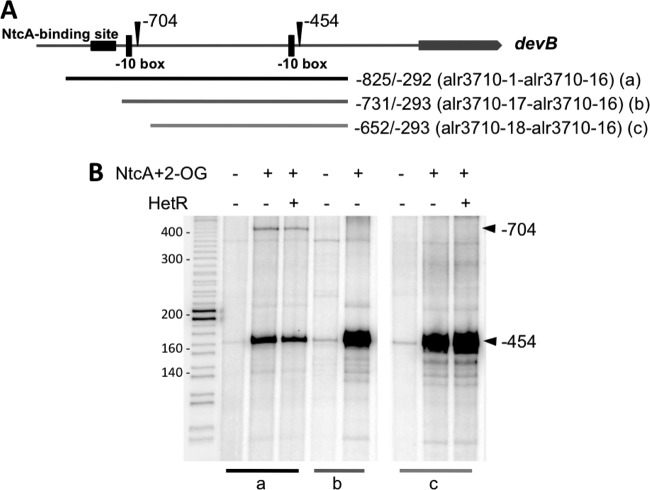
The −704 TSP of the devB operon is preceded by a canonical NtcA-activated promoter, including a −10 box and a consensus NtcA-binding site found 22 nt upstream from the −10 box (14) (see Fig. 7 below). We have previously shown that NtcA and 2-OG are necessary and sufficient for in vitro transcription from this promoter using the cloned Anabaena RNAP holoenzyme including the main sigma factor SigA (34). Here, we have assayed in vitro transcription, and the effect of purified NtcA protein, with different DNA fragments including the two promoters or only the proximal devB promoter. As expected, when a DNA fragment including the distal promoter was used, a transcript of 411 nt (the distance between the −704 TSP and the end of the fragment) was obtained, provided that NtcA and 2-OG were present, irrespective of the presence or absence of HetR (Fig. 3, fragment a).
Fig 7.
DNA sequence of the promoter region of the devB gene. The regions of NtcA interaction, as deduced from DNase I footprinting, between the two identified TSPs are indicated with shading, and some hypersensitive positions are indicated with arrowheads. Arrows denote the TSPs, and horizontal lines denote A+T stretches (see the text). Putative −10 boxes, the distal promoter NtcA-binding site, and the sequence upstream of the proximal promoter resembling an NtcA-binding site (see the text) are also indicated.
Fig 3.
Effect of NtcA and HetR on in vitro transcription from the two promoters of the devB gene. (A) Diagram of the DNA fragments (a to c) used in in vitro transcription assays. Approximate locations of the TSPs at positions −454 and −704, the putative −10 promoter boxes, and a consensus NtcA-binding site are indicated. (B) In vitro runoff transcription assays were performed with the three DNA fragments of the devB genomic region represented in panel A in the presence or absence of NtcA plus 2-OG and HetR. NtcA (130 nM), supplemented with 2-OG (0.6 mM) and HetR (28 nM) as indicated, was incubated with the DNA fragment before addition of RNAP (27 nM) and SigA (88 nM). Arrowheads indicate the full transcript encompassing positions from the corresponding TSP to the end of the DNA fragment used. Size markers (in nucleotides) are indicated to the left.
The −454 TSP of devB is separated by 5 nt of a TAATGA sequence that may represent a −10 box. No other promoter determinant or consensus NtcA-binding site could be recognized upstream from this TSP (see Fig. 7). Surprisingly, when different DNA fragments encompassing the −454 TSP and positions upstream from it (either including the distal promoter or not) were used, NtcA (in the presence of 2-OG) was again necessary and sufficient for effective production of transcripts initiated at the proximal TSP (161 nt from the TSP to the end of the fragment). When a fragment including only the proximal TSP was used, HetR had a small, but reproducible, positive effect on transcription (Fig. 3, see results with fragment c). It was also observed that transcription from the proximal TSP was more efficient when the two fragments lacking the distal promoter, either the one lacking the consensus NtcA-binding site (fragment b) or the one lacking the whole promoter and the −704 TSP (fragment c), were used (Fig. 3).
Binding of NtcA to DNA upstream from the proximal TSP of devB.
Given that NtcA was required for effective in vitro transcription from this TSP and although no consensus NtcA-binding site is present upstream from the −454 TSP of devB, we investigated direct binding of NtcA to the DNA region between the two devB TSPs. Different DNA fragments of this region were used in EMSA and DNase I footprinting assays with purified NtcA in the presence or absence of 2-OG. In EMSA, a DNA fragment including the sequences between the two devB TSPs but excluding the consensus NtcA-binding site (Fig. 4A, fragment a) was effectively retarded by purified NtcA protein in a concentration-dependent manner (Fig. 4B) and 2-OG had a positive effect on retardation (Fig. 4C). Two retarded bands could be detected, with the slower one requiring higher concentrations of NtcA and showing a stronger dependence on 2-OG. Addition of an excess of an unlabeled DNA fragment including a consensus NtcA-binding site (Fig. 4A, fragment b) or of unlabeled fragment a (Fig. 4A) decreased the fraction of the fragment that was retarded, whereas an excess of an unrelated fragment (PpsbA) had no effect (Fig. 4D). Thus, binding of NtcA to the DNA between the two devB TSPs appears to be specific.
In DNase I footprinting assays, NtcA induced an altered digestion pattern in both strands in the DNA region between the two devB TSPs. Two main windows of changes were observed, with some positions exhibiting hypersensitivity to digestion (Fig. 5). No consistent changes were found outside this stretch using other DNA fragments that included the rest of the DNA region between the two TSPs (not shown). Taken together, results of EMSA and DNase I footprinting indicate a direct interaction of NtcA, possibly at two sites, with DNA in the proximal promoter of devB.
Fig 5.
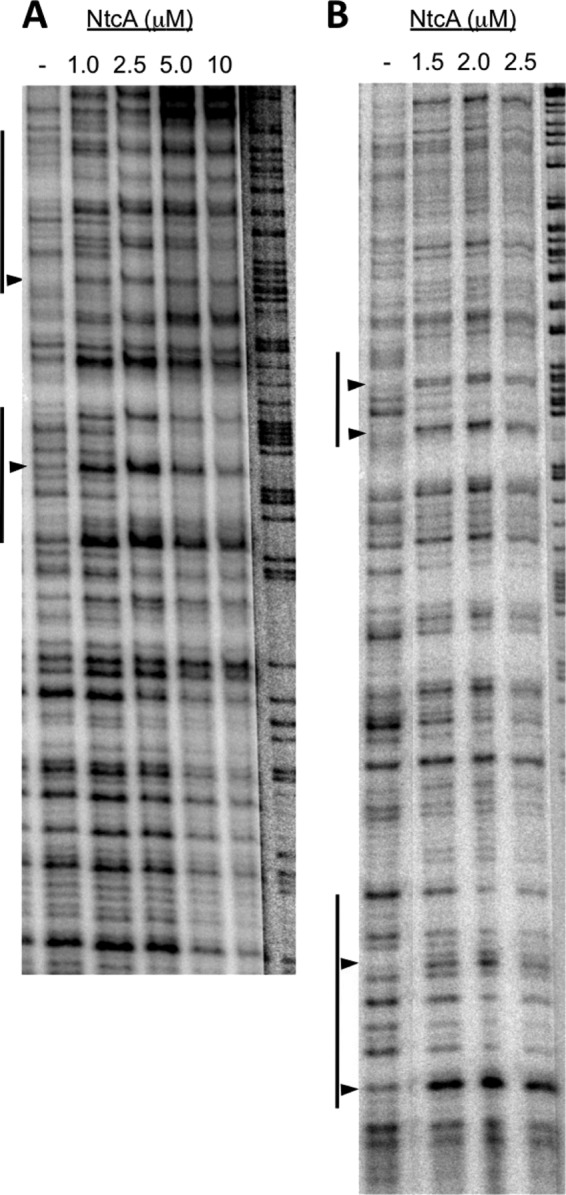
DNase I footprinting of NtcA on DNA sequences between the two TSPs of the devB gene. DNase I footprinting assays were carried out with purified NtcA and DNA fragments of the devB promoter region that were amplified by PCR using primers alr3710-17 (32P labeled) and alr3710-26 (unlabeled) (upper strand, panel A) or alr3710-17 (unlabeled) and alr3710-19 (32P labeled) (lower strand, panel B) in the presence of 0.6 mM 2-OG. Except for NtcA, final concentrations of all components were the same in the different reaction mixtures. Vertical lines denote DNA stretches showing changes due to the presence of NtcA. Arrowheads point to some hypersensitive positions.
Effect of HetR on NtcA binding to DNA in the proximal devB promoter.
Given the positive effect of HetR on in vivo transcription from the proximal TSP of devB, we tested whether purified HetR protein interacted with DNA sequences upstream from −454. HetR alone had no noticeable effect on DNA mobility in EMSA (not shown) or on the pattern of DNase I digestion (Fig. 6). However, when it was added together with NtcA in DNase I footprinting assays, we found a small but reproducible effect of HetR potentiating the protection that could be observed with NtcA alone (Fig. 6, see windows indicated with vertical lines). This effect was observed with different molar ratios of NtcA and HetR, and although it increased with increasing concentrations of HetR (compare lanes with 71 and 286 nM HetR in the presence of NtcA), the observation of footprint signals required a threshold concentration of NtcA, which itself produced a footprint in the absence of HetR. These results suggest that HetR has a positive effect on NtcA binding to the tested DNA.
Fig 6.
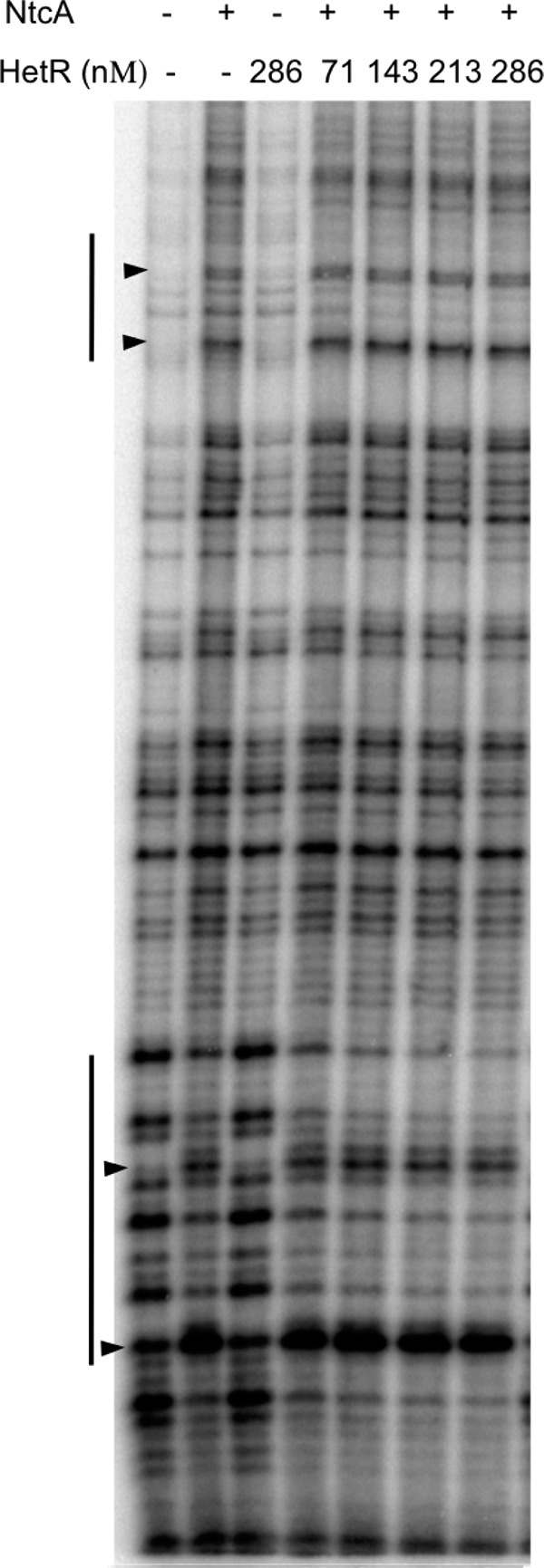
Effect of HetR on DNase I footprinting of NtcA on DNA sequences between the two TSPs of the devB gene. DNase I footprinting assays were carried out with purified NtcA (1 μM) and HetR, as indicated, and a DNA fragment of the devB promoter region that was amplified by PCR using primers alr3710-17 (unlabeled) and alr3710-19 (32P labeled) (lower strand) in the presence of 0.6 mM 2-OG. Except for NtcA and HetR, final concentrations of all components were the same in the different reaction mixtures. Vertical lines denote windows of protection. Arrowheads point to some hypersensitive positions.
DISCUSSION
In the model cyanobacterium Anabaena sp. strain PCC 7120, the differentiation of the N2 fixation-specialized heterocysts involves the activation of many genes following a precise temporal sequence as well as cell specificity along the filament. Many of these genes bear several consecutive promoters differentially affected by the global N regulator NtcA and the heterocyst differentiation regulator HetR. Deciphering the molecular mechanism of activation of each of those promoters appears key to the understanding of the spatiotemporal specificity of gene activation during heterocyst differentiation. However, given the mutual dependence of ntcA and hetR activation (28), it is difficult to distinguish by in vivo studies which of the regulators plays a direct role in a given promoter. Here, we have investigated, by primer extension analyses and gfp transcriptional fusions, the specificity of the expression and its in vivo requirement for NtcA and HetR and have used a defined in vitro transcription system to examine a direct role of NtcA and HetR on the two promoters of the devBCA operon.
At the whole-filament level (primer extension analyses), the distal promoter directing transcription initiation at −704 exhibits in vivo a stringent requirement for NtcA and is not significantly affected by a lack of HetR. Consistently, in vitro transcription from this TSP is observed in the presence of the Anabaena RNAP and purified NtcA with its effector 2-OG. Upstream from this TSP, a consensus NtcA-binding site that is separated by 22 nucleotides from the −10 promoter box is found. Thus, the distal promoter of devBCA represents a canonical class II NtcA-activated promoter. With GFP as a reporter, in vivo expression from this upstream promoter is detected in all cells of the filament with a requirement for NtcA, but expression is increased in proheterocysts dependent on HetR. Thus, the effect of HetR on the localized expression of this promoter may be indirect due to the HetR-dependent increase in NtcA expression that takes place specifically in the differentiating cells (29). On the other hand, the in vivo accumulation of RNA molecules with a 5′ end corresponding to position −454 has a strong requirement for NtcA and is impaired in hetR mutants. No promoter determinant, except from a putative −10 box with the sequence TAATGA, can be recognized upstream from this −454 TSP. No consensus NtcA-binding site can be identified upstream from this position either. With GFP as a reporter, activation of this downstream promoter is confined to the differentiating cells with a requirement for NtcA and HetR. Thus, the in vivo requirement for NtcA of transcription from the proximal devB promoter could, at least in part, respond to an indirect effect exerted through HetR.
Surprisingly, in spite of the lack of consensus NtcA-binding sites, in vitro transcript production from the proximal TSP, even in the absence of the distal promoter and its associated NtcA-binding site, requires NtcA. Moreover, EMSA and DNase I footprinting analyses showed direct binding of NtcA to DNA sequences between the proximal and the distal TSPs of devB. These results indicate that NtcA directly activates transcription of devB from the proximal promoter.
Binding of NtcA likely takes place at two different DNA sites (Fig. 7). The proximal region of NtcA interaction includes the sequence GTCATCTAAGTTGC, which resembles the consensus of NtcA-binding sites (bold letters denote nucleotides matching the consensus of NtcA-binding sites). This sequence is centered 42.5 nucleotides upstream from the TSP, a position of NtcA-binding sites in class II NtcA-activated promoters.
Concerning the in vitro effect of HetR, although no binding of purified HetR alone to the DNA sequences between the two devB TSPs was detected, when added together with NtcA, HetR could potentiate NtcA binding to DNA in the devB proximal promoter. Taken together, our results are consistent with transcription activation from the devB proximal promoter taking place by NtcA bound at the −42.5 sequence, aided by interaction with an upstream bound NtcA molecule and/or by HetR. This resembles the situation of the NtcA homolog CRP that, at certain promoters dependent on the Sxy factor, binds to low-affinity, degenerated CRP-binding sites, the so-called CRP-S sites, and activates transcription aided by the Sxy protein (9). In some of these promoters, A+T-rich sequences upstream from or in the upstream end of CRP-S sites, resembling UP elements, have been implicated in promoter activation (10). Notably, several A+T runs can be found upstream of the putative suboptimal NtcA-binding regions in the devB proximal promoter (Fig. 7). Other factors have been described to contribute to the binding of CRP to DNA and transcription activation, as is the case for MelR at the E. coli melAB promoter (38; for a review, see reference 3). In the case of HetR, protein-protein contacts, involving NtcA and/or RNAP, would be consistent with the crystallographic structure of the protein that shows flexible flap domains proposed to represent contact sites for DNA and/or other proteins (22). It is worth pointing out that although the in vitro effects of HetR on both NtcA binding to DNA and transcription activation at the proximal devB promoter are small, they could be stronger in vivo depending on the amount of the NtcA and HetR proteins or on a specific DNA topology. Also, the possibility that the cloned HetR protein that we are using is not fully active, if a posttranslational modification of HetR took place in Anabaena, should be taken into account. In summary, the available data are consistent with the notion that the proximal devB promoter is a suboptimal NtcA-activated promoter in which HetR can act as a coactivator improving NtcA interaction with DNA, with the physiological effect of confining the activity of this promoter to conditions of maximum NtcA and HetR levels.
Considering globally the expression of the devBCA operon from its dual promoter region, upon ammonium deprivation, the upstream canonical class II promoter would be activated by NtcA at a low level in all cells of the filament, thereafter increasing in the differentiating cells in response to the localized increase in NtcA levels that results from localized increase in the HetR levels and the mutual activation loop of the hetR and ntcA genes. The proximal devB promoter would also be directly activated by NtcA, with a possible positive direct effect of HetR. Thus, through direct effects of NtcA on both promoters, an indirect effect of HetR in the distal one, and a direct effect of HetR in the proximal one, both promoters would contribute to locating the expression of the devBCA operon to the differentiating heterocysts.
ACKNOWLEDGMENTS
We thank Alicia Orea, Vicente Mariscal, and Victoria Merino-Puerto for assistance with confocal microscopy.
The work was supported by grants BFU2007-60457 and BFU2010-17980, cofinanced by FEDER, from the Ministerio de Ciencia e Innovación (Spain).
Footnotes
Published ahead of print 30 March 2012
REFERENCES
- 1.Aldea MR, Kumar K, Golden JW. 2008. Heterocyst development and pattern formation, p 75–90 In Winans SC, Bassler BL. (ed), Chemical communication among bacteria. ASM Press, Washington, DC [Google Scholar]
- 2.Ausubel FM, et al. 2012. Current protocols in molecular biology. Greene Publishing & Wiley-Interscience, New York, NY [Google Scholar]
- 3.Barnard A, Wolfe A, Busby S. 2004. Regulation of complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102–108 [DOI] [PubMed] [Google Scholar]
- 4.Black TA, Cai Y, Wolk CP. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77–84 [DOI] [PubMed] [Google Scholar]
- 5.Buikema WJ, Haselkorn R. 1991. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 5:321–330 [DOI] [PubMed] [Google Scholar]
- 6.Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199–213 [DOI] [PubMed] [Google Scholar]
- 7.Cai Y, Wolk CP. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Y, Wolk CP. 1997. Anabaena sp. strain PCC 7120 responds to nitrogen deprivation with a cascade-like sequence of transcriptional activations. J. Bacteriol. 179:267–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameron ADS, Redfield RJ. 2006. Non-canonical CRP sites control competence regulons in Escherichia coli and many other γ-proteobacteria. Nucleic Acids Res. 34:6001–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron ADS, Redfield RJ. 2008. CRP and transcription activation at CRP-S sites. J. Mol. Biol. 383:313–323 [DOI] [PubMed] [Google Scholar]
- 11.Elhai J, Wolk CP. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119–138 [DOI] [PubMed] [Google Scholar]
- 12.Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiedler G, Arnold M, Hannus S, Maldener I. 1998. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 27:1193–1202 [DOI] [PubMed] [Google Scholar]
- 14.Fiedler G, Muro-Pastor AM, Flores E, Maldener I. 2001. NtcA-dependent expression of the devBCA operon, encoding a heterocyst-specific ATP-binding cassette transporter in Anabaena spp. J. Bacteriol. 183:3795–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores E, Herrero A. 2010. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 8:39–50 [DOI] [PubMed] [Google Scholar]
- 16.Frías JE, Flores E, Herrero A. 1994. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 14:823–832 [DOI] [PubMed] [Google Scholar]
- 17.Herrero A, Muro-Pastor AM, Flores E. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero A, Muro-Pastor AM, Valladares A, Flores E. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469–487 [DOI] [PubMed] [Google Scholar]
- 19.Higa KC, Callahan SM. 2010. Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. PCC 7120. Mol. Microbiol. 77:562–574 [DOI] [PubMed] [Google Scholar]
- 20.Huang X, Dong Y, Zhao J. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. U. S. A. 101:4848–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ionescu D, Voss B, Oren A, Hess WH, Muro-Pastor AM. 2010. Heterocyst-specific transcription of NsiR1, a non-coding RNA encoded in a tandem array of direct repeats in cyanobacteria. J. Mol. Biol. 398:177–188 [DOI] [PubMed] [Google Scholar]
- 22.Kim Y, et al. 2011. Structure of transcription factor HetR required for heterocyst differentiation in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 108:10109–10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Igual R, Flores E, Herrero A. 2010. Inactivation of a heterocyst-specific invertase indicates a principal role of sucrose catabolism in heterocysts of Anabaena sp. J. Bacteriol. 192:5526–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luque I, Flores E, Herrero A. 1994. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 13:2862–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meeks JC, Elhai J. 2002. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol. Mol. Biol. Rev. 66:94–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohamed A, Jansson C. 1989. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 13:693–700 [DOI] [PubMed] [Google Scholar]
- 27.Muro-Pastor AM, Flores E, Herrero A. 2009. NtcA-regulated heterocyst differentiation genes hetC and devB from Anabaena sp. strain PCC 7120 exhibit a similar tandem promoter arrangement. J. Bacteriol. 191:5765–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muro-Pastor AM, Valladares A, Flores E, Herrero A. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 44:1377–1385 [DOI] [PubMed] [Google Scholar]
- 29.Olmedo-Verd E, Muro-Pastor AM, Flores E, Herrero A. 2006. Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 188:6694–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olmedo-Verd E, Valladares A, Flores E, Herrero A, Muro-Pastor AM. 2008. Role of two NtcA-binding sites in the complex ntcA gene promoter of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 190:7584–7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Partensky F, Hess WR, Vaulot D. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1–61 [Google Scholar]
- 33.Risser DD, Callahan SM. 2007. Mutagenesis of hetR reveals amino acids necessary for HetR function in the heterocystous cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 189:2460–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valladares A, Flores E, Herrero A. 2008. Transcription activation by NtcA and 2-oxoglutarate of three genes involved in heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 190:6126–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valladares A, et al. 2011. Specific role of the cyanobacterial PipX factor in the heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 193:1172–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vázquez-Bermúdez MF, Flores E, Herrero A. 2002. Analysis of binding sites for the nitrogen-control transcription factor NtcA in the promoters of Synechococcus nitrogen-regulated genes. Biochim. Biophys. Acta 1578:95–98 [DOI] [PubMed] [Google Scholar]
- 37.Vázquez-Bermúdez MF, Herrero A, Flores E. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 512:71–74 [DOI] [PubMed] [Google Scholar]
- 38.Wade JT, Belyaeva TA, Hyde EI, Busby SJW. 2001. A simple mechanism for co-dependence on two activators at an Escherichia coli promoter. EMBO J. 20:7160–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei T-F, Ramasubramanian TS, Golden JW. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 176:4473–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolk CP, Ernst A, Elhai J. 1994. Heterocyst metabolism and development, p 769–823 In Bryant DA. (ed), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 41.Zhao M-X, et al. 2010. Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate. Proc. Natl. Acad. Sci. U. S. A. 107:12487–12492 [DOI] [PMC free article] [PubMed] [Google Scholar]