Abstract
To successfully establish an infection, Acinetobacter baumannii must overcome the iron starvation and oxidative stress imposed by the human host. Although previous studies have shown that ATCC 19606T cells acquire iron via the acinetobactin-mediated siderophore system, little is known about intracellular iron metabolism and its relation to oxidative stress in this pathogen. Screening of an insertion library resulted in the isolation of the ATCC 19606T derivative 1644, which was unable to grow in iron-chelated media. Rescue cloning and DNA sequencing showed that the insertion inactivated a gene coding for an NfuA Fe-S cluster protein ortholog, without any effect on the expression of the acinetobactin system. The nfuA mutant was also more sensitive to hydrogen peroxide and cumene hydroperoxide than the parental strain. The iron chelation- and oxidative-stress-deficient responses of this mutant were corrected when complemented with either the ATCC 19606T parental allele or the Escherichia coli MG1655 nfuA ortholog. Furthermore, electron paramagnetic resonance (EPR) and inductively coupled plasma-atomic emission spectroscopy (ICP-AES) analyses showed that the ATCC 19606T NfuA ortholog has iron-binding properties compatible with the formation of [Fe-S] cluster protein. Ex vivo and in vivo assays using human epithelial cells and Galleria mellonella, respectively, showed that NfuA is critical for bacterial growth independent of their capacity to acquire iron or the presence of excess of free iron. Taken together, these observations indicate that the A. baumannii NfuA ortholog plays a role in intracellular iron utilization and protection from oxidative-stress responses that this pathogen could encounter during the infection of the human host.
INTRODUCTION
Iron-sulfur [Fe-S]-containing proteins represent a major pool of iron found within the cell. These proteins play a role in a broad range of biological processes, such as electron transfer reactions, substrate binding, nonredox catalysis, sulfur donation, and sensing of redox status for regulatory processes (13). The cellular concentrations of iron and sulfide needed for spontaneous formation of [Fe-S] cluster complexes are toxic for cells. Therefore, cells have developed intricate biosynthetic pathways to assemble these clusters. Three distinct [Fe-S] biosynthesis pathways have been identified in bacteria: Isc (iron-sulfur cluster), Suf (sulfur mobilization), and Nif (nitrogen fixation) (13, 40). The Isc system appears to be the housekeeping [Fe-S] cluster assembly system within both prokaryotic and eukaryotic systems, while the Suf system appears to be in place for the adaptation of [Fe-S] cluster formation under iron starvation and oxidative-stress conditions (3, 28, 33). The Nif system is generally specific to the maturation of [Fe-S] proteins in nitrogen-fixing organisms, such as Azotobacter vinelandii (4), while both the Isc and Suf [Fe-S] assembly pathways are present in bacteria such as Escherichia coli.
The basics of [Fe-S] cluster assembly for all three pathways can be divided into three main steps: formation of elemental sulfur, iron and sulfur cluster assembly, and cluster insertion into apoproteins (40). All pathways involve a cysteine desulfurase and a U-type and/or A-type scaffold protein capable of assembling both [2Fe-2S] and [4Fe-4S] clusters (3). U-type scaffold proteins, such as NifU and IscU, preferentially bind [2Fe-2S] clusters while A-type scaffold proteins, such as SufA and IscA, can bind [2Fe-2S] monomeric clusters, which then mature to [4Fe-4S]-containing proteins, as it is in the case of aconitase (23, 37). More recent studies led to the observation that E. coli (1) and A. vinelandii (4) produce an additional protein involved in [Fe-S] biogenesis that was named NfuA. Interestingly, this protein has a hybrid structure with a conserved C-terminal Nfu domain but a degenerate N-terminal A-type domain, which lacks critical Cys residues. This protein proved to work as an intermediate [Fe-S] carrier and plays a role in iron starvation and oxidative stress when tested in vitro (1, 4).
Acinetobacter baumannii is the most prevalent Acinetobacter species associated with human infections, mainly in hospitalized and compromised patients (6, 29). The clinical importance of this pathogen could be associated with its ability to form biofilms on medically relevant materials (38), to persist in a variety of environments, including those that are severely desiccated (19), and to grow under iron-limiting conditions such as those imposed by the human host (7, 10, 41). In spite of this progress, there is a significant gap in the understanding of basic metabolic processes expressed by A. baumannii and their involvement in the pathogenesis of the infections this bacterium causes in the human host, a situation that is further compounded by the continuous emergence of multidrug- and pan-drug-resistant isolates (17). Therefore, it is important to study some of these processes not only to understand basic aspects of the physiology of this bacterium but also to exploit them as potential targets to develop more effective and convenient strategies to prevent and treat the infections A. baumannii causes in humans worldwide.
In this work, we describe the analysis of the A. baumannii ATCC 19606T NfuA ortholog, which was identified by screening a mutant library for derivatives impaired in their ability to grow under iron-chelated conditions. The 1644 nfuA isogenic insertion derivative showed a deficient response to iron chelation and oxidative stress, defects that were corrected after genetic complementation with the parental allele or a copy of the bona fide E. coli MG1655 ortholog. The ATCC 19606T NfuA, which displays chemical properties compatible with an [Fe-S] cluster protein, was critical for the ability of A. baumannii to persist in the presence of human epithelial cells and kill Galleria mellonella caterpillars. Taken together, all these observations indicate that the A. baumannii NfuA protein plays key roles in intracellular iron metabolism and virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. Luria-Bertani (LB) broth and agar (32) were used to maintain all bacterial strains. M9 minimal medium (25) was used for growth under chemically defined conditions. Iron-rich and iron-deficient conditions were obtained by adding FeCl3 and the synthetic iron chelator 2,2′-dipyridyl (DIP), respectively, to LB or M9 minimal medium. The MIC of the iron chelator DIP was determined in LB broth and M9 minimal medium containing increasing concentrations of this chelator. Sensitivity to oxidation was determined by the ability of bacterial cells to grow in the presence of increasing concentrations of H2O2 or cumene hydroperoxide. Cell growth was determined spectrophotometrically at 600 nm after overnight incubation (12 to 14 h) at 37°C in a shaking incubator set at 200 rpm.
Table 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Relevant characteristica | Source/reference |
|---|---|---|
| Strains | ||
| A. baumannii | ||
| 19606T | Clinical isolate, type strain | ATCC |
| 19606T s1 | basD::aph, 19606T acinetobactin-production deficient derivative; Kmr | (10) |
| 19606T 1644 | nfuA::EZ::TN<R6Kγori/KAN-2> derivative of 19606T; Kmr | This work |
| 1644-944 | 1644 insertion mutant harboring pMU944; Apr | This work |
| 1644-952 | 1644 insertion mutant harboring pMU952; Apr | This work |
| E. coli | ||
| MG1655 | F− λ− ilvG rfb-50 rph-1 | F. R. Blattner |
| DH5α | Used for DNA recombinant methods | Gibco-BRL |
| EC100D+ | pir+, host for plasmid rescue maintenance | Epicentre |
| TOP10 | Used for DNA recombinant methods | Invitrogen |
| BL21(DE3) | λDE3, T7 RNA polymerase | Invitrogen |
| Plasmids | ||
| pCRBlunt II | PCR cloning vector; Kmr Zeor | Invitrogen |
| pWH1266 | Shuttle vector; Apr Tetr | (20) |
| pET100D | T7 expression vector; Apr | Invitrogen |
| pMU903 | Plasmid rescue of self-ligation of EcoRI-digested DNA from mutant 1644; Kmr | This work |
| pMU937 | pCRBlunt II harboring nfuA from 19606T; Kmr | This work |
| pMU941 | pET100D harboring the nfuA coding region; Apr | This work |
| pMU944 | pWH1266 harboring nfuA from 19606T; Apr Tets | This work |
| pMU949 | pCRBlunt II harboring nfuA from MG1655; Kmr | This work |
| pMU952 | pWH1266 harboring nfuA from MG1655; Apr Tets | This work |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Tetr, tetracycline resistance; Tets, tetracycline sensitivity; and Zeor, zeocin resistance.
General DNA procedures.
Total DNA was isolated either by ultracentrifugation in CsCl density gradients (24) or by using a miniscale method adapted from previously published research (5). Plasmid DNA was isolated using commercial kits (Qiagen). DNA digests were performed with restriction enzymes as indicated by the supplier (New England BioLabs) and size fractionated by agarose gel electrophoresis (32).
Transposition mutagenesis and rescue cloning of interrupted genes.
A. baumannii ATCC 19606T was mutagenized using the EZ::TN<R6Kγori/KAN-2> Tnp transposition mutagenesis system (Epicentre) and electroporation as described previously (9). Transformants that grew after plating on LB agar containing 40 μg/ml kanamycin (Km) were transferred onto LB agar plates containing 100 μM DIP using a toothpick. Derivatives displaying impaired growth under iron-chelated conditions after incubation at 37°C for 24 h were further analyzed. The genomic regions harboring the insertion of the EZ::TN<R6Kγori/KAN-2> transposon were rescued by self-ligation of EcoRI-digested chromosomal DNA and electroporation into E. coli EC100D pir+ cells. Plasmid DNA (pMU903) isolated from a colony that grew on LB agar containing 40 μg/ml Km was used as a template to determine the nucleotide sequence of the genomic DNA flanking the transposon element with the primers complementary to this insertion element that were supplied with the mutagenesis kit. Further extension of nucleotide sequences was done using custom-designed primers 3642 and 3643 (Table 2) using standard BigDye-based automated DNA sequencing (Applied Biosystems). Sequences were examined and assembled using Sequencher (Gene Codes). Nucleotide and amino acid sequences were analyzed with DNASTAR, BLAST, and analysis tools available through the ExPASy Molecular Biology Server (http://www.expasy.ch/).
Table 2.
Primers used in this study
| No. | Nucleotide sequencea |
|---|---|
| 3273 | 5′-AGGCTATGCATTAGGCAC-3′ |
| 3353 | 5′-TGCTGGTAAAGCTGCTAC-3′ |
| 3545 | 5′-TACAGAAAGCTGGTGCATGG-3′ |
| 3546 | 5′-TGCACCATTTGTGCCTGTAG-3′ |
| 3642 | 5′-GATCAGACGGTTCATGAC-3′ |
| 3643 | 5′-CACCACTTCTTCTGGTGC-3′ |
| 3651 | 5′-GGATCCGATCAGACGGTTCATGAC-3′ |
| 3652 | 5′-GGATCCTCACTTGCTCTTCTGCTG-3′ |
| 3673 | 5′-CACCATGTCGACTGAGAACACC-3′ |
| 3674 | 5′-TTTGAAATAAGCACCTTC-3′ |
| 3681 | 5′-GGATCCCGTTATCACGCTGGTTGC-3′ |
| 3682 | 5′-GGATCCCACGACTGATACCCACTA-3′ |
| 3712 | 5′-CACCATCTGCACAAGAGT-3′ |
Underlined sequences indicate added BamHI restriction sites.
Genetic complementation of an nfuA mutant.
The A. baumannii ATCC 19606T nfuA gene was PCR amplified from the parental strain genome with Phusion DNA polymerase (New England BioLabs) and the primers 3651 and 3652, which include BamHI restriction sites (Table 2). The nfuA gene from E. coli MG1655 was amplified from the chromosome of this strain with Phusion DNA polymerase and the primers 3681 and 3682, which include BamHI restriction sites (Table 2). The blunt-ended amplicons were ligated into pCR-Blunt II and transformed into E. coli TOP10. The cloned ATCC 19606T and MG1655 nfuA genes were subcloned from pMU937 and pMU949, respectively, into the shuttle vector pWH1266 as BamHI restriction fragments to generate the cognate derivatives pMU944 and pMU952, respectively. The recombinant derivatives were electroporated as described before (9) into the A. baumannii ATCC 19606T 1644 derivative harboring a transposon insertion within the nfuA gene. Transformants that grew after overnight incubation at 37°C on LB agar containing 500 μg/ml ampicillin were tested for their ability to grow under iron-limiting conditions and in the presence of peroxides. The presence of pMU944 and pMU952 in the complemented strains was confirmed by restriction analysis of plasmid DNA isolated from cells grown in LB broth containing 100 μg/ml ampicillin.
Overproduction and purification of recombinant NfuA.
The nfuA coding region was PCR amplified using ATCC 19606T total DNA as a template, Phusion DNA polymerase, and the primers 3673 and 3674 (Table 2). The amplicon was ligated into pET100D and transformed into E. coli TOP10. Plasmid DNA from an ampicillin-resistant colony (pMU941) was isolated, sequenced with the T7 forward and reverse primers provided with the kit (Invitrogen) to verify the proper orientation of the insert, and transformed into E. coli BL21(DE3). A transformant was used to overexpress the ATCC 19606T N-terminal His-tagged NfuA recombinant derivative as described previously (35). Soluble protein was isolated by Ni2+ affinity column chromatography from cell lysates prepared as recommended by the manufacturer's protocol (Pierce). The identity of the overexpressed protein was confirmed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry of the protein band extracted and analyzed as described before (27). Purified protein was used to raise anti-NfuA polyclonal antibodies in rabbits as reported before (8). LB broth overnight cultures were used to prepare whole-cell lysates as well as total membrane and cytoplasmic fractions as described previously (8). The production of NfuA was examined by Western blotting with specific polyclonal antibodies, detecting the immunocomplexes by chemiluminescence with horseradish peroxidase (HRP)-labeled protein A (8). Differential production of NfuA was determined by densitometry of Western blots as described before (31).
Detection of siderophore compounds and production of the BauA acinetobactin receptor.
The presence of phenolic extracellular compounds in M9 culture supernatants was detected with the Arnow colorimetric assay (2). The production of the siderophore acinetobactin was tested with siderophore utilization bioassays, using the ATCC 19606T acinetobactin-deficient derivative s1 as a reporter strain (10), and high-pressure liquid chromatography (HPLC) analysis. Briefly, supernatants of M9 minimal medium cultures, which reached the same optical density at 600 nm (OD600) after overnight incubation at 37°C in a shaking incubator, were filtered with 0.45 mm cellulose acetate filter units (Spin-X centrifuge filter units; Costar, Cambridge, MA) and examined with an Agilent 1100 LC instrument using a Vydac C-8, 5 μm, 250 mm by 4.6 mm reversed-phase column (Grace Davison Discovery Sciences, Deerfield, IL). The mobile phases were water and acetonitrile containing 0.13% and 0.1% trifluoroacetic acid, respectively. The gradient exposures were as follows: 17% acetonitrile for 5 min; 17% to 30% acetonitrile, with increases in concentration occurring within 30 min; and then 30% for 15 min. Detection was at 317 nm with a flow rate of 0.5 ml/min. The production of the acinetobactin outer membrane receptor protein BauA was examined by Western blotting with specific polyclonal antibodies as described before (10).
Transcriptional analysis of gene expression.
Three independent 1-ml cultures of ATCC 19606T cells grown overnight at 37°C in LB or LB supplemented with 100 μM DIP were combined and used to extract total RNA using hot phenol as described before (26). The total RNA samples were further purified with the RNeasy kit with on-the-column RNase-free DNase treatment (Qiagen). Gene transcription was examined with the Qiagen real-time one-step QuantiTect SYBR green quantitative reverse transcription (qRT)-PCR system by following the manufacturer's recommendations. A 124-bp bauA internal fragment was amplified with primers 3273 and 3353 while primers 3643 and 3712 were used to amplify a 140-bp nfuA internal fragment. A 155-bp recA internal fragment was amplified with primers 3545 and 3546 to serve as an internal control of a constitutively expressed gene. A Bio-Rad iCycler was used under the following conditions: an RT cycle of 50°C for 30 min and 95°C for 15 min, followed by 35 amplification cycles of 94°C for 15 s, 50°C for 30 s, and 72°C for 30 s. The nfuA and bauA transcript levels were normalized to recA levels detected under iron-chelated and nonchelated conditions. This analysis was done in triplicate using samples containing no template or no reverse transcriptase as negative controls.
EPR and metal content analysis.
Electron paramagnetic resonance (EPR) spectroscopy was performed on the purified recombinant NfuA protein at X-band using a Bruker EMX CW-EPR spectrometer, equipped with an ER-4102 (TE102) resonator. Temperature was maintained with an Oxford Instruments ESR-900 cryostat and temperature controller (±0.05 K). The NfuA protein was prepared at a concentration of 250 μM in equilibrium buffer (50 mM NaH2PO4, 0.3 M NaCl, 10 mM imidazole, 10 mM Tris [pH 8.0]), with 20% glycerol (vol/vol) as a glassing agent. The EPR spectrum of purified recombinant NfuA (see Fig. 2) represents the average of 8 scans, using 5-G field modulation (100 kHz), a receiver gain of 105, a microwave frequency of 9.38 GHz (2 mW), and a time constant/conversion time of 82 ms, at a temperature of 10 K. For power-saturation measurements, the same conditions were used, while varying the temperature and microwave power.
Fig 2.
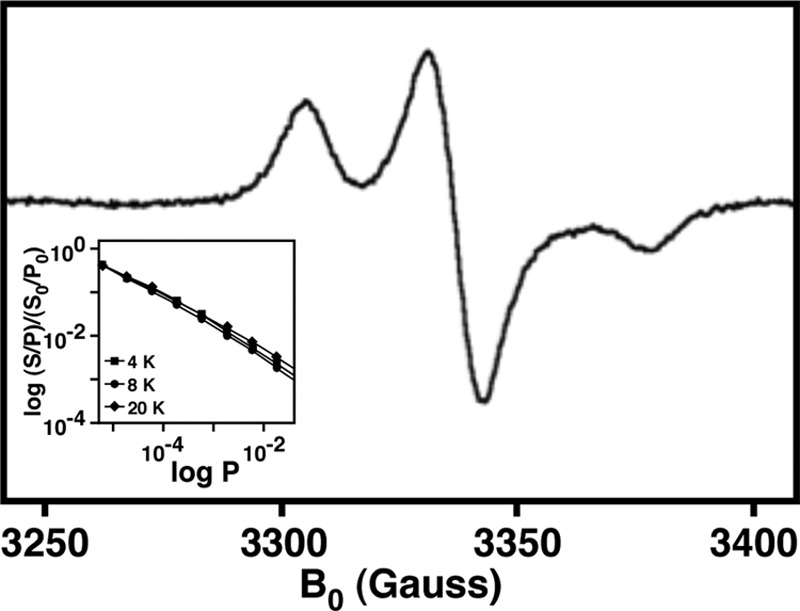
X-band EPR of the A. baumannii ATCC 19606T NfuA recombinant protein. (Inset) Power saturation behavior as a function of temperature.
Inductively coupled plasma-atomic emission spectroscopy (ICP-AES) was used to determine the metal content of as-isolated recombinant NfuA diluted to 6.5 μM in 0.15 M NaCl. Calibration curves were generated using serial dilutions of a Fisher Fe standard. The emission lines at 238.20 and 239.56 nm were compared to ensure the lowest possible detection limits.
A549 infection assays.
A549 human alveolar epithelial cells were cultured and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum at 37°C in the presence of 5% CO2 as previously described (16). Twenty-four-well tissue culture plates were seeded with approximately 104 cells per well and then incubated for 16 h. Bacterial cells were grown 24 h in LB broth at 37°C with shaking at 200 rpm, collected by centrifugation at 15,000 rpm for 10 min, washed, resuspended, and diluted in modified Hanks' balanced salt solution (mHBSS; same as HBSS but without glucose). It is important to note that A. baumannii does not grow in mHBSS when incubated overnight at 37°C in a shaking incubator. Therefore, the colony counts reported reflect the ability of bacteria to persist and grow intracellularly and extracellularly because of nutrients provided by the epithelial cells, which undergo apoptosis after infection (15). The A549 monolayers were either singly infected with 103 cells of the ATCC 19606T parental strain or the 1644 nfuA isogenic mutant or coinfected with 5.0 × 102 cells of the parental strain and the nfuA mutant. Inocula were estimated spectrophotometrically at OD600 and confirmed by plate count. Infected monolayers were incubated 24 h in mHBSS at 37°C in 5% CO2. The tissue culture supernatants were collected, the A549 monolayers were lysed with sterile distilled H2O, and lysates were added to the cognate tissue culture supernatants. Bacteria were collected from the resulting suspensions by centrifugation, resuspended in 1 ml sterile distilled H2O, serially diluted, and then plated on nutrient agar. Samples from coinfection experiments were also plated on nutrient agar containing 50 μg/ml Km to determine 1644 cell counts. After overnight incubation at 37°C, the CFU were counted, and the CFU/ml for each sample were calculated and recorded. Counts were compared using the Student t test; P values of ≤0.05 were considered significant. Replicate experiments were performed using fresh biological samples each time (n = 12).
G. mellonella killing assays.
Bacterial cells previously grown in LB broth were collected by centrifugation and suspended in phosphate-buffered saline solution (PBS). The number of bacterial cells was estimated spectrophotometrically at OD600 and diluted in PBS to appropriate concentrations. All bacterial inocula were confirmed by plating serial dilutions on LB agar and determining colony counts after overnight incubation at 37°C. Ten freshly received final-instar G. mellonella larvae (Grubco, Fairfield, OH) weighing 0.250 to 0.350 g were randomly selected and used in killing assays as we recently described (15). Larvae were injected with 5-μl inocula containing 1 × 105 bacterial cells ± 0.25 log of each tested strain. Each test series included control groups of noninjected larvae or larvae injected with sterile PBS or PBS containing 100 μM FeCl3. The test groups included larvae infected with the parental strain ATCC 19606T or the 1644 nfuA isogenic mutant, which were injected in the absence or presence of 100 μM FeCl3. After injection, the larvae were incubated at 37°C in darkness, assessing death at 24-h intervals over 6 days (15). The experiments were repeated three times using 10 larvae per experimental group, and the resulting survival curves were plotted using the Kaplan-Meier method (21). P values of ≤0.05 were considered statistically significant for the log rank test of survival curves (SAS Institute Inc., Cary, NC).
RESULTS
Isolation and characterization of an iron starvation-deficient derivative.
Screening of the A. baumannii ATCC 19606T EZ::TN<R6Kγori/KAN-2> insertion library generated previously yielded more than 20 derivatives displaying diminished growth on LB agar plates containing 100 μM 2,2′-dipyridyl (DIP) (9, 10) compared with the parental strain. Initial characterization of some of these derivatives resulted in the identification of the t6 and t7 derivatives affected in acinetobactin transport functions (9, 10). Further analysis of some of the uncharacterized iron utilization-deficient mutants resulted in the identification of the derivative 1644. This mutant harbors an EZ::TN<R6Kγori/KAN-2> insertion 26 nucleotides downstream of the initiation codon of the predicted 212-amino-acid coding region annotated as ACIB1v1_270041 (BaumannoScope project information; https://www.genoscope.cns.fr/agc/microscope/about/collabprojects.php?P_id=8) (Fig. 1A). In silico analysis showed that this 23-kDa predicted protein contains N-terminal amino acid sequences related to the SufA (COG0316) and IscA (TIGR02011) domains and C-terminal amino acid sequences related to the Nfu domain (pfam01106) (Fig. 1B). The CXXC motif, which is essential for the in vivo function of NfuA in E. coli and A. vinelandii (1, 4), is also present in the C-terminal region of the ATCC 19606T predicted protein. In contrast, and as it was found in the E. coli NfuA protein (1), the ATCC 19606T predicted protein lacks the three N-terminal conserved C residues described in type-A scaffold proteins. Overall, the predicted amino acid composition of this protein is highly similar (47% identity, 66% similarity) to that of the E. coli NfuA protein, also known as YhgI and GntY, which participates in the formation of [Fe-S] clusters and plays a role in cell responses to iron chelation and oxidative stress (1). Based on these observations and the fact that the 1644 insertion mutant was isolated because of its iron utilization-defective phenotype, this ATCC 19606T protein was designated NfuA.
Fig 1.
(A) Genetic organization of the A. baumannii ATCC 19606T DNA region harboring the nfuA gene. The horizontal arrows and their direction indicate the predicted location of coding regions and their direction of transcription, respectively. The vertical arrow indicates the transposon insertion site in the ATCC 19606T 1644 insertion derivative. Numbers on top of the horizontal line indicate DNA sizes in bp. (B) Predicted amino acid sequence and domains of NfuA. The black bar represents the N terminus SufA/IscA domain. The gray bar represents the C terminus Nfu domain. The two conserved cysteine residues located in the Nfu domain are underlined and in bold.
Sequence analysis of the ATCC 19606T chromosomal region surrounding nfuA showed that upstream and transcribed in the opposite direction of this gene there is a predicted open reading frame (ORF) coding for a protein that contains the pfam 01694 domain, which is present in members of the rhomboid protein family (Fig. 1A). This family includes integral-membrane proteases found in bacteria and eukaryotes, the function of which is not completely understood although they may be involved in protein translocation and bacterial cell signaling (14). Downstream of nfuA and separated by a 172-nucleotide intergenic region there is a predicted gene coding for a 754-amino acid protein significantly similar (E values less than 1 × e−5) to TonB-dependent/ligand-gated outer membrane proteins related to bacterial siderophore receptors (Fig. 1A). BLAST searches showed that the ATCC 19606T NfuA protein is highly related to orthologs potentially produced by a wide range of bacteria, with the predicted products of A. baumannii and Acinetobacter baylyi ADP1 being the top matches. These searches also showed that there is synteny between the three genes shown in Fig. 1A and those present in the chromosome of the A. baumannii strains AYE, ACICU, AB0057, ATCC 17978, and SDF as well as the genome of Acinetobacter spp. DR1 and A. baylyi ADP1.
The ATCC 19606T NfuA protein binds a [Fe-S] cluster.
The EPR spectrum of purified recombinant NfuA, shown in Fig. 2, is consistent with that expected for an [Fe-S] cluster (11). The rhombic spectrum is characterized by g = [2.03, 2.01, 1.99], which represents the three-dimensional distribution of unpaired electron spin in the [Fe-S] cluster. Based on power saturation measurements at multiple temperatures (Fig. 2, inset) and comparisons to preceding studies of NfuA from E. coli and A. vinelandii, we can assign the behavior of the ATCC 19606T recombinant NfuA to that of a [4Fe-4S] ferredoxin-type cluster (36). The iron content of the NfuA protein of 5.5% ± 2% determined by ICP-AES is further suggestive of the protein's ability to bind a [Fe-S] cluster as purified by Ni chromatography.
Cell location and differential production of NfuA.
Western blotting of whole-cell lysate proteins with polyclonal anti-NfuA antibodies showed that the iron starvation-deficient phenotype of the 1644 mutant is associated with the absence of a protein band that displays a 29-kDa relative mass (Fig. 3A), which is larger than the theoretical mass deduced from the DNA sequence. This difference is similar to that of the E. coli NfuA ortholog that displays a relative mass 3 kDa larger than the theoretical size (1). Analysis of different subcellular fractions showed that the cytoplasmic sample contains all detectable NfuA (data not shown), a location that agrees with that predicted using standard bioinformatic tools, although it was annotated as a putative membrane-bound protein in the genome of the A. baumannii AYE, SDF, and ATCC 17978 strains (34, 39).
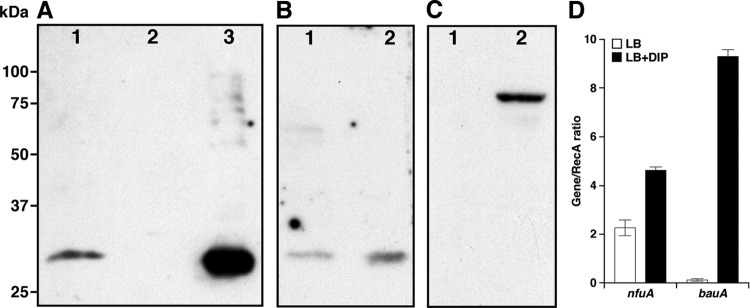
Fig 3.
Transcriptional and translational expression of nfuA and bauA. (A) Whole-cell lysate proteins samples prepared from cells of the parental ATCC 19606T strain (lane 1) or the nfuA 1644 mutant harboring no plasmid (lane 2) or transformed with the complementing plasmid pMU944 (lane 3) cultured in LB broth were probed with anti-NfuA antibodies. (B) ATCC 19606T whole-cell lysate protein samples prepared from cells cultured in LB broth containing either 100 μM FeCl3 (lane 1) or 100 μM DIP (lane 2) were probed with anti-NfuA antibodies. (C) The same protein samples used in panel B were probed with anti-BauA antiserum to detect the production of the BauA acinetobactin receptor. The electrophoretic mobility of molecular weight markers is shown on the left-hand side of panel A. (D) RNA isolated from bacteria grown overnight (12 to 14 h) at 37°C in a shaking incubator in LB without or with the addition of 100 μM DIP was used as the template for qRT-PCR using nfuA- and bauA-specific primers. Transcription of recA was used as a constitutively expressed internal control. The error bars show the standard errors (SE) of the means of an assay done in triplicate.
Probing of nitrocellulose blots containing size-fractionated whole-cell lysate proteins with the anti-NfuA antiserum showed that the presence of 100 μM DIP in the culture medium results in a 3-fold increase, as determined by densitometry, in NfuA production compared with cells cultured under iron-rich conditions (Fig. 3B). Western blot analysis of the same protein samples with anti-BauA antibodies showed that although iron repressed, the control of NfuA production by excess of free iron is more relaxed than that of BauA, which is detected only in cells cultured in the presence of 100 μM DIP (Fig. 3, compare panels B and C). This observation is further supported by the fact that qRT-PCR analysis of RNA extracted from ATCC 19606T cells cultured in LB, which has enough free iron to repress iron-regulated genes (27), showed that the addition of DIP to the medium resulted in a 2.02-fold increase in nfuA transcription (Fig. 3D). This value matches the 2.94-fold difference in transcription observed for this gene by global transcriptomic analysis of ATCC 17978 cells cultured under iron-rich and iron-chelated conditions (12). On the other hand, the presence of 100 μM DIP in the medium resulted in a 66-fold increase in bauA transcription compared with the expression of this gene in cells grown under nonchelated conditions, an outcome that is in accordance with the Western blot results shown in Fig. 3C.
The ATCC 19606T NfuA protein plays a role in in vitro iron starvation and oxidative-stress responses.
The iron starvation deficiency of the insertion derivative 1644, which was initially detected during the screening of the ATCC 19606T EZ::TN<R6Kγori/KAN-2> insertion library, was further confirmed by testing the effect of increasing concentrations of DIP on bacterial growth. Figure 4A shows that, compared with the ATCC 19606T parental strain, the growth of the 1644 nfuA mutant is significantly reduced as the DIP concentration in LB broth increased, with almost no bacterial growth when the broth contained 150 μM DIP. A similar response was obtained when cells were cultured in M9 minimal medium, although the iron chelation effects were detected at lower DIP concentrations as expected, considering the defined composition of this medium (data not shown). The iron utilization-deficient phenotype of the 1644 mutant was corrected to wild-type levels when LB broth samples containing 100 μM or 150 μM DIP were each supplemented with 50 μM FeCl3 (Fig. 4B), a response that confirms the iron starvation-deficient phenotype of this isogenic derivative. The role of the nfuA gene in the phenotype of the 1644 mutant was further confirmed by the fact that its transformation with pMU944, a derivative of pWH1266 shuttle vector harboring a copy of the nfuA parental allele, was enough to restore its growth in chelated LB broth (Fig. 4A) as well as the production of a 29-kDa protein band immunologically related to NfuA (Fig. 3A). Furthermore, genetic complementation of the 1644 mutant with pMU952, a pWH1266 derivative harboring a copy of the E. coli MG1655 nfuA gene, was also enough to correct the severe growth defect of this mutant observed when cultured under iron-chelated conditions (Fig. 4A). In contrast, compared to the parental strain, the 1644 mutant displayed a rather modest delay in initial growth but reached a cell density similar to that of ATCC 19606T after 24-h incubation in LB broth without any selective pressure (Fig. 4C). A similar effect on growth was observed when M9 broth was used to culture bacteria (data not shown).
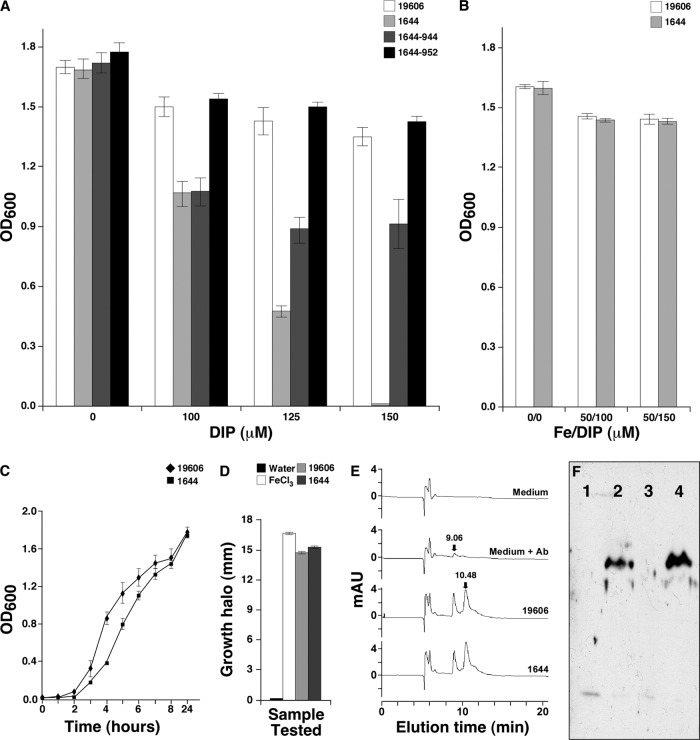
Fig 4.
Effect of nfuA inactivation on bacterial growth, and the expression of iron acquisition-related functions and bacterial growth. (A) Growth of the ATCC 19606T parental strain and the 1644 nfuA mutant either harboring no plasmid or transformed with pMU944 (1644-944) or pMU952 (1644-952), carrying the parental and MG1655 nfuA orthologs, respectively, in LB broth containing increasing concentrations of DIP. (B) Growth of the ATCC 19606T parental strain and the 1644 nfuA mutant in LB broth without any addition or supplemented with 50 μM FeCl3 plus either 100 μM or 150 μM DIP. OD600 readings reported in both panels were taken after overnight incubation (12 to 14 h) at 37°C in a shaking incubator. (C) Growth curves of the ATCC 19606T parental strain and the 1644 isogenic nfuA mutant in LB broth. (D) Growth of A. baumannii ATCC 19606T s1 cells under iron-chelated conditions around filter disks impregnated with distilled water, FeCl3, or cell-free supernatants from ATCC 19606T or 1644 cultures incubated overnight (12 to 14 h) in M9 minimal medium containing 50 μM DIP. Error bars in panels A to D represent 1 SE. (E) HPLC profiles of sterile M9 minimal medium (medium) or culture supernatants from the ATCC 19606T parental strain (19606) or the 1644 nfuA mutant (1644). Sterile M9 minimal medium spiked with purified acinetobactin (medium + Ab) was used as a positive control. The elution times for acinetobactin (9.06 min) and dihydroxybenzoic acid (10.48 min) are indicated on top of the cognate peaks. (F) Immunoblot detection of BauA in whole lysates prepared from ATCC 19606T (lanes 1 and 2) and 1644 (lanes 3 and 4) cells cultured under iron-rich (lanes 1 and 3) or iron-chelated (lanes 2 and 4) conditions.
Arnow colorimetric tests (not shown) and siderophore utilization bioassays using the ATCC 19606T s1 derivative (Fig. 4D), which does not produce acinetobactin but expresses all functions needed for its internalization (10), as the indicator strain showed that the ATCC 19606T 1644 mutant produces catechol and acinetobactin, respectively. The production of acinetobactin by the nfuA mutant was further confirmed by HPLC analysis of culture supernatants, which contained an amount of acinetobactin similar to that detected in the culture supernatant of the ATCC 19606T parental strain (Fig. 4E). Western blotting of whole-cell lysate proteins with specific polyclonal antibodies against BauA, the acinetobactin outer membrane receptor (10), showed that the parental strain and the 1644 mutant produce comparable levels of this protein only under iron-chelated conditions (Fig. 4F). Taken together, these results indicate that the iron starvation deficiency of the A. baumannii ATCC 19606T 1644 insertion derivative is not due to the lack of expression of a fully active acinetobactin-mediated iron uptake system (10) but rather due to a defect in intracellular iron metabolism only when bacteria are cultured under iron-chelated conditions.
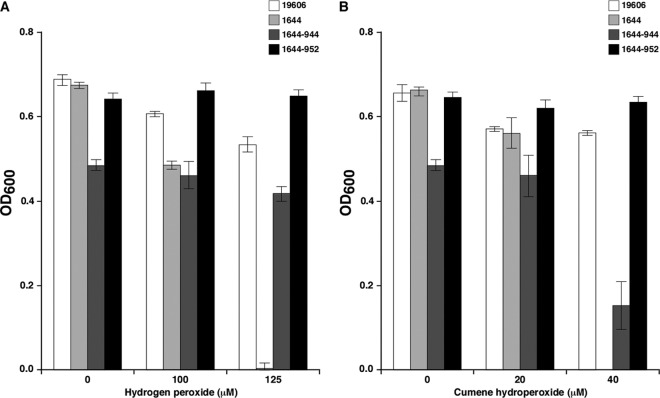
The growth of the ATCC 19606T 1644 nfuA insertion mutant was also severely affected when cultured in M9 minimal broth containing increasing concentrations of hydrogen peroxide or cumene hydroperoxide compared to the ATCC 19606T parental strain (Fig. 5, panels A and B). As was observed with the response to iron chelation, the oxidative-stress-defective response of the 1644 mutant was corrected by genetic complementation with plasmid copies of the ATCC 19606T or MG1655 nfuA gene, although a better restoration response was obtained with the MG1655 gene than with the parental copy. In spite of this difference, it is apparent that nfuA also plays a key role in the ability of A. baumannii to persist and grow in the presence of cell-damaging peroxides.
Fig 5.
Effect of peroxides on bacterial growth. Growth of the ATCC 19606T parental strain and the 1644 nfuA mutant either harboring no plasmid or transformed with pMU944 (1644-944) or pMU952 (1644-952), carrying the parental and MG1655 nfuA orthologs, respectively, in M9 minimal medium containing increasing concentrations of hydrogen peroxide (A) or cumene hydroperoxide (B). OD600 readings reported in both panels were taken after overnight incubation (12 to 14 h) at 37°C in a shaking incubator. Error bars represent 1 SE.
Taken together, all these results indicate that the A. baumannii and E. coli NfuA orthologs perform similar functions by allowing bacterial cells to overcome iron starvation and oxidative stress under laboratory conditions.
NfuA virulence role.
The role of NfuA in the virulence of the ATCC 19606T strain was tested using ex vivo and in vivo experimental models. The former model showed that there is a significant reduction in the number of bacteria recovered from A549 human alveolar epithelial cells infected with the 1644 nfuA mutant compared with the number of bacteria recovered from monolayers infected with the parental strain (P = 0.0102) (Fig. 6A). Coinfection experiments using inocula containing equal amounts of parental and mutant bacteria showed a similar outcome with almost no colony counts recovered on nutrient agar plates containing Km after A549 monolayers were infected for 24 h (P = 0.0002) (Fig. 6A). These findings indicate that, although the 1644 nfuA mutant expresses all functions needed for production and utilization of acinetobactin, its virulence defect cannot be corrected by the presence of the parental strain, which produces and secretes acinetobactin during the infection of A549 cells (15). These observations further support our hypothesis that the iron utilization-deficient phenotype of the ATCC 19606T 1644 nfuA mutant is due to a defect in intracellular steps that follow acinetobactin-mediated acquisition of iron from the extracellular environment.
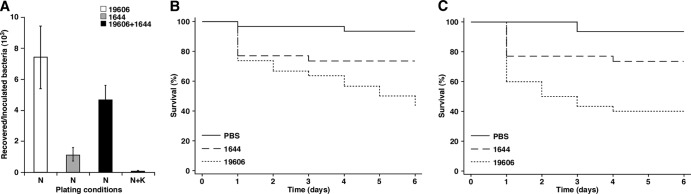
Fig 6.
Role of NfuA in virulence. (A) A549 monolayers were infected for 24 h with 1 × 103 cells of the ATCC 19606T parental strain (19606) or cells of the NfuA mutant (1644). Monolayers were also coinfected with 5 × 102 cells of the parental strain mixed with an equal number of cells of the NfuA mutant (19606 + 1644). Bacterial counts were obtained after A549 cell lysates were plated on nutrient agar (N) or nutrient agar containing 40 μg/ml Km (N + K) to determine total bacterial and 1644 mutant cell counts, respectively. The bacterial counts are represented as the ratio between the CFU/ml of recovered bacteria and the CFU/ml of infecting bacteria after 24 h of incubation. Error bars represent SE. (B) G. mellonella caterpillars were injected with 1 × 105 bacteria of the ATCC 19606T parental strain (19606) or the nfuA insertion derivative 1644 in the absence (panel B) or the presence (panel C) of 100 μM Fe3Cl. As negative controls, caterpillars were injected with comparable volumes of PBS or PBS plus 100 μM Fe3Cl. Caterpillar death was determined daily for 6 days after incubation at 37°C in darkness.
The G. mellonella infection model showed that more than 50% of the injected caterpillars were killed 6 days after infection with the parental strain, while less than 30% of the larvae died when infected with the 1644 nfuA mutant (Fig. 6B). These changes in virulence between ATCC 19606T and the isogenic derivative 1644 are significantly different (P = 0.0299). The addition of 100 μM FeCl3 to the inocula did not result in significant changes in the number of infected caterpillars killed at the end of the experiment by the parental strain and the 1644 nfuA insertion mutant compared with the data obtained without the addition of an external source of free iron (Fig. 6, compare panels B and C). However, the presence of additional free inorganic iron in the infecting inocula increased the killing rates during the early days of infections, as we recently observed during the analysis of ATCC 19606T iron utilization mutants (15). Taken together, these observations indicate that nfuA plays a role in the ability of the ATCC 19606T to kill G. mellonella larvae, although the defect caused by mutating this gene could not be corrected by simply adding an extracellular source of iron. This response, which parallels that obtained with the A549 ex vivo model, strongly indicates that the defect of the nfuA mutant relates to its inability to deal with the oxidative response mounted by the host rather than its capacity to acquire iron from it during the infection process.
DISCUSSION
This report describes the characterization of the A. baumannii ATCC 19606T NfuA ortholog, which was initially identified by the diminished ability of the isogenic nfuA::EZ::TN<R6Kγori/KAN-2> insertion derivative 1644 to grow under iron-chelated conditions but not when the media contained enough free iron to promote bacterial growth. This phenotype is congruent with the predicted amino acid sequence and structural similarities of the ATCC 19606T NfuA protein to the E. coli and A. vinelandii NfuA orthologs, which are [Fe-S] proteins containing A-type and Nfu-type scaffold domains (1, 4). The functional role of this A. baumannii ATCC 19606T protein in intracellular iron metabolism is further supported by the capacity of the E. coli MG1655 allele to correct the defect of the ATCC 19606T 1644 mutant, although the plasmid copy of the latter seems to be more effective than the parental ATCC 19606T allele. This could be due to differences in the affinity for binding [Fe-S] clusters between these two proteins or different protein production from the complementing plasmids harboring the ATCC 19606T or MG1655 alleles. The latter possibility is supported by the observation, after proper DNA sequencing analysis, that the ATCC 19606T nfuA gene is cloned downstream of the pWH1266 Tetr promoter element in the pMU944 complementing plasmid. In contrast, the MG1655 nfuA allele is located in the opposite orientation in the pMU952 derivative and most likely transcribed from its native promoter. These observations suggest that overexpression of nfuA could have a deleterious effect in cell viability, perhaps by increasing intracellular iron limitation. Nevertheless, all these results indicate with a high degree of confidence that an NfuA ortholog works as a [Fe-S] protein complex in A. baumannii ATCC 19606T, which is critical in iron metabolism only under particular stress conditions. These conclusions do not only apply to this particular strain since genomic data show that other A. baumannii isolates and other members of the Acinetobacter genus also have the potential of producing a highly related NfuA ortholog.
Interestingly, the purified NfuA recombinant protein contains bound iron, as it was isolated from E. coli BL21 lysates according to ICP-AES and EPR analyses. However, it is not clear at the moment whether the amount of bound iron corresponds to a [2Fe-2S] or a [4Fe-4S] cluster. Nevertheless, this observation contrasts with the finding that most proteins involved in [Fe-S] biosynthesis are often purified in the apoform (18), with the E. coli NfuA protein forming iron complexes only under anaerobic conditions (1). The detection of iron already bound to the A. baumannii ATCC 19606T NfuA ortholog could also indicate that this protein is a potentially important intracellular iron source when cells are growing under limiting conditions. Such a possibility is supported by the differential expression of nfuA, which we and others have observed (12), and production of its translation product, as well as by the fact that the nfuA iron utilization-deficient phenotype can be corrected by the presence of free inorganic iron in the culture medium. Furthermore, the fact that this derivative expresses a fully functional acinetobactin-mediated iron acquisition system, which is the only high-affinity system expressed by this strain when cultured under laboratory conditions (10), indicates that the growth defect of the ATCC 19606T 1644 nfuA mutant is in intracellular metabolic processes that follow iron uptake from the extracellular environment.
It is also apparent from our data that the capacity of A. baumannii ATCC 19606T cells to resist oxidative stress imposed by the presence of organic and inorganic peroxides in the culture media depends on the production of the NfuA ortholog. However, this response is different from that observed in E. coli, where inactivation of the nfuA gene did not alter the hydrogen peroxide response of the MG1655 SA002 mutant (1). This difference could be due to the composition of medium used to culture the bacteria; we detected deleterious oxidative effects when cells were cultured in M9 minimal medium but not with cells incubated with LB broth. Interestingly, we recently showed that LB has enough free iron to downregulate the acinetobactin-mediated iron acquisition system (27). Taken together, all these observations indicate that NfuA plays a role in the response of A. baumannii to oxidative damage caused by peroxides when cells are actively acquiring iron from the environment. This response is appropriate considering the conditions this pathogen encounters during the infection of the human host. These observations are also in agreement with the hypothesis that NfuA proteins are Fe-S carriers that participate in the maturation of a subset of intracellular proteins that are critical for particular cell stress responses but not for central cellular processes (30).
While the studies that were conducted using standard bacteriological media and laboratory conditions showed that the failure of the 1644 nfuA mutant to grow under iron-chelated conditions could be corrected by the addition of inorganic iron to the media, the ex vivo and in vivo observations reported here demonstrate the contrary. Coinfection of A549 human alveolar epithelial cells, which is one of the most likely targets during the A. baumannii lung infections, with the 1644 nfuA mutant and 19606T parental strain did not restore the ability of the mutant to grow under the iron-restricted conditions imposed by this immortalized cell line. We recently showed that ATCC 19606T intracellular bacteria express the acinetobactin-mediated iron acquisition system, which cross-feeds and rescues the growth of the s1 derivative that does not produce but uses acinetobactin when externally supplied (15). The addition of inorganic iron to the inocula used to infect G. mellonella caterpillars, which mount a complex host response to bacterial infections (22), also failed to correct the significantly diminished capacity of 1644 nfuA mutant cells to kill infected worms. This outcome contrasts with our recent observations, showing that the addition of FeCl3 to the inocula injected into the worms corrects the virulence defects of A. baumannii mutants unable to acquire iron via the acinetobactin-mediated system (15).
Taken together, all these results indicate that, while NfuA could play a critical role in the response to iron availability and oxidative stress when bacterial cells are cultured in vitro, this [Fe-S] ortholog seems to be mainly involved in the ability of bacteria to resist the oxidative responses of infected human cells and G. mellonella caterpillars. This is a significant observation, considering that very little is known regarding the role of NfuA proteins in biological systems, although some hints are becoming more apparent as we have a better understanding of the biochemistry and physiology of the [Fe-S] in different bacterial systems (30).
In conclusion, the results of our study provide strong evidence for the critical protective role NfuA plays in the persistence of A. baumannii under some of the stress conditions it could encounter during host infections. Understanding the role of NfuA in iron metabolism and virulence provides new insights into the biochemistry and physiology of this metal and [Fe-S] clusters. These novel insights could facilitate the development of much needed alternative therapeutics required to treat recalcitrant infections caused by emerging A. baumannii multidrug-resistant isolates.
ACKNOWLEDGMENTS
This work was supported by funds from Public Health grant AI070174, from grants NSF 0420479 and CHE0839233, and from Miami University research funds. T.M.P. was supported by funds from the NSF URM award DBI-0731634.
We are grateful to the Miami University Center for Bioinformatics and Functional Genomics for its support and assistance with automated DNA sequencing and nucleotide sequence analysis and C. Nwugo, Department of Microbiology, Miami University, for his assistance in protein identification by MALDI-TOF spectrometry. We thank F. R. Blattner, University of Wisconsin-Madison, for providing the E. coli MG1655 strain.
Footnotes
Published ahead of print 30 March 2012
REFERENCES
- 1. Angelini S, et al. 2008. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biochem. 283:14084–14091 [DOI] [PubMed] [Google Scholar]
- 2. Arnow L. 1937. Colorimetric determination of the components of 3,4-dihydroxyphenylalanine-tyrosine mixtures. J. Biol. Chem. 118:531–537 [Google Scholar]
- 3. Bandyopadhyay S, Chandramouli K, Johnson MK. 2008. Iron-sulfur cluster biosynthesis. Biochem. Soc. Trans. 36:1112–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bandyopadhyay S, et al. 2008. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J. Biol. Chem. 283:14092–14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barcak JG, Chandler MS, Redfield RJ, Tomb JF. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321–432 [DOI] [PubMed] [Google Scholar]
- 6. Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951 [DOI] [PubMed] [Google Scholar]
- 7. Dorsey CW, Beglin MS, Actis LA. 2003. Detection and analysis of iron uptake components expressed by Acinetobacter baumannii clinical isolates. J. Clin. Microbiol. 41:4188–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dorsey CW, Tolmasky ME, Crosa JH, Actis LA. 2003. Genetic organization of an Acinetobacter baumannii chromosomal region harbouring genes related to siderophore biosynthesis and transport. Microbiology 149:1227–1238 [DOI] [PubMed] [Google Scholar]
- 9. Dorsey CW, Tomaras AP, Actis LA. 2002. Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl. Environ. Microbiol. 68:6353–6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dorsey CW, et al. 2004. The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiology 150:3657–3667 [DOI] [PubMed] [Google Scholar]
- 11. Dunham WR, Sands RH. 2003. g-Strain, ENDOR, and structure of active centers of two-iron ferredoxins. Biochem. Biophys. Res. Commun. 312:255–261 [DOI] [PubMed] [Google Scholar]
- 12. Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. 2011. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fontecave M. 2006. Iron-sulfur clusters: ever-expanding roles. Nat. Chem. Biol. 2:171–174 [DOI] [PubMed] [Google Scholar]
- 14. Freeman M. 2008. Rhomboid proteases and their biological functions. Annu. Rev. Genet. 42:191–210 [DOI] [PubMed] [Google Scholar]
- 15. Gaddy JA, et al. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect. Immun. 80:1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77:3150–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garnacho-Montero J, Amaya-Villar R. 2010. Multiresistant Acinetobacter baumannii infections: epidemiology and management. Curr. Opin. Infect. Dis. 23:332–339 [DOI] [PubMed] [Google Scholar]
- 18. Gupta V, et al. 2009. Native Escherichia coli SufA, coexpressed with SufBCDSE, purifies as a [2Fe-2S] protein and acts as an Fe-S transporter to Fe-S target enzymes. J. Am. Chem. Soc. 131:6149–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Houang ET, Sormunen RT, Lai L, Chan CY, Leong AS. 1998. Effect of desiccation on the ultrastructural appearances of Acinetobacter baumannii and Acinetobacter lwoffii. J. Clin. Pathol. 51:786–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51 [DOI] [PubMed] [Google Scholar]
- 21. Kaplan EL, Meier P. 1958. Nonparametric estimation from incomplete data. J. Am. Stat. Assoc. 53:457–481 [Google Scholar]
- 22. Kavanagh K, Reeves EP. 2004. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 28:101–112 [DOI] [PubMed] [Google Scholar]
- 23. Lu J, Yang J, Tan G, Ding H. 2008. Complementary roles of SufA and IscA in the biogenesis of iron-sulfur clusters in Escherichia coli. Biochem. J. 409:535–543 [DOI] [PubMed] [Google Scholar]
- 24. Meade HM, Long SR, Ruvkum GB, Brown SE, Ausubel FM. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 26. Mussi MA, et al. 2010. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J. Bacteriol. 192:6336–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nwugo CC, Gaddy JA, Zimbler DL, Actis LA. 2011. Deciphering the iron response in Acinetobacter baumannii: a proteomics approach. J. Proteomics 74:44–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Outten FW, Djaman O, Storz G. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52:861–872 [DOI] [PubMed] [Google Scholar]
- 29. Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Py B, Barras F. 2010. Building Fe-S proteins: bacterial strategies. Nat. Rev. Microbiol. 8:436–446 [DOI] [PubMed] [Google Scholar]
- 31. Rhodes ER, Tomaras AP, McGillivary G, Connerly PL, Actis LA. 2005. Genetic and functional analyses of the Actinobacillus actinomycetemcomitans AfeABCD siderophore-independent iron acquisition system. Infect. Immun. 73:3758–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambrook J, Russell DW. 2001. Molecular cloning. A laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Shepard EM, Boyd ES, Broderick JB, Peters JW. 2011. Biosynthesis of complex iron-sulfur enzymes. Curr. Opin. Microbiol. 15:319–327 [DOI] [PubMed] [Google Scholar]
- 34. Smith MG, et al. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Studier F, Rosenberg A, Dunn J, Dubendorff J. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60–89 [DOI] [PubMed] [Google Scholar]
- 36. Tamarit J, Mulliez E, Meier C, Trautwein A, Fontecave M. 1999. The anaerobic ribonucleotide reductase from Escherichia coli. The small protein is an activating enzyme containing a [4Fe-S]2+ center. J. Biol. Chem. 274:31291–31296 [DOI] [PubMed] [Google Scholar]
- 37. Tan G, Lu J, Bitoun JP, Huang H, Ding H. 2009. IscA/SufA paralogues are required for the [4Fe-4S] cluster assembly in enzymes of multiple physiological pathways in Escherichia coli under aerobic growth conditions. Biochem. J. 420:463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484 [DOI] [PubMed] [Google Scholar]
- 39. Vallenet D, et al. 2008. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu XM, Moller SG. 2008. Iron-sulfur cluster biogenesis systems and their crosstalk. Chembiochem 9:2355–2362 [DOI] [PubMed] [Google Scholar]
- 41. Zimbler DL, et al. 2009. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals 22:23–32 [DOI] [PubMed] [Google Scholar]