Abstract
Escherichia coli strains that cause disease outside the intestine are known as extraintestinal pathogenic E. coli (ExPEC) and include pathogens of humans and animals. Previously, the genome of avian-pathogenic E. coli (APEC) O1:K1:H7 strain O1, from ST95, was sequenced and compared to those of several other E. coli strains, identifying 43 genomic islands. Here, the genomic islands of APEC O1 were compared to those of other sequenced E. coli strains, and the distribution of 81 genes belonging to 12 APEC O1 genomic islands among 828 human and avian ExPEC and commensal E. coli isolates was determined. Multiple islands were highly prevalent among isolates belonging to the O1 and O18 serogroups within phylogenetic group B2, which are implicated in human neonatal meningitis. Because of the extensive genomic similarities between APEC O1 and other human ExPEC strains belonging to the ST95 phylogenetic lineage, its ability to cause disease in a rat model of sepsis and meningitis was assessed. Unlike other ST95 lineage strains, APEC O1 was unable to cause bacteremia or meningitis in the neonatal rat model and was significantly less virulent than uropathogenic E. coli (UPEC) CFT073 in a mouse sepsis model, despite carrying multiple neonatal meningitis E. coli (NMEC) virulence factors and belonging to the ST95 phylogenetic lineage. These results suggest that host adaptation or genome modifications have occurred either in APEC O1 or in highly virulent ExPEC isolates, resulting in differences in pathogenicity. Overall, the genomic islands examined provide targets for further discrimination of the different ExPEC subpathotypes, serogroups, phylogenetic types, and sequence types.
INTRODUCTION
Avian-pathogenic Escherichia coli (APEC) strains cause avian colibacillosis, the most significant infectious bacterial disease of poultry worldwide (36). Since avian colibacillosis is an extraintestinal disease, APEC strains are commonly classified as extraintestinal pathogenic E. coli, or ExPEC (15). ExPEC strains also include uropathogenic E. coli (UPEC), neonatal meningitis E. coli (NMEC), and septicemia-associated E. coli, which cause extraintestinal disease in humans and other mammalian hosts (15). Regardless of their immediate source host, all ExPEC strains share certain virulence attributes enabling their extraintestinal lifestyle, including production of adhesins, toxins, protectins, siderophores, iron transport systems, and invasins (16). Identification of such traits among APEC strains has fostered development of a refined definition of the APEC pathotype and has led to interest in the potential of some APEC strains to infect nonavian hosts (9, 20, 22, 34).
Although many ExPEC strains possess sets of virulence factors suggesting a particular host- or syndrome-specific ExPEC subpathotype, some ExPEC strains, such as strain APEC O1, harbor traits of multiple subpathotypes (18). Previously, we reported that, as a group, APEC strains overlap substantially with human UPEC and NMEC strains according to serogroups, phylogenetic groups, and virulence genotypes (18, 22). Comparison of the completed genome sequences strain APEC O1 and human uropathogenic E. coli strains, all belonging to the serotype 95 (ST95) multilocus sequence typing complex, found that they were remarkably similar (18). This suggests that such isolates may have the potential to cause different forms of disease in both human and animal hosts (18, 27, 28). Additionally, in silico comparison of APEC O1 to all sequenced E. coli strains revealed that some of the human ExPEC strains, including both UPEC CFT073 and NMEC RS218, were more similar to APEC O1 than to other human ExPEC isolates (18). Although these genomic similarities suggested that APEC strains such as APEC O1 overlap UPEC or NMEC, slight genomic differences between APEC O1 and the other sequenced ExPEC strains of similar inferred phylogeny led to the hypothesis that strains belonging to similar serogroups and sequence types still might differ substantially.
Accordingly, here we examined further the distribution of previously unstudied APEC O1 genomic islands among populations of avian and human ExPEC strains. Additionally, we determined the ability of APEC O1 to cause disease in murine models of human ExPEC-caused septicemia and meningitis relative to human ExPEC reference strains.
MATERIALS AND METHODS
Bacterial strains.
APEC isolates (n = 452) were defined as E. coli isolated from visceral lesions of commercial broiler chickens or turkeys clinically diagnosed with avian colibacillosis. These isolates originated from various avian host species, sites within these birds, forms of colibacillosis, and farms within at least 22 U.S. states (19, 34, 35). Avian fecal E. coli isolates (n = 106) were obtained from cloacal swabs of apparently healthy birds from various locations in the United States. UPEC strains (n = 200) were isolated from cases of human urinary tract infection (UTI) and were kindly provided by Paul Carson (Meritcare Hospital, Fargo, ND) (34). NMEC strains were isolated from the cerebrospinal fluid of newborns diagnosed with meningitis. These isolates were obtained from a collection within the Netherlands Reference Laboratory for Bacterial Meningitis (Amsterdam, the Netherlands) (14). Isolates were serogrouped by researchers of the E. coli Reference Center (Pennsylvania State University, University Park, PA) and screened (as described below) for APEC O1 genomic islands (Table 1). Organisms were stored at −80°C in brain heart infusion broth (BHI) (Difco Laboratories, Detroit, MI) containing 20% (vol/vol) glycerol with limited passage until use.
Table 1.
APEC O1 genomic islands examined in this study
| Island | Start (bp) | Stop (bp) | Descriptiona | Size (bp)b |
|---|---|---|---|---|
| 1 | 242000 | 273000 | PAI IIAPEC O1 near tRNA-Asp, contains a type VI secretion system | 31,001 |
| 2 | 293675 | 315797 | PAI IIIAPEC O1 near tRNA-Thr; contains vat | 22,123 |
| 3 | 327212 | 339885 | ExPEC island | 12,674 |
| 4 | 346253 | 354806 | ExPEC island | 8,554 |
| 5 | 368417 | 376332 | ExPEC island | 7,916 |
| 8 | 918972 | 957034 | Prophage | 38,063 |
| 13_14 | 1456357 | 1522180 | Prophage and ExPEC island containing cdt locus | 65,824 |
| 17 | 2252024 | 2283233 | Prophage | 31,210 |
| 20 | 2738686 | 2780664 | Prophage | 41,979 |
| 30 | 3847603 | 3861568 | ExPEC island containing auf fimbrial operon | 13,966 |
| 40 | 4709466 | 4771169 | Ethanolamine utilization island near tRNA-Phe | 61,704 |
| 42 | 4935532 | 5001633 | ExPEC island containing ibeA | 66,102 |
PAI, pathogenicity island.
Size data include backbone genes flanking the genomic island.
Virulence gene and phylogenetic typing.
Test and control organisms were examined for the presence of 162 genes previously identified on APEC O1 genomic islands (18) by the use of multiplex PCR (see Table S1 in the supplemental material). The genes sought spanned 12 different APEC O1 genomic islands, previously defined for their presence in APEC O1 and absence from E. coli K-12 MG1655. Multiplex PCR was carried out as previously described (22). PCR-based phylogenetic typing was performed previously according to the methods and interpretive approach of Clermont et al. (7).
Mouse sepsis model.
Isolates were tested for extraintestinal virulence using an established mouse model of systemic sepsis (12, 32). Approximately 108 CFU of logarithmic-growth-phase organisms (from shaking broth cultures), suspended in saline, were injected subcutaneously into the nape of the neck of female Swiss-Webster mice (mean weight, 23 gm; range, 20 to 30 gm). Mice were observed twice daily over the following 3 days for health status, which was scored on a 5-step scale (1 = healthy, 2 = minimally ill, 3 = moderately ill, 4 = severely ill, 5 = dead), with the worst score on a given day being used as the score for that day. Mice were euthanatized if observed in stage 4 illness or at the end of the 72-h observation period, whichever came first. Mice euthanatized on day 1 or 2 received a score of 5 for the subsequent day(s). The mean of the 3 daily health status scores was used to summarize quantitatively each mouse's infection experience over the 3-day observation period. In addition, mice were scored as “dead” or “alive,” with any mouse with a daily status score of 4 or 5 qualifying as “dead,” since, in pilot experiments, all mice that reached stage 4 illness died spontaneously either later that day or that night (not shown).
On each day of mouse injections, the test strains included equal numbers of representatives of all of the matched strain groups. Bacterial strains were administered to 5 mice each, in a random sequence. Strains included APEC O1, UPEC CFT073 (positive control) (23), and E. coli K-12 MG1655 (negative control) (4). Test strains that exhibited a disproportionate variability of effect among the first 5 mice challenged were tested subsequently in 5 additional mice.
To confirm specificity of effect, in pilot experiments, postmortem quantitative cultures of cardiac puncture blood and spleen homogenates were grown at the time of euthanasia or for mice found dead. Consistently, healthy mice had sterile or very-low-count cultures, whereas cultures from ill mice were uniformly positive and yielded heavier growth, with bacterial densities correlating significantly with clinical severity of disease (not shown). Likewise, for each of several dozen mice challenged with various strains, postmortem spleen and blood isolates matched the corresponding challenge strain according to random amplified polymorphic DNA profiling (1), whereas the different challenge strains exhibited distinct profiles (not shown).
Meningitis model.
APEC O1, positive-control NMEC strain RS218, and negative-control E. coli strain DH5α were assessed for their abilities to induce septicemia and meningitis in 5-day-old rats, as previously described (17). Each experimental group contained at least 20 rats, and each experiment was performed twice on separate occasions. When they were 5 days old, specific-pathogen-free Sprague-Dawley rats were inoculated via intraperitoneal injection with 200 CFU of a single strain suspended in phosphate-buffered saline (PBS). At 18 h postinoculation, 25 μl of blood was drawn from each rat via the tail vein and plated on MacConkey agar to determine the concentration of the strain in the blood. Rats were subsequently euthanized, and 10 μl of cerebrospinal fluid (CSF) was removed via cisternal puncture and plated on MacConkey agar to determine the concentration of each strain in the CSF.
Genomic comparisons.
For genomic island comparisons between APEC O1 and sequenced E. coli genomes, BLASTN was used. Each island was compared to whole genome sequences available from NCBI. Genomic islands displaying nucleotide similarity (between APEC O1 and sequenced E. coli) of 90% or greater were considered positive matches and were depicted as percent coverage across the island of interest above the cutoff similarity value of 90%. For genomic comparisons between APEC O1 and NMEC IHE3034, BLASTP was used with a cutoff value of 90% similarity to identify the subsets of unique proteins in each genome (29).
Biostatistics.
For each gene sought via multiplex PCR, Fisher's exact test (two-tailed) was used to test the null hypothesis of equal proportions between populations, with step-down permutation multiplicity adjustments used to address the inflation of the type I error rate associated with large numbers of tests (45). Hierarchical clustering analysis was performed using JMP 7.0 to identify correlations between sequenced genomes based upon APEC O1 island prevalences, between the groups (source group, phylogenetic group, and serogroup) examined based upon gene prevalences, and between the genes sought among the isolates examined (46).
Differences in mean status values for the mouse sepsis model were tested for statistical significance using the Mann-Whitney U test, where a P value of <0.05 was considered significant.
RESULTS AND DISCUSSION
Avian species-specific genomic islands.
The presence of 81 genes belonging to 12 APEC O1 genomic islands (Fig. 1 and 2) among 828 ExPEC and fecal E. coli isolates was sought by multiplex PCR (Table 1). Very few of the targeted genes and their associated genomic islands differed in overall prevalence in comparisons of APEC to human ExPEC. Specifically, only genes belonging to island GI13_14 were significantly (P < 0.05) more prevalent among isolates of avian origin (see Table S2 in the supplemental material) than among human ExPEC isolates. GI13_14 is a 65.8-kb island containing the cdt locus, encoding cytolethal distending toxin type IV, which has been established as an ExPEC virulence factor. It is thought that E. coli acquired the cdt type IV genes by phage transduction (42). The prevalence of the cdt locus was low (0% to 2% per group) among all isolates examined, without significant between-group differences. In contrast, the prophage-associated genes of this island were much more prevalent and were significantly concentrated among APEC (25% to 35% prevalence) and avian fecal E. coli (AFEC) (20% to 58% prevalence) compared with UPEC (4% to 14% prevalence) and NMEC (8% to 14% prevalence) isolates. Therefore, while cdt is a recognized but uncommon pan-ExPEC virulence factor, the adjacent prophage-associated genes in GI13_14 could be useful markers for tracking or identifying specific ExPEC isolates of avian origin.
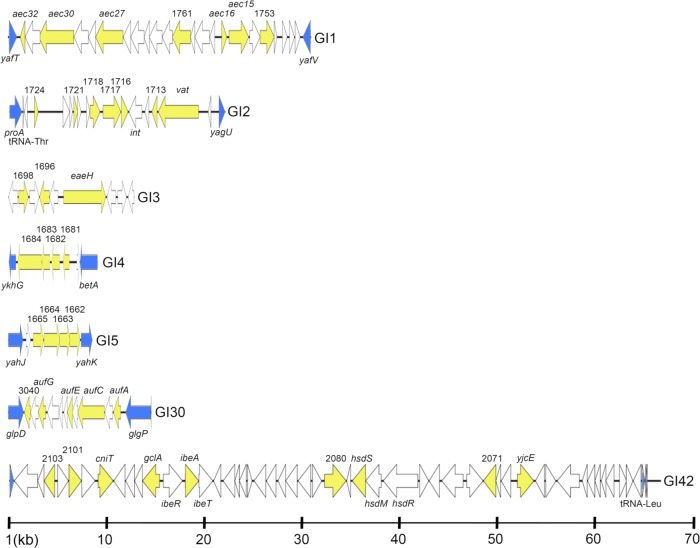
Fig 1.
Linear maps of APEC O1 genomic islands examined in this study. Blue arrows indicate boundary genes of the genomic island. Yellow arrows indicate APEC O1 genes sought via multiplex PCR.
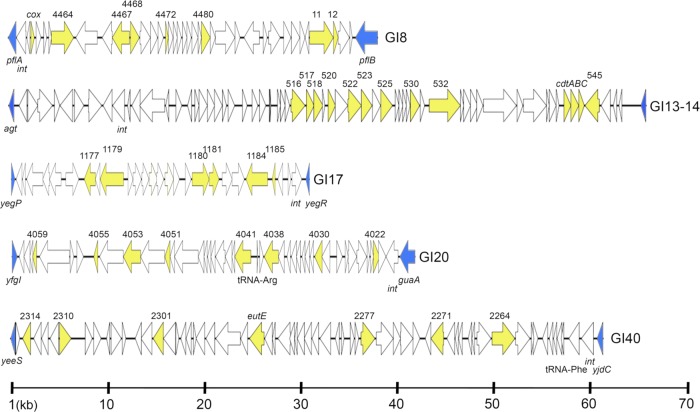
Fig 2.
Linear maps of APEC O1 prophage-associated genomic islands examined in this study. Blue arrows indicate boundary genes of the genomic island. Yellow arrows indicate APEC O1 genes sought via multiplex PCR.
Human ExPEC-associated genomic islands.
In contrast to the avian species-associated GI13_14, several genes and their associated genomic islands were significantly more prevalent among human ExPEC isolates than among isolates of avian origin. GI1 is a 31-kb region adjacent to the tRNA-Asp locus, encoding a putative type VI secretion system in APEC O1. This region was also present in the sequenced genomes of O18 strains UTI89 (UPEC) and IHE3034 (NMEC) and of non-O18 strains S88 (NMEC) and 536 (UPEC) (Fig. 3). Among the populations examined, genes of GI1 were most prevalent among NMEC (71 to 77%), only slightly less prevalent among UPEC (40 to 60%), and significantly less prevalent among APEC (8 to 16%) and AFEC (1 to 12%) isolates. Other type VI secretion systems have been described for APEC strains and shown to be important for pathogenesis (8). However, a derivative of UPEC CFT073 in which its type VI secretion system within the metV island had been inactivated was not attenuated in a mouse model of UTI (25). The strong association of this uncharacterized type VI secretion system with serogroup O1 and O18 NMEC isolates suggests both its potential usefulness as a marker for NMEC and a possible role in NMEC pathogenesis, which warrants exploration.
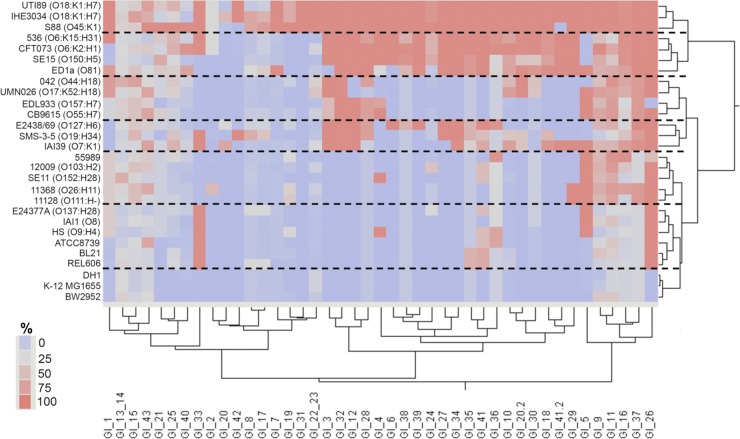
Fig 3.
Presence of all APEC O1 genomic islands among 28 sequenced E. coli strains. Two-way unsupervised hierarchical clustering was performed to identify associations in genomic island content (left to right) among the sequenced genomes (top to bottom). Dashed lines separate clusters comprising highly similar strains. The color scale depicts percent coverage of each APEC O1 genomic island detected in each sequenced strain at 90% or greater nucleotide similarity.
GI2 contains the vacuolating autotransporter gene vat, which is carried on a 22-kb pathogenicity island within APEC O1. The vat gene has previously been shown to be essential for the virulence of certain APEC strains (31). vat has also been significantly associated with human E. coli isolates collected from patients with bacteremia (13) and human UPEC strains belonging to phylogenetic groups B2 and D (30, 33). The prevalence of both vat and its adjacent transcriptional regulator was highest among UPEC and NMEC (63 to 82%) strains, whereas among APEC strains, although vat was highly prevalent (71%), its adjacent transcriptional regulator occurred significantly less frequently (31%). This likely reflects the ability of our vat primers to also detect the tsh gene, which is known to occur among APEC strains on ColV plasmids and to lack an adjacent transcriptional regulator (20). The prevalence of the remainder of GI2 was low (0% to 12%) among all groups, supporting the hypothesis of the occurrence of vat in a different genetic context in most ExPEC strains.
GI3 is a 12-kb island containing a putative adhesion/attaching and effacing gene with similarity to the eaeH gene of diarrheagenic E. coli (43). Among sequenced E. coli strains, genes of GI3 were present in ExPEC, commensal E. coli, E. coli O157:H7, and some diarrheagenic isolates. According to PCR analysis, genes of GI3 were highly (79% to 97%) prevalent among UPEC and NMEC isolates; however, whereas eaeH was highly (92%) prevalent among APEC isolates, other genes associated with GI3 were found at a significantly lower (28% to 47%) prevalence, a pattern mimicked by AFEC isolates. Since nucleotide variations within the eaeH gene are known to affect its involvement in pathogenesis, further work would be required to determine if eaeH in APEC O1 actually contributes to its virulence.
GI4 is an 8-kb island that encodes a putative autotransporter/adhesion predicted to have an autotransporter barrel domain. GI4 was highly conserved among sequenced ExPEC and some sequenced commensal isolates. Its prevalence was relatively high (69% to 84%) among NMEC and UPEC strains but significantly lower among APEC (46%) and AFEC (12% to 16%) isolates. Its possible role in ExPEC pathogenesis has not yet been investigated.
GI5 is a 7.9-kb island containing a putative sugar ABC transport system (Fig. 1), the predicted protein sequence of which shares homology with systems involved in the transport of monosaccharides such as arabinose, galactose, and ribose. Genes of GI5 were highly prevalent among NMEC (78% to 84%) but significantly less prevalent among UPEC, APEC, and AFEC (32% to 46%) isolates, suggesting their potential usefulness as markers for the NMEC pathotype.
GI30 is a 13.9-kb island that contains the auf fimbrial operon and is highly conserved among sequenced ExPEC isolates. The auf genes within GI30 were highly prevalent among NMEC (67 to 88%), somewhat less prevalent among UPEC (59%), and significantly less prevalent among APEC (27%) and AFEC (7%) isolates. In UPEC strain CFT073, this island has been shown to be functional but has not been shown to contribute to mouse urinary tract colonization (5). In contrast, its possible role in NMEC pathogenesis has yet to be assessed. Regardless of its importance in virulence, the auf locus appears to be potentially useful as another marker of NMEC and UPEC.
GI40 is a 61-kb island containing a putative ethanolamine utilization system plus multiple hypothetical genes of unknown function (Fig. 4). It is located adjacent to tRNA-Phe and contains an integrase gene. The island's structure in other sequenced genomes did not resemble that of APEC O1, as the prevalences of the genes sought that represent this island differed (Fig. 2). The Eut system has been shown to form a microcompartment within the bacterial cell that acts to sequester ethanolamine metabolism (40). This system could be advantageous to a pathogenic strain residing in the host's gut by providing additional nitrogen sources to the bacterium (3). The prevalences of genes of this system and its corresponding island were generally low among all groups but were significantly higher among UPEC (16% to 19%) and NMEC (11% to 14%) than among isolates of avian origin (0% to 4%).
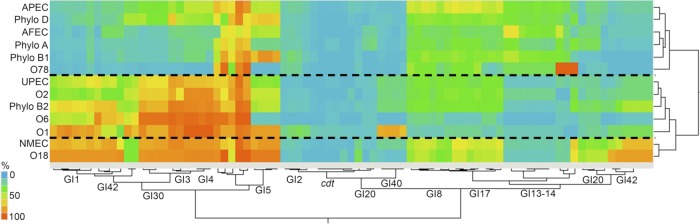
Fig 4.
Prevalence of the studied genomic islands of APEC O1 within various subgroups of Escherichia coli. Two-way unsupervised hierarchical clustering was done based upon percent prevalence of each genomic island sought (left to right) among different E. coli pathotypes, phylogenetic groups, and serogroups (top to bottom). Dashed lines separate clusters comprising highly similar groups.
GI42 is a 66.1-kb island, adjacent to tRNA-Leu, containing ibeAR (“invasion of brain endothelium”) and a putative restriction modification system (Fig. 1). IbeA is a recognized NMEC virulence factor (6) and also is involved in biofilm formation and avian fibroblast invasion (44). The ibeA gene has previously been shown to be a component of the GimA locus, thought to be an ancestral component of strains belonging to phylogenetic group B2 that has evolved via reductive evolution (10). The presence of a portion of GimA in APEC O1 supports this hypothesis. Here, the upstream portion of this region, containing a putative restriction modification system, was generally more prevalent than the downstream GimA-like region harboring ibeAR. Genes of the GimA-like portion of this region were significantly more prevalent among NMEC strains (48% to 72%) than within the other groups (6% to 18%).
In addition to GI13_14, three other prophage-associated islands were also sought. GI8 is a 38-kb island containing prophage-associated genes; its prevalences were similar among APEC and NMEC (42 to 53%) and significantly lower among the AFEC and UPEC (21 to 30%) isolates. GI17 is a 31.2-kb island containing prophage-associated genes; these were significantly more prevalent among APEC and NMEC (37 to 55%) than among AFEC and UPEC (21 to 28%) isolates. Genes of GI20 did not occur at high prevalence among any of the isolate groups.
Serogroup- and phylogenetic group-associated genomic islands.
The prevalences of the genes and associated genomic islands were also examined relative to serogroup and phylogenetic group (Fig. 4; see also Table S3 in the supplemental material). Several genomic islands, including GI1, GI3, GI4, GI30, and GI42, were significantly more prevalent among isolates belonging to phylogenetic group B2, which includes APEC O1 and most ExPEC strains, compared to groups A, B1, and D. In contrast, genomic islands GI3, GI4, and GI5 were significantly more prevalent among isolates belonging to phylogenetic groups B2 and D compared to groups A and B1. Of the most prevalent serogroups in this study, certain genomic islands were highly (>75%) prevalent among them, including GI1 (O1, O18), GI3 (O1, O2, O6, O18), GI4 (O1, O2, O6, O18), GI5 (O1, O18), GI30 (O1, O6, O18), GI40 (O1), and GI42 (O18). Interestingly, although O78 is the most common APEC serogroup (32), none of the studied APEC O1 genomic islands were highly prevalent among the serogroup O78 isolates. This suggests that avian O78 isolates are distinct from APEC O1 and might possess a different gene repertoire enabling success in avian species. Aside from their avian pathogenicity, isolates belonging to this serogroup have also been implicated in human septicemia and therefore may represent another ExPEC group with zoonotic potential (36).
Correlations among genes.
Unsupervised hierarchical clustering was used to identify groups of genes that occurred together among the isolates studied (Table 2). In most cases, genes belonging to the same APEC O1 GI clustered together. In some cases, genes from multiple APEC O1 GIs clustered together. Co-occurring APEC O1 genes included those of APEC O1 islands GI2, GI3, GI5, and GI30 (linking vat, the auf operon, and the putative ABC transporter system); GI13_14 and GI42; and GI8 and GI17 (prophage-associated islands). Clustering of the isolates examined revealed a number of isolate clusters containing different subsets of the above-mentioned gene clusters. Notably, most of the clusters containing O1 and O18 isolates possessed a large number of gene clusters. Overall, the hierarchical clustering suggested that O1 and O18 isolates are relatively conserved regarding their APEC O1 GI-associated content and contain a large number of the genes sought in this study.
Table 2.
Co-occurring gene clusters among isolates examined
| Genes or gene cluster(s) includeda | APEC O1 islands included | |
|---|---|---|
| Gene cluster | ||
| 1 | 1712 (vat), 1713, 3030 (aufA), 3032 (aufC), 3034 (aufE), 1683, 1681, 1684, 1683, 1696, 1698 | GI2, GI3, GI5, GI30 |
| 2 | 1753, 1761, 1772, 1767, 1770, 1757, 1756 | GI1 |
| 3 | 2078 (hsdS), 2099 (cniT), 545 | GI13_14, GI42 |
| 4 | 2093 (ibeA), 2101, 2103 (dhaK), 2095 (gclA) | GI42 |
| 5 | 2071, 2068 (yjcE) | GI42 |
| 6 | 1662, 1665, 1664, 1663 | GI5 |
| 7 | 2277, 2271, 2291 (eutE), 2264 | GI40 |
| 8 | 4022, 4038, 4030, 4059 | GI20 |
| 9 | 2301, 2310 | GI40 |
| 10 | 532 (ynaA), 530, 522, 525, 520, 516 (trkG), 517 | GI13_14 |
| 11 | 4480, 4464, 4467, 4468, 11, 12, 1185, 1181, 1180, 1184, 1177, 1179 | GI8, GI17 |
| Isolate cluster | ||
| 1 | None | |
| 2 | 8 | |
| 3 | 6 | |
| 4 | 6, 10 | |
| 5 | 6, 10, 11 | |
| 6 | 11 | |
| 7 | 1, 2, 3, 4, 5, 6, 8 | |
| 8 | 1, 2, 3, 4, 5, 6, 8, 11 | |
| 9 | 1, 2, 3, 4, 5, 6, 11 | |
| 10 | 1, 4, 11 | |
| 11 | 1, 6, 9, 11 | |
| 12 | 1, 2, 7, 9, 11 | |
| 13 | 1, 4 | |
| 14 | 1 | |
| 15 | 1, 2, 5, 9 |
Gene cluster numbers refer to APEC O1 gene numbers. Isolate cluster numbers refer to hierarchical genotyping clusters (data not shown).
Assessment of APEC O1 virulence.
Since APEC O1 belongs to ST95 and shares similarities with some NMEC strains in relation to its virulence gene-related content, phylogeny, and serogroup, it was assessed for its ability to cause sepsis in the mouse and meningitis in the neonatal rat (Table 3). APEC O1 was significantly less virulent than CFT073 in the mouse sepsis model (P < 0.05), although it was still significantly more virulent than negative-control strain MG1655. Furthermore, APEC O1 was avirulent in the neonatal rat meningitis model compared to the archetypic NMEC RS218 strain (11), as it was unable to induce bacteremia in the neonatal rat or to invade the central nervous system of the neonatal rat.
Table 3.
Examination of APEC O1 in models of avian and human disease
| Parameter | APEC O1 | Positive bacterial control (strain) | Negative bacterial control (strain) |
|---|---|---|---|
| Mean status score in mouse sepsis assay | 2.5a | 4.6 (CFT073) | 1.0 (MG1655) |
| Mean log10 CFU/ml in blood in rat meningitis assay | 0b | >3.5 (RS218) | 0 (DH5α) |
| Mean log10 CFU/ml in cerebrospinal fluid in rat meningitis assay | 0b | >4.1 (RS218) | 0 (DH5α) |
Mean values for APEC O1 were significantly (P < 0.01) higher than negative-control DH5α values but significantly lower than positive-control values.
Mean values for APEC O1 were significantly (P < 0.01) lower than positive-control values but not significantly different from negative-control values.
That APEC O1 was not fully virulent in the mouse sepsis model and was avirulent in the meningitis model suggests that it differs from other ST95 strains in virulence potential (2, 26, 41). This is interesting, because APEC O1 exhibits characteristics that are highly similar to those of other O1 and O18 NMEC isolates, including its possession of multiple recognized NMEC virulence factors, including the K1 capsule and genes encoding IbeA, OmpA, and TraJ (24). Therefore, it was expected that APEC O1 would be able to induce meningitis in the neonatal rat. Its inability to induce septicemia or meningitis suggested that genomic differences between APEC O1 and other NMEC strains account for these phenotypic differences. Since the genome sequence was available for NMEC strain IHE3034, an O18:K1:H7 strain belonging to the same sequence type (ST95) as APEC O1 (26), we compared these two strains for gross chromosomal genomic differences (gene presence or absence). We found 431 genes unique to APEC O1 and 325 genes unique to IHE3034 (data not shown). The majority of these genes were prophage-associated sequences; however, one notable genomic island was identified in IHE3034 that was absent in APEC O1, containing the sfa fimbrial operon and iroBCDEN siderophore system. Since APEC O1 contains iroBCDEN on its plasmid pAPEC-O1-ColBM, the only notable difference between APEC O1 and IHE3034 with regard to virulence-associated gene content was sfa. In a recent study of virulence-related functions of the sfaX(II) gene of strain IHE3034, sfaX(II) inhibited type 1 fimbrial expression and decreased motility and flagellum production (38). Additionally, a second novel regulatory gene, sfaY(II), was recently identified within IHE3034's sfa operon (39). This, coupled with the fact that sfa is highly prevalent among NMEC isolates (22), suggests that sfa might play a key role in meningovirulence and could help to explain APEC O1's inability to cause meningitis. Further work, including examination of potential polymorphisms within structural genes and regulatory regions and whether gene acquisition versus reductive evolution resulted in host adaptation within strains of the ST95 lineage, is required to elucidate these differences.
Conclusions.
This study advances our knowledge of APEC O1, a strain of avian origin belonging to a serogroup (O1) and ST (ST95) known for their involvement in both avian and human extraintestinal disease. Our genomic comparisons confirm that APEC O1 is representative of APEC strains belonging to the O1 serogroup but different from those belonging to the O78 serogroup, to which most APEC strains belong. Thus, it is representative of the subset of APEC strains that share similarities with human ExPEC. Of the APEC O1 islands examined, few loci were identified that could serve as unique markers of APEC strains, strengthening the argument that APEC O1 and similar strains of avian origin share significant genetic overlap with human ExPEC. Since the islands studied here were not previously characterized, we know little about their role in pathogenesis. Nonetheless, several of them deserve future attention because of their strong associations with certain pathotypes. Despite their gross similarities, evident from this work is that numerous genetic and phenotypic differences exist among strains within the ST95 lineage, underscoring the need for more-discriminating genomic analyses to better define these differences and their functional implications, including pathogenesis and host adaptation. Such studies would increase our understanding of the genome modifications that occur during host adaptation and their impact on an ExPEC strain's virulence potential.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lodewijik Spanjaard, Netherlands Reference Laboratory for Bacterial Meningitis, for providing strains for use in the study. Data analyses were carried out using tools available through the Minnesota Supercomputing Institute.
This material is based in part on work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J.); the Roy J. Carver Charitable Trust; and USDA NIFA awards 2008-35600-418805 and 0826675 (L.K.N., T.J.J., and S.K.).
Footnotes
Published ahead of print on 30 March 2012.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Berg DE, Akopyants NS, Kersulyte D. 1994. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol. Cell Biol. 5:13–24 [Google Scholar]
- 2. Bert F, et al. 2010. Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis. J. Clin. Microbiol. 48:2709–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bertin Y, et al. 2011. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13:365–377 [DOI] [PubMed] [Google Scholar]
- 4. Blattner FR, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474 [DOI] [PubMed] [Google Scholar]
- 5. Buckles EL, et al. 2004. Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster. Infect. Immun. 72:3890–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Che X, et al. 2011. Involvement of IbeA in meningitic Escherichia coli K1-induced polymorphonuclear leukocyte transmigration across brain endothelial cells. Brain Pathol. 21:389–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Pace F, et al. 2010. The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect. Immun. 78:4990–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ewers C, et al. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297:163–176 [DOI] [PubMed] [Google Scholar]
- 10. Homeier T, Semmler T, Wieler LH, Ewers C. 2010. The GimA locus of extraintestinal pathogenic E. coli: does reductive evolution correlate with habitat and pathotype? PLoS One 5:e10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang SH, et al. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 63:4470–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson JR, et al. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141–1150 [DOI] [PubMed] [Google Scholar]
- 13. Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. 2008. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J. Clin. Microbiol. 46:3906–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson JR, Oswald E, O'Bryan TT, Kuskowski MA, Spanjaard L. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J. Infect. Dis. 185:774–784 [DOI] [PubMed] [Google Scholar]
- 15. Johnson JR, Russo TA. 2002. Extraintestinal pathogenic Escherichia coli: “the other bad E coli.” J. Lab. Clin. Med. 139:155–162 [DOI] [PubMed] [Google Scholar]
- 16. Johnson JR, Russo TA. 2005. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int. J. Med. Microbiol. 295:383–404 [DOI] [PubMed] [Google Scholar]
- 17. Johnson TJ, et al. 2010. Sequence analysis and characterization of a transferable hybrid plasmid encoding multidrug resistance and enabling zoonotic potential for extraintestinal Escherichia coli. Infect. Immun. 78:1931–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson TJ, et al. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189:3228–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson TJ, et al. 2009. Examination of the source and extended virulence genotypes of Escherichia coli contaminating retail poultry meat. Foodborne Pathog. Dis. 6:657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson TJ, Siek KE, Johnson SJ, Nolan LK. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188:745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson TJ, et al. 2008. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 46:3987–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson TJ, et al. 2008. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 74:7043–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kao J-S, Stucker DM, Warren JW, Mobley HLT. 1997. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect. Immun. 65:2812–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim KS. 12 October 2006, posting date. Meningitis-associated Escherichia coli. In Böck A, et al. (ed.), EcoSal---Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: http://www.ecosal.org [Google Scholar]
- 25. Lloyd AL, Henderson TA, Vigil PD, Mobley HL. 2009. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J. Bacteriol. 191:3469–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moriel DG, et al. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9072–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moulin-Schouleur M, et al. 2007. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J. Clin. Microbiol. 45:3366–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moulin-Schouleur M, et al. 2006. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J. Clin. Microbiol. 44:3484–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Overbeek R, et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parham NJ, et al. 2005. Distribution of the serine protease autotransporters of the Enterobacteriaceae among extraintestinal clinical isolates of Escherichia coli. J. Clin. Microbiol. 43:4076–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parreira VR, Gyles CL. 2003. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect. Immun. 71:5087–5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Picard B, et al. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Restieri C, Garriss G, Locas MC, Dozois CM. 2007. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl. Environ. Microbiol. 73:1553–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodriguez-Siek KE, et al. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151:2097–2110 [DOI] [PubMed] [Google Scholar]
- 35. Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Nolan LK. 2005. Characterizing the APEC pathotype. Vet. Res. 36:241–256 [DOI] [PubMed] [Google Scholar]
- 36. Ron EZ. 2006. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr. Opin. Microbiol. 9:28–32 [DOI] [PubMed] [Google Scholar]
- 37. Saif YM, Fadly AM. 2008. Diseases of poultry, vol 12 Blackwell Publishing, Ames, IA [Google Scholar]
- 38. Sjöström AE, et al. 2009. The SfaXII protein from newborn meningitis E. coli is involved in regulation of motility and type 1 fimbriae expression. Microb. Pathog. 46:243–252 [DOI] [PubMed] [Google Scholar]
- 39. Sjöström AE, et al. 2009. Analysis of the sfaX(II) locus in the Escherichia coli meningitis isolate IHE3034 reveals two novel regulatory genes within the promoter-distal region of the main S fimbrial operon. Microb. Pathog. 46:150–158 [DOI] [PubMed] [Google Scholar]
- 40. Tanaka S, Sawaya MR, Yeates TO. 2010. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327:81–84 [DOI] [PubMed] [Google Scholar]
- 41. Tivendale KA, et al. 2010. Avian pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect. Immun. 78:3412–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tóth I, et al. 2009. Cytolethal distending toxin type I and type IV genes are framed with lambdoid prophage genes in extraintestinal pathogenic Escherichia coli. Infect. Immun. 77:492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tzipori S, et al. 1995. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect. Immun. 63:3621–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang S, et al. 2011. Effects of ibeA deletion on virulence and biofilm formation of avian pathogenic Escherichia coli. Infect. Immun. 79:279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Westfall PH, Tobias RD, Rom D, Wolfinger RD, Hochberg Y. 1999. Multiple comparisons and multiple tests using the SAS system. SAS Institute, Inc., Cary, NC [Google Scholar]
- 46. Ye C, Liu J, Ren F, Okafo N. 2000. Design of experiment and data analysis by JMP (SAS institute) in analytical method validation. J. Pharm. Biomed. Anal. 23:581–589 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.