Abstract
Alnumycin A is an aromatic polyketide with a strong resemblance to related benzoisochromanequinone (BIQ) antibiotics, such as the model antibiotic actinorhodin. One intriguing difference between these metabolites is that the positions of the benzene and quinone rings are reversed in alnumycin A in comparison to the BIQ polyketides. In this paper we demonstrate that inactivation of either the monooxygenase alnT gene or the flavin reductase alnH gene results in the accumulation of a novel nonquinoid metabolite, thalnumycin A (ThA), in the culture medium. Additionally, two other previously characterized metabolites, K1115 A and 1,6-dihydroxy-8-propylanthraquinone (DHPA), were identified, which had oxidized into quinones putatively nonenzymatically at the incorrect position in the central ring. None of the compounds isolated contained correctly formed pyran rings, which suggests that on the alnumycin pathway quinone biosynthesis occurs prior to third ring cyclization. The regiochemistry of the two-component monooxygenase system AlnT/AlnH was finally confirmed in vitro by using ThA, FMN, and NADH in enzymatic synthesis, where the reaction product, thalnumycin B (ThB), was verified to contain the expected p-hydroquinone structure in the lateral ring.
INTRODUCTION
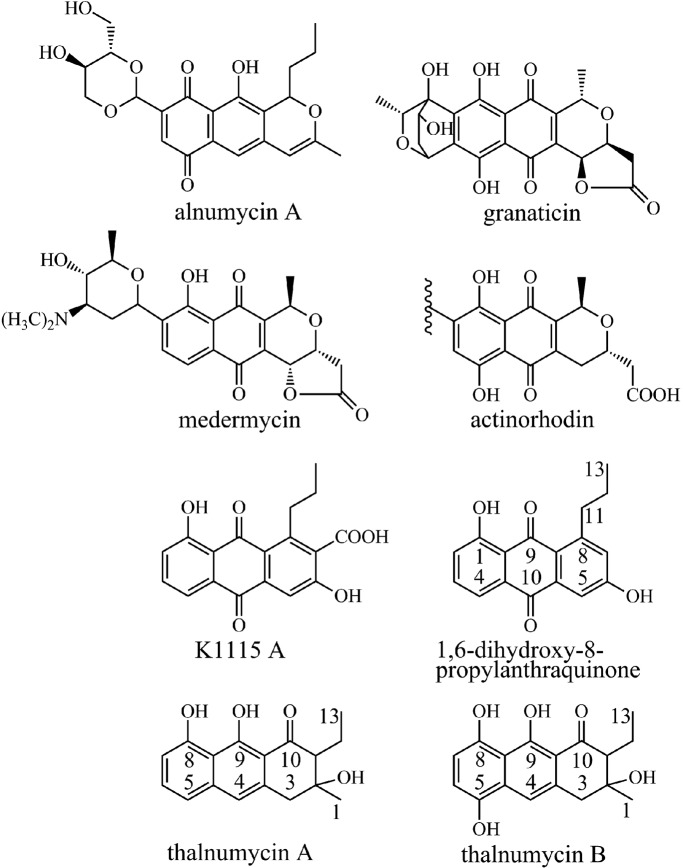
Bacteria of the genus Streptomyces have been a treasure trove in the discovery of biologically active natural products, many of which have found applications in medicine and agriculture (33). Aromatic polyketides, a distinct subclass of natural products, have been extensively used as antibiotics (e.g., tetracycline) (2) and in cancer chemotherapy (e.g., doxorubicin) (9, 15). Alnumycin A (Fig. 1) is a polyketide that has been shown to exhibit several biological activities, including antibiotic, cytostatic, gyrase inhibitory, and topoisomerase inhibitory activities (3, 4). From a structural point of view, alnumycin A is intriguing due to its relation to the closely related benzoisochromanequinones (BIQ), such as granaticin, medermycin, and the model antibiotic actinorhodin (Fig. 1). The polyketide aglycone is similar, but distinctly unique, while the most curious feature is the carbohydrate-like 4′-hydroxy-5′-hydroxymethyl-2′,7′-dioxane moiety.
Fig 1.
Structures and carbon numbering of the metabolites relevant to the study.
Aromatic polyketides are biologically synthesized via dedicated pathways, which often encompass as many as 20 to 30 individual steps (7, 16). Early stages of aromatic polyketide biosynthesis resemble fatty acid synthesis in bacteria, in which acetyl and malonyl coenzyme A (CoA) precursors are combined in repeated Claisen condensations to form a carbon chain. However, unlike in fatty acid synthesis, polyketide synthases do not reduce the elongating carbon chain during synthesis, and subsequently a long polyketide is produced. This highly reactive polyketide chain is subsequently reduced, aromatized, and cyclized in a controlled manner, resulting in the formation of the various polyaromatic carbon skeletons. Differences in the stereo- and regiochemistries of cyclization and further tailoring reactions, such as hydroxylations, reductions, and methylations, explain the vast number of compounds belonging to this class of natural products.
In order to understand how the structural diversity of BIQ metabolites is reflected in the composition of the biosynthetic enzymes, the alnumycin gene cluster has previously been cloned and expressed heterologously in Streptomyces albus (35). Molecular genetic studies have clarified that the biosynthesis is initiated from a butyryl starter unit (46), while cyclization of the third ring is catalyzed by enzymes belonging to entirely different enzyme families than those found from the actinorhodin and granaticin pathways (35). Recently, the novel pathway leading to the attachment and formation of the sugar-like 4′-hydroxy-5′-hydroxymethyl-2′,7′-dioxane moiety was characterized in vivo and in vitro (34).
Many aromatic polyketides, such as the anthracyclines, the isochromanequinones, and the angucyclines, are p-quinones (16). Since this arrangement does not directly arise from the polyketide backbone upon cyclization, biosynthesis usually includes a tailoring step, which introduces a hydroxyl group in the para-arrangement in relation to a hydroxyl originating from the polyketide backbone. The polyphenolic dihydroquinones thus formed may readily oxidize nonenzymatically to the final p-quinone structures. Typically, enzymes catalyzing these steps belong to one of three enzyme families: the flavoprotein hydroxylases (22, 25), the so-called cofactorless monooxygenases (10, 14, 38), and the two-component flavin-dependent monooxygenases (36, 44). In effect, the actinorhodin gene cluster harbors genes for both of the two latter classes, whose products are able to catalyze quinone formation (36). The alnumycin biosynthetic cluster (35), on the other hand, only contains genes for the putative two-component monooxygenase system, alnT and alnH, which encode a flavin-dependent monooxygenase and a flavin reductase, respectively. The AlnT/AlnH pair is homologous to the ActVA-ORF5/ActVB from the actinorhodin pathway, respectively, which has been studied in recent years using molecular genetics (36) and biochemistry (42–44). The lack of a cofactorless oxygenase homologous to ActVA-ORF6 on the alnumycin pathway suggests a closer evolutionary relationship to the granaticin (20) and medermycin (21) (Fig. 1) gene clusters, which both encode genes only for the two-component system.
The aim of the work described in this paper was to uncover the molecular basis for formation of the p-quinone arrangement in the lateral instead of the central ring in alnumycin A. We demonstrate that alnT and alnH affect the same biosynthetic step, as both single-gene deletion mutants produced the same novel naphthodiol derivative, thalnumycin A (ThA). Use of this compound as a substrate for AlnT in the presence of AlnH, FMN, and NADH yielded a novel p-hydroquinone product, thalnumycin B (ThB), which conclusively verified that the two-component monooxygenase system is responsible for formation of the p-quinone in the lateral ring of alnumycin A. The isolation of ThA and two other previously known metabolites, K1115 A (32, 35) and 1,6-dihydroxy-8-propylanthraquinone (DHPA) (19), from knockout mutants indicated that, unlike in the biosynthesis of BIQs, the cyclization of the third ring takes place only after formation of the p-quinone.
MATERIALS AND METHODS
Strains and culture conditions.
The alnumycin producer Streptomyces sp. CM020 was obtained from Galilaeus Oy. S. albus (6) was used as a heterologous host for metabolite production. The strains were maintained on ISP4 (Difco) and MS plates (18), while tryptic soy broth (Oxoid) and NoS-soyE1 liquid media (39) were used for plasmid isolation and metabolite production, respectively. Amberlite XAD-7 (1 g/50 ml; Rohm & Haas) was added for absorption of metabolites from the cultivation media. Cultures were shaken at 28°C, 300 rpm, for 5 days. Apramycin (Fluka) was used for selection of Streptomyces in solid and liquid media at 50 μg/ml for plasmid-containing strains. Escherichia coli strain TOP10 (Invitrogen) was used as a cloning host and for overexpression of alnT and alnH. E. coli K-12 and ET123567/pUZ8002 (24) were used for heterologous recombination and conjugation of the manipulated cosmids into S. albus, respectively. The strains were cultured in LB medium for DNA isolation or 2× yeast extract-tryptone (YT) medium for protein production. Ampicillin (Sigma) and apramycin were used for selection of plasmids and cosmids, respectively, in E. coli cultivations at 50 μg/ml.
General DNA techniques, cloning, and sequencing.
Streptomyces sp. CM020 genomic DNA was isolated according to standard procedures (18). The cosmid pAlnuori containing the complete alnumycin biosynthetic cluster has been previously described (35). Isolation of plasmid DNA from E. coli was performed with the QIAprep spin miniprep kit (Qiagen). Restriction enzymes were purchased from Fermentas, while DNA fragments generated by restriction enzyme digestions and PCR were recovered from agarose gels by using the QIAquick gel extraction kit (Qiagen). For all PCR amplifications, Phusion DNA polymerase (Finnzymes) was used according to the manufacturer's recommendations. Oligonucleotide primers were obtained from and DNA sequencing was performed by Eurofins MWG Operon.
Construction of the AlnT and AlnH overexpression plasmids.
Both the alnT and alnH genes were amplified from Streptomyces sp. CM020 genomic DNA. The primers used for amplification of alnH were alnHforB (BglII site in bold), 5′-CTAAGATCTAACCGCACCGACCGGCG, and alnHrevH (HindIII site in bold), 5′-CTAAAGCTTCAGGGACGCGGCAGGC. The PCR product generated was cloned as a BglII-HindIII fragment into a modified pBAD/HisB vector (23), resulting in pBAD/HisBdalnH. The construction of pBAD/HisBdalnT was executed in a similar manner. The primers used for amplification of alnT were alnTBfor (BglII site in bold), 5′-CAGAGATCTCGCGAACGCACGCCGC, and alnTErev (EcoRI site in bold), 5′-CAGGAATTCCGCTACCGGGCCGCTG. The product was cloned in the same vector as a BglII-EcoRI fragment. Both constructs were verified by sequencing.
Inactivation of alnH and alnT.
The open reading frames (ORFs) alnH and alnT were deleted from the cosmid pAlnuori by using the λ Red recombinase system (8) through a two-step homologous recombination process. The primers used for inactivation consisted of a 50-nucleotide (nt) region homologous to the target sequence followed by a 20-nt priming sequence (in bold) for amplification of the Cmr gene with flanking FRT (FLP recognition target) sites. Elimination of the resistance gene was subsequently executed using the helper plasmid pFLP2 (17) expressing the FLP recombinase enzyme. The primers used for inactivation of alnH were alnHfcamfor, 5′-GCACCGACCGGCGGGCCGTCGCCGACGTCGAACCCTCTCGGGTCCGTGAGGTGTAGGCTGGAGCTGCTTC, and alnHfcamrev, 5′-TCAGGGACGCGGCAGGCGGGGGTGTTCCGGGTGGCGCAGCTGCTGGAAGGATGGGAATTAGCCATGGTCC. As a result, the gene was inactivated, leaving a 58-bp fragment from the 5′ end, a 103-bp scar sequence, and 50 bp from the 3′ end in place of alnH, resulting in pAoriΔalnH. Likewise, the inactivation of alnT was performed using the primers alnTfcamfor, 5′-GTGCGCGAACGCACGCCGCTGACGGCATGGCCGCCTGGAGCGCGACGTCCGTGTAGGCTGGAGCTGCTTC, and alnTfcamrev, 5′-CTACCGGGCCGCTGCCGGACTGCTGGTCTGCGTGGTCTCCGTGGTCCACAATGGGAATTAGCCATGGTCC. After the excision of the resistance gene, alnT was replaced by a 50-bp fragment from the 5′ end, a 103-bp scar sequence, and 50 bp from the 3′ end, resulting in pAoriΔalnT. After conjugation into S. albus, the mutated cosmids were reisolated, transformed back into E. coli TOP10, and verified by restriction enzyme analysis.
Expression and purification of AlnH and AlnT.
Recombinant AlnH protein was produced in 500 ml 2×YT medium (in a 2-liter Erlenmeyer flask) supplemented with 100 μg/ml ampicillin and inoculated with a 5-ml preculture of E. coli TOP10/pBADHisBdalnH. Induction was performed at an optical density at 600 nm (OD600) of 0.6 with l-arabinose to 0.02% (final concentration). After induction, the culture was incubated at 22°C with 250 rpm shaking for 12 h. The cell pellets, harvested by centrifugation, were resuspended in wash buffer (50 mM Na-phosphate buffer [pH 7.2], 100 mM NaCl, 10% [wt/vol] glycerol, 20 mM imidazole) and broken using a French press. The cell extract was purified using a 1-ml HisTrap FF column (GE Healthcare) on an Äktaprime fast-performance liquid chromatography (FPLC) system with a linear gradient. The elution buffer used was the same as the wash buffer, except that it contained 400 mM imidazole and 5 mM β-mercaptoethanol. AlnH was further purified by anion exchange using a 6-ml Resource Q column (GE Healthcare) on an Äktaprime FPLC system with a linear gradient (16 column volumes). The wash buffer (50 mM Tris-HCl [pH 8.0] and 5% glycerol) and elution buffer differed only in salt content (1.5 M NaCl in the elution buffer). After the purification step (see Fig. S1 in the supplemental material), a buffer exchange was performed by using a PD-10 desalting column (GE Healthcare) with 50 mM Na-phosphate buffer (pH 7.2), 100 mM NaCl, 5 mM, β-mercaptoethanol, and 50% (wt/vol) glycerol. AlnH was stored at −20°C, and activity was retained at least for 6 months in storage. Recombinant AlnT protein was obtained by inoculating a 15-liter fermentor (Bioengineering) with 150 ml preculture of E. coli TOP10/pBADHisBdalnT. Induction was performed at an OD600 of 0.8 with a final concentration of 0.02% l-arabinose. Once induced, the cells were incubated at 20°C with 250 rpm stirring for 12 h. The cells were harvested by centrifugation and then stored at −20°C. The cell pellets were thawed, resuspended in wash buffer (50 mM Na-phosphate buffer [pH 8.0], 35 mM imidazole, 500 mM NaCl, 10% [wt/vol] glycerol, 0.5 mM dithiotreitol), 1 mM phenylmethanesulfonylfluoride, and 1 mM benzamidine and broken using a French press. For purification of AlnT, after an initial wash, a second 10-column volume wash step was performed with 5% elution buffer in wash buffer prior to elution of AlnT using 100% elution buffer. The elution buffer contained 500 mM imidazole in the wash buffer. As described above, the pooled, concentrated fractions (see Fig. S2 in the supplemental material) were exchanged into 25 mM Tris-HCl (pH 7.5) with 10% (wt/vol) glycerol. The AlnT obtained was stored at −85°C. Enzyme concentrations were determined at 280 nm by using absorption coefficients calculated with ProtParam (13).
Enzyme reaction conditions.
The enzyme assay mixture included 100 mM phosphate buffer (pH 7.0), 5 μM FMN, 1 mM NADH, approximately 200 μM ThA, and 0.25% (vol/vol) dimethyl sulfoxide in a final volume of 200 μl. AlnT and AlnH were added to the reaction mixture at 8 μM and 0.4 μM (final concentrations), respectively, and the reaction mixture was incubated at room temperature for 10 min. Two successive 50-μl chloroform extractions were performed. The extracted compounds were then vacuum concentrated and resuspended in acetone for high-performance liquid chromatography (HPLC) analysis. For the negative-control reaction, AlnT was incubated in boiling water for 15 min before addition to the reaction mixture.
Spectroscopic analysis of NADH consumption by AlnH.
Kinetic measurements of NADH and NADPH consumption were performed in triplicate 200-μl reaction mixtures on a 96-well plate (Multiskan GO; Thermo Scientific) by measuring absorbance at 340 nm. Concentrations of NADH, NADPH, FMN, and FAD were determined using the extinction coefficients 6,290 M−1 cm−1 (340 nm), 6,220 M−1 cm−1 (340 nm), 12,500 M−1 cm−1 (450 nm) (27), and 11,300 M−1 cm−1 (450 nm) (27), respectively. The reaction assay was performed in 20 mM Tris-HCl (pH 7.5) with varied concentrations of FMN, NADH, FAD, or NADPH. All other components were at non-rate-limiting concentrations (3 times the determined Km values). Kinetic values were obtained by a nonlinear regression analysis of the initial reaction rates using Origin Pro (version 8.0). All reagents were purchased from Sigma except FMN (TCI Europe).
Analysis of metabolites and reaction mixtures.
The XAD-7 resin from the cultivations of both S. albus/pAoriΔalnH and S. albus/pAoriΔalnT was separated from the culture by repeated washing with tap water and decanting. The compounds bound to the XAD-7 were extracted using acetonitrile for subsequent HPLC analysis using an SCL-10Avp HPLC system equipped with an SPD-M10Avp (Shimadzu) diode array detector. A 5-μm LiChroCART 250-4 RP-18 column (Merck) was used with a 25-min gradient from 40% acetonitrile in 0.1% formic acid to 100% acetonitrile at 0.5 ml/min for metabolite analysis. Extracts from reaction mixtures were analyzed using the same HPLC system but with a 3.5-μm Sunfire C18 (Waters) column, which was used with a 25-min gradient from 15% acetonitrile in 0.1% formic acid to 100% acetonitrile at 0.25 ml/min.
Purification of metabolites.
For large-scale purification of ThA and DHPA, S. albus/pAoriΔalnH was cultivated for 5 days. One day prior to harvesting, XAD-7 was added to the culture medium. The cell and XAD pellet were collected via centrifugation and stored at −20°C until extraction. Once thawed, the pellet was resuspended in tap water and the XAD-7 was separated as described above. Metabolites were extracted using acetonitrile. The compounds were fractionated with a 0-to-100% methanol gradient in CHCl3 using normal-phase column chromatography (silica gel 60, 0.040 to 0.063 mm; Merck). For the final polishing step for ThA and DHPA, fractions obtained from the silica column were applied to a preparative LiChroCART 250-10 RP-18 column (10 μm; Merck) with an L-6200A HPLC system (Merck Hitachi) and eluted with aqueous 0.1% formic acid and a 50-to-100% acetonitrile gradient at 2.5 ml/min. For large-scale purification of ThB, the reaction volume was scaled up to 1.5 ml and a total of 17 reactions were carried out using, in total, approximately 2.2 mg ThA. Two successive chloroform extractions (500 μl for every reaction mixture) were performed. The extracted compounds were then vacuum concentrated and resuspended in acetone for purification by a preparative HPLC step using the Merck L-6200A apparatus and a 5-μm Sunfire prep C18 250- by 10-mm column (Waters) with a 25-min gradient from 15% acetonitrile with 0.1% formic acid to 100% acetonitrile at 2.5 ml/min. The fractions containing ThB were extracted by addition of water and chloroform and subsequently vacuum dried. The approximate yield of ThB was 1.6 mg. The HPLC buffers, water, and chloroform were bubbled with nitrogen gas for at least 10 min prior to use. Nitrogen gas was used to fill the vacuum after drying ThB to prevent unwanted oxidation. Nuclear magnetic resonance (NMR) measurements were conducted immediately on the day of enzymatic synthesis at 283 K, since several degradation products of ThB could be observed already on the next day by proton NMR and HPLC.
MS and NMR measurements.
The analysis of samples for mass determination was performed with a high-resolution HPLC-electron spray ionization-mass spectrometry (MS) system using a MicrOTOF-Q mass spectrometer (Bruker) with 4-kV capillary voltage, 573 K dry heater temperature, and a nebulizer pressure of 1.6 × 105 Pa. An Agilent Technologies 1200 series HPLC system equipped with a diode array detector and Sunfire C18 column (Waters) with a 25-min gradient from 15% to 100% acetonitrile in 0.1% formic acid at 0.25 ml/min was used. NMR spectra were collected with a Bruker Avance 500-MHz spectrometer for both ThA and ThB.
RESULTS
Sequence analysis of AlnH and AlnT.
A BLAST search of all nonredundant proteins as well as a CD search (28) with the AlnH sequence in GenBank uncovered a large number of proteins homologous to the FlaRed enzyme superfamily (NCBI accession number cl00801).These enzymes contain a flavin reductase domain or a domain typically associated with flavoprotein oxygenases. The sequence identified with highest similarity was a putative flavin reductase from Streptomyces sp. SA3_actF (GenBank accession number ZP_07976974.1; 57% sequence identity). The well-characterized NADH-dependent FMN reductase ActVB (accession number NP_629242.1 [36, 43, 44]), involved in the biosynthesis of actinorhodin, showed 49% sequence identity. A multiple sequence alignment with four closely related sequences is shown in Fig. S3 of the supplemental material.
Proteins homologous to AlnT belonged to the acyl-CoA dehydrogenase (ACAD) superfamily (NCBI accession number cl09933). The sequence identified with highest similarity was a putative oxygenase (GenBank accession number AEM44243.1) from an uncultured bacterium (54% identity). More relevant in terms of putative function were the angucyclinone hydroxylase LndZ5 (37) from Streptomyces globisporus (53% identity) and ActVA-ORF5 (36, 42–44) from Streptomyces coelicolor A3(2) (48% identity), involved in biosynthesis of the aromatic polyketides landomycin and actinorhodin, respectively. Sequence comparison with the monooxygenase component of p-hydroxyphenylacetate hydroxylase (see Fig. S4 in the supplemental material) from Acinetobacter baumannii (1) identified conserved residues in AlnT that may be involved in catalysis (H396) and formation of hydrophobic interactions (W210) or hydrogen bonds (S171) with the flavin. According to the classification reported by van Berkel et al. (45), AlnT and AlnH form a class D flavoprotein monooxygenase.
Inactivation of alnT and alnH.
The genes alnT and alnH reside in different operons approximately 15.3 kbp apart within the alnumycin gene cluster (35). In order to experimentally verify that the gene products formed a two-component monooxygenase system and were involved in the same biosynthetic step, both genes were individually inactivated from cosmid pAlnuori as described in Materials and Methods by using RED/ET recombineering (8) in E. coli K-12. The resulting cosmids pAoriΔalnT and pAoriΔalnH, containing the alnT and alnH deletions, respectively, were introduced into the heterologous host S. albus through intergeneric conjugation from E. coli ET12567/pUZ8002 (24). Since both alnT and alnH were situated as last genes in their respective operon structures, the gene inactivation experiments were unlikely to stimulate any downstream effects.
Analysis of metabolites produced by the mutant strains.
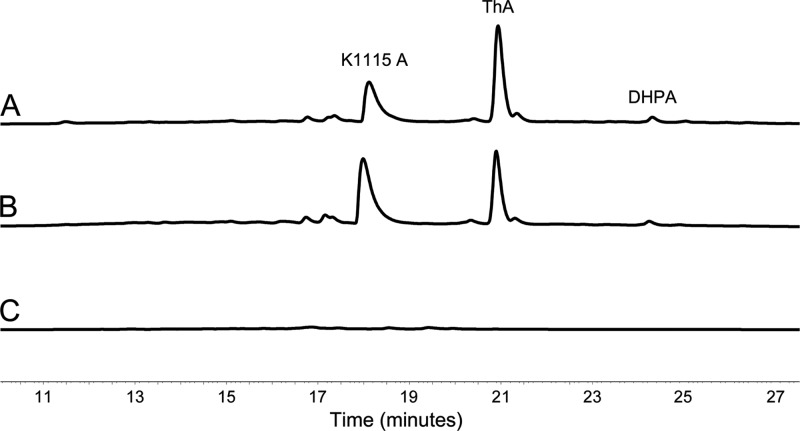
The strains S. albus/pAoriΔalnT and S. albus/pAoriΔalnH were cultivated for 5 days, and the metabolites produced were analyzed by HPLC. Encouragingly, the production profiles of the strains were identical, and three novel metabolites, not detected from cultivations of the parental strain S. albus/pAlnuori, were observed (Fig. 2). One of the compounds was readily identified as K1115 A (Fig. 1), which has been previously obtained through inactivation of the aldo-ketoreductase aln4 (35). For structure elucidation of the two remaining metabolites, the strain S. albus/pAoriΔalnT was grown on a large scale, and the compounds produced were purified by open column chromatography and preparative HPLC.
Fig 2.
Analysis by HPLC monitoring of metabolites produced by the recombinant strains. The chromatogram traces are shown at 410 nm for S. albus/pAoriΔalnH containing K1115 A, DHPA, and ThA (A), S. albus/pAoriΔalnT containing K1115 A, DHPA, and ThA (B), and S. albus (C).
The second compound was confirmed as the decarboxylated form of K1115 A, DHPA (Fig. 1), which has previously been isolated from the marine Streptomyces sp. FX-58 (19). The structure elucidation was based on 1H and 13C NMR spectra (Table 1), and high-resolution MS measurements in the negative mode ([M − H]− observed 281.0848, calculated 281.0814). Consistent with this assessment, the UV-Vis spectrum of DHPA was identical to that of K1115 A (see Fig. S5 in the supplemental material).
Table 1.
NMR spectral data for DHPA, ThA, and ThBa
| Position | NMR spectral data |
|||||
|---|---|---|---|---|---|---|
| DHPA |
ThA |
ThB |
||||
|
1H (ppm) |
13C (ppm) |
1H (ppm) |
13C (ppm) |
1H (ppm) |
13C (ppm)b | |
| 1 | 162.5 | 1.38, s, 3H | 26.2 | 1.36, s, 3H | 26.3 | |
| 1-OH | 13.07, s | |||||
| 2 | 7.28, dd, 1.2, 8.4 | 124.8 | 73.8 | |||
| 2-OH | Exchange | Exchange | ||||
| 3 | 7.59, dd, 7.5, 8.5 | 135.6 | 3.18, dd, 16.5, 1.5 (A) | 40.9 | 3.23, d, 16.7 (A) | 41.5 |
| 3.00, d, 16.5 (B) | 3.06, d, 16.7 (B) | |||||
| 3a | 133.7 | |||||
| 4 | 7.75, dd, 1.2, 7.5 | 118.8 | 6.99, d, 1.5 | 118.3 | 7.42, s | 113.2 |
| 4a | 132.8 | 139.4 | ||||
| 5 | 7.65, d, 2.6 | 112.6 | 7.13, dd, 0.9, 8.2 | 118.0 | ||
| 5-OH | 9.16, s, brd | |||||
| 6 | 150.9 | 7.46, dd, 7.7, 8.3 | 132.4 | 6.98, d, 8.4 | 115.4 | |
| 6-OH | 5.81, s, brb | |||||
| 7 | 7.02, d, 2.6 | 124.1 | 6.83, dd, 0.9, 7.7 | 110.8 | 6.61, d, 8.4 | 110.1 |
| 8 | 140.2 | 158.0 | ||||
| 8-OH | 9.72, d, 1.0 | 8.65, s, brd | ||||
| 8a | 124.2 | 112.9 | ||||
| 9 | 189.7 | 165.3 | ||||
| 9-OH | 15.92, d, 1.0 | Exchange | ||||
| 9a | 117.0 | 108.6 | ||||
| 10 | 183.0 | 206.6 | ||||
| 10a | 137.9 | |||||
| 11 | 3.20, m, 2H | 38.1 | 2.50, ddd, 10.2, 3.6, 1.0 | 60.7 | 2.53, m | 61.4 |
| 12 | 1.69, m, 2H | 23.9 | 1.80, m, (A) | 21.6 | 1.90, m | 21.6 |
| 1.59, m, (B) | 1.58, m | |||||
| 13 | 1.05, t, 3H | 14.3 | 1.11, t, 3H | 13.1 | 1.06, t, 3H | 13.2 |
Chemical shifts are reported relative to tetramethyl silane, which was used as an internal standard. Measurements were conducted in CDCl3 (DHPA) and acetone-d6.
Values were obtained indirectly via heteronuclear single quantum coherence spectroscopy measurements.
For structure elucidation of the third metabolite, the molecular formula C17H17O4 was inferred from the high-resolution mass spectrometry data ([M − H]− observed 285.1123, calculated 285.1127). A literature search revealed that the 1H and 13C spectra (Table 1) for the compound were similar to those reported for HU235, a metabolite isolated from heterologous expression of the R1128 pathway (29). Significantly, only a single carbonyl carbon signal at 206.6 was present in the 13C NMR spectra, indicating that the compound isolated did not contain a quinone ring. The structure was finally verified by heteronuclear single quantum coherence and heteronuclear multiple bond correlation techniques (see Fig. S6 in the supplemental material), which demonstrated that the metabolite was a novel polyketide denoted as ThA (Fig. 1).
In vitro activity of AlnT/AlnH and enzymatic synthesis of ThB.
Both AlnH and AlnT were produced in E. coli TOP10 as N-terminally histidine-tagged recombinant proteins that contained an additional AHHHHHHHRS sequence and purified to near homogeneity (see Fig. S1 and S2 in the supplemental material) as described in Materials and Methods. Based on sequence analysis, it was likely that AlnH was an NADH-dependent flavin reductase. In order to confirm this supposition, AlnH was incubated together with FMN and an excess of NADH. Monitoring of the reaction at 340 nm, specific for NADH, showed a clear decrease in absorbance over time, indicating oxidation of the cofactor to NAD+. Activity was also seen with FAD, although FMN appeared to be the preferred flavin species. Slow consumption of NADPH was also detected with FMN but not with FAD (Table 2). Similar to the closely related flavin reductase homologue ActVB (11, 12), NADH and FMN appeared to be the preferred cofactors (Table 2).
Table 2.
Apparent kinetic parameters for the reductase activities of AlnH and ActVB
| Enzyme | Substrate | Km (μM) | kcat (s−1) | Cosubstrate | Reference |
|---|---|---|---|---|---|
| AlnH | FMN | 1.15 ± 0.08 | 0.56 ± 0.01 | NADH | This study |
| FAD | 12.3 ± 1.5 | 0.64 ± 0.02 | NADH | ||
| NADH | 45.5 ± 2.8 | 1.05 ± 0.02 | FMN | ||
| NADPH | 2,687 ± 1,480 | 0.93 ± 0.38 | FMN | ||
| NADPH | No activity | FAD | |||
| ActVB | FMN | 1.0 ± 0.1 | 9.2 ± 0.4 | NADH | 8 |
| FAD | 8.7 ± 0.6 | 8.2 ± 0.7 | NADH | 9 | |
| NADH | 6.6 ± 0.5 | 9.2 ± 0.4 | FMN | 9 | |
| FMN | 39.6 | 2.7 | NADH | 9 |
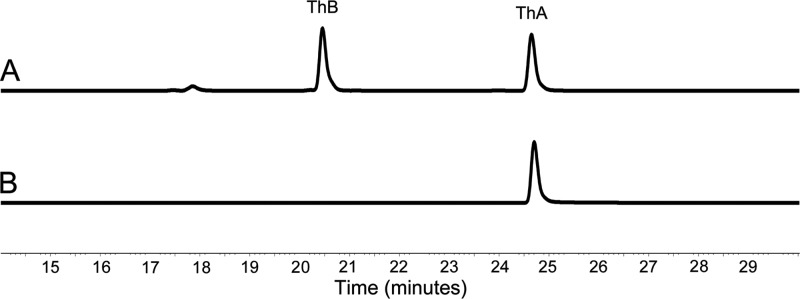
None of the metabolites isolated from the knockout strains contained correctly cyclized pyran rings, which indicates that quinone formation is likely to occur at an earlier stage during polyketide folding and that the natural substrate for AlnT is likely to be an unstable and highly reactive bicyclic intermediate of the pathway. Therefore, in order to demonstrate the regiochemistry of the putative monooxygenase component AlnT, various substrate analogues were screened in a reaction together with AlnH, AlnT, FMN, and NADH. While none of the synthetic compounds tested (e.g., dithranol and 1,8-dihydroxynaphthalene) proved promising, HPLC analysis identified a new peak when ThA was used as a substrate (Fig. 3A). The appearance of the product was dependent on the presence of all of the reaction mixture components mentioned above and, importantly, a control reaction using heat-deactivated AlnT that did not lead to product formation (Fig. 3B). The UV-Vis spectrum of the reaction product was similar to that of ThA but displayed a bathochromic shift of 26 nm, which was consistent with our expectation of a hydroxylation product (see Fig. S5 in the supplemental material). This assumption was verified by high-resolution MS measurements in the negative mode ([M − H]− observed 301.1082, calculated 301.1076), which confirmed the molecular formula, C17H17O5. Despite the instability of the compound, multiple enzymatic reactions provided sufficient material for structure elucidation by 1H NMR and detection of selected carbon signals indirectly via heteronuclear single-quantum correlation spectroscopy. The relatively large coupling constant (3J, 8.4 Hz) between H-6 and H-7 indicated that the protons were vicinal, which thus excluded the m-position for the hydroxyl group at the lateral ring. The distinction between the o- and p-positions was accomplished by detection of the relatively large downfield change in the chemical shift for H-4 (Table 1), which confirmed that the product ThB was the p-hydroquinone (Fig. 1).
Fig 3.
Analysis by HPLC of the AlnT/AlnH reaction. The chromatogram traces are shown at 430 nm for conversion of ThA into ThB in the presence of AlnT, AlnH, FMN, and NADH (A) and the control reaction with ThA in the presence of heat-inactivated AlnT, AlnH, FMN, and NADH (B).
DISCUSSION
The great chemical diversity of aromatic polyketides is brought about via selection of starter units and the number of extension cycles used in the initial polyketide synthesis, variations in cyclization patterns, and through the action of numerous tailoring enzymes (7, 16). Oxidative modifications are arguably one of the most common means of generating diversity in polyketide metabolites, together with methylations, glycosylations, and reductions. This holds especially true in the biosynthesis of polyaromatic metabolites, since many of the pathway intermediates can also react with molecular oxygen nonenzymatically, which could lead to the formation of undesirable shunt products (31). Therefore, in order to assert control over correct regiochemistry, particularly in p-quinone formation, a specific tailoring reaction is requisite.
Enzymes belonging to three different families have been identified to be responsible for p-quinone biosynthesis in various pathways, as indicated above. Gene clusters involved in the biosynthesis of BIQ metabolites harbor gene products homologous to the much-studied p-hydroxyphenyl acetate hydroxylase C1/C2 (1, 5, 40). Of these, ActVA-ORF5 and ActVB have been experimentally demonstrated to form a two-component monooxygenase system and to catalyze quinone formation in the biosynthesis of actinorhodin (36, 44). ActVA-ORF5 has been shown to be the actual FMN-dependent monooxygenase, while ActVB is a flavin:NADH oxidoreductase that provides reduced FMN for ActVA-ORF5. Analysis of the alnumycin A biosynthetic gene cluster revealed that the most likely candidates for production of the quinone arrangement in the lateral ring were the putative monooxygenase AlnT and the flavin reductase AlnH.
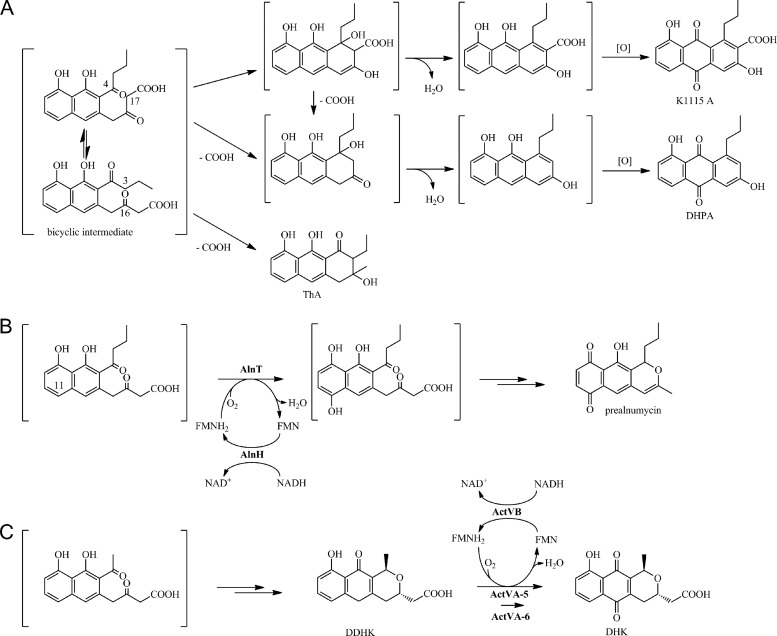
Consistent with the assumption, the production profiles of alnT and alnH deletion mutants were highly similar, indicating that the gene products might be involved in the same biosynthetic step. Somewhat surprisingly, endogenous flavin reductases of S. albus were not able to complement the alnH mutation. Structural elucidation of the metabolites, however, revealed that all of the compounds lacked correct third ring cyclization, which suggested that the substrate for AlnH/AlnT would be a highly reactive bicyclic intermediate of the pathway. Retrosynthesis of K1115 A and DHPA (Fig. 4A) implies that the compounds isolated might have originated from an aldol condensation (41), in this case probably nonenzymatic, between carbons C-4 and C-17 (polyketide numbering) of the bicyclic intermediate. The timing of the decarboxylation of DHPA is more challenging to assess, but as β-keto acid decarboxylation is a well-established reaction in chemistry (26), it may be that the loss of the terminal carboxy group takes place prior to cyclization. In any case, subsequent dehydration and nonenzymatic oxidation of the intermediates would yield the final quinone products (Fig. 4A). In a similar manner, formation of ThA is likely to proceed via β-keto acid decarboxylation and C-3/C-16 cyclization of the same bicyclic intermediate (Fig. 4A).
Fig 4.
Model pathways for formation of the shunt products K1115 A, DHPA, and ThA in S. albus/pAoriΔalnH and S. albus/pAoriΔalnT (A), biosynthesis of prealnumycin in Streptomyces sp. CM020 (B), and pathway towards formation of the model antibiotic actinorhodin in S. coelicolor (36) (C). Abbreviations: DDHK, 6-deoxydihydrokalafungin; DHK, dihydrokalafungin.
We propose that in the alnumycin pathway, AlnH and AlnT form a two-component monooxygenase system that is responsible for hydroxylation of position C-11 (polyketide numbering) of the bicyclic intermediate of the pathway (Fig. 4B). Specifically, our data demonstrate that AlnH is a flavin:NADH oxidoreductase, which is required to provide FMNH2 for AlnT for the actual monooxygenation reaction. The reduced FMN could next react with molecular oxygen, according to the rules established for classical flavin chemistry (30), and form an FMN peroxide intermediate that would be capable of hydroxylating the substrate. Although the probable natural substrate of the reaction is unstable and thus not available for in vitro studies, the enzymatic synthesis of ThB yielded strong evidence for the regiochemistry of the reaction and indicated that the hydroxylation would occur in the lateral ring in the para-position with regard to the hydroxyl group originating from the polyketide backbone.
Our results conclusively verify that both the timing and regiochemistry of alnumycin quinone formation differ from the equivalent reactions in the actinorhodin pathway (36) (Fig. 4C), which was somewhat unexpected due to the high sequence similarity of the monooxygenase components AlnT and ActVA-ORF5. It is interesting, however, that based on sequence similarity, AlnT is in effect more closely related to LndZ5, which is responsible for hydroxylation of the equivalent lateral ring in the biosynthesis of the larger four-ring angucycline antibiotic landomycin (37), than to ActVA-ORF5. It is plausible that an earlier hydroxylation step is required in the alnumycin pathway to prevent unwarranted nonenzymatic quinone formation, as seen in the structures of K1115 A and DHPA. The results presented here also raise an interesting question regarding the timing of quinone formation in the biosynthesis of medermycin and granaticin and whether they follow the paradigms that have now been established for alnumycin (Fig. 4B) and actinorhodin (Fig. 4C) biosynthesis.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Graduate School in Informational and Structural Biology (ISB) and the Academy of Finland (grant numbers 121688, 136060, and 127844).
We thank Olli Martiskainen for assistance with high-resolution mass spectrometry and Maria Aromaa for assistance with chromatographic separations.
Footnotes
Published ahead of print 30 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alfieri A, et al. 2007. Structure of the monooxygenase component of a two-component flavoprotein monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 104:1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bahrami F, Morris DL, Pourgholami MH. 2012. Tetracyclines: drugs with huge therapeutic potential. Mini Rev. Med. Chem. 12:44–52 [DOI] [PubMed] [Google Scholar]
- 3. Bieber B, Nüske J, Ritzau M, Gräfe U. 1998. Alnumycin: a new naphthoquinone antibiotic produced by an endophytic Streptomyces sp. J. Antibiot. (Tokyo) 51:381–382 [DOI] [PubMed] [Google Scholar]
- 4. Björn B, Jörg N. 1998. Alnumycin useful as antibiotic. German patent DE19745914.
- 5. Chaiyen P, Suadee C, Wilairat P. 2001. A novel two-protein component flavoprotein hydroxylase. Eur. J. Biochem. 268:5550–5561 [DOI] [PubMed] [Google Scholar]
- 6. Chater KF, Wilde LC. 1980. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J. Gen. Microbiol. 116:323–334 [DOI] [PubMed] [Google Scholar]
- 7. Das A, Khosla C. 2009. Biosynthesis of aromatic polyketides in bacteria. ACC Chem. Res. 42:631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duggan ST, Keating GM. 2011. Pegylated liposomal doxorubicin: a review of its use in metastatic breast cancer, ovarian cancer, multiple myeloma and AIDS-related Kaposi's sarcoma. Drugs 71:2531–2558 [DOI] [PubMed] [Google Scholar]
- 10. Fetzner S. 2007. Cofactor-independent oxygenases go it alone. Nat. Chem. Biol. 3:374–375 [DOI] [PubMed] [Google Scholar]
- 11. Filisetti L, Fontecave M, Niviere V. 2003. Mechanism and substrate specificity of the flavin reductase ActVB from Streptomyces coelicolor. J. Biol. Chem. 278:296–303 [DOI] [PubMed] [Google Scholar]
- 12. Filisetti L, Valton J, Fontecave M, Nivière V. 2005. The flavin reductase ActVB from Streptomyces coelicolor: characterization of the electron transferase activity of the flavoprotein form. FEBS Lett. 579:2817–2820 [DOI] [PubMed] [Google Scholar]
- 13. Gasteiger E, et al. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607 In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ [Google Scholar]
- 14. Grocholski T, et al. 2010. Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism. Biochemistry 49:934–944 [DOI] [PubMed] [Google Scholar]
- 15. Hanušová V, Boušová I, Skálová L. 2011. Possibilities to increase the effectiveness of doxorubicin in cancer cells killing. Drug Metab. Rev. 43:540–557 [DOI] [PubMed] [Google Scholar]
- 16. Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. 2007. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 24:162–190 [DOI] [PubMed] [Google Scholar]
- 17. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 18. Hopwood DA, et al. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 19. Huang YF, Tian L, Sun Y, Pei YH. 2006. Two new compounds from marine Streptomyces sp. FX-58. J. Asian Nat. Prod Res. 8:495–498 [DOI] [PubMed] [Google Scholar]
- 20. Ichinose K, et al. 1998. The granaticin biosynthetic gene cluster of Streptomyces violaceoruber Tü22: sequence analysis and expression in a heterologous host. Chem. Biol. 5:647–659 [DOI] [PubMed] [Google Scholar]
- 21. Ichinose K, Ozawa M, Itou K, Kunieda K, Ebizuka Y. 2003. Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology 149:1633–1645 [DOI] [PubMed] [Google Scholar]
- 22. Kallio P, et al. 2011. Flavoprotein hydroxylase PgaE catalyzes two consecutive oxygen-dependent tailoring reactions in angucycline biosynthesis. Biochemistry 50:5535–5543 [DOI] [PubMed] [Google Scholar]
- 23. Kallio P, Sultana A, Niemi J, Mäntsälä P, Schneider G. 2006. Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. J. Mol. Biol. 357:210–220 [DOI] [PubMed] [Google Scholar]
- 24. Kieser T, Bibb M, Buttner M, Chater K, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 25. Koskiniemi H, et al. 2007. Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis. J. Mol. Biol. 372:633–648 [DOI] [PubMed] [Google Scholar]
- 26. Logue MW, Pollack RM, Vitullo VP. 1975. Nature of the transition state for the decarboxylation of β-keto acids. J. Am. Chem. Soc. 97:6868–6869 [Google Scholar]
- 27. Macheroux P. 1999. UV-visible spectroscopy as a tool to study flavoproteins. Methods Mol. Biol. 131:1–7 [DOI] [PubMed] [Google Scholar]
- 28. Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marti T, Hu Z, Pohl NL, Shah AN, Khosla C. 2000. Cloning, nucleotide sequence, and heterologous expression of the biosynthetic gene cluster for R1128, a non-steroidal estrogen receptor antagonist. Insights into an unusual priming mechanism. J. Biol. Chem. 275:33443–33448 [DOI] [PubMed] [Google Scholar]
- 30. Massey V. 2000. The chemical and biological versatility of riboflavin. Biochem. Soc. Trans. 28:283–296 [PubMed] [Google Scholar]
- 31. McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. 1995. Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature 375:549–554 [DOI] [PubMed] [Google Scholar]
- 32. Naruse N, Goto M, Watanabe Y, Terasawa T, Dobashi K. 1998. K1115 A, a new anthraquinone that inhibits the binding of activator protein-1 (AP-1) to its recognition sites. II. Taxonomy, fermentation, isolation, physico-chemical properties and structure determination. J. Antibiot. (Tokyo) 51:545–552 [DOI] [PubMed] [Google Scholar]
- 33. Newman DJ, Cragg GM. 2007. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70:461–477 [DOI] [PubMed] [Google Scholar]
- 34. Oja T, et al. 2 April 2012, posting date Biosynthetic pathway toward carbohydrate-like moieties of alnumycins contains unusual steps for C-C bond formation and cleavage. Proc. Natl. Acad. Sci. U. S. A. doi:10.1073/pnas.1201530109 [DOI] [PMC free article] [PubMed]
- 35. Oja T, et al. 2008. Characterization of the alnumycin gene cluster reveals unusual gene products for pyran ring formation and dioxan biosynthesis. Chem. Biol. 15:1046–1057 [DOI] [PubMed] [Google Scholar]
- 36. Okamoto S, Taguchi T, Ochi K, Ichinose K. 2009. Biosynthesis of actinorhodin and related antibiotics: discovery of alternative routes for quinone formation encoded in the act gene cluster. Chem. Biol. 16:226–236 [DOI] [PubMed] [Google Scholar]
- 37. Ostash B, et al. 2004. Generation of new landomycins by combinatorial biosynthetic manipulation of the LndGT4 gene of the landomycin E cluster in S. globisporus. Chem. Biol. 11:547–555 [DOI] [PubMed] [Google Scholar]
- 38. Sciara G, et al. 2003. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 22:205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siitonen V, et al. 2012. Identification of late-stage glycosylation steps in the biosynthetic pathway of the anthracycline nogalamycin. Chembiochem 13:120–128 [DOI] [PubMed] [Google Scholar]
- 40. Sucharitakul J, Chaiyen P, Entsch B, Ballou DP. 2006. Kinetic mechanisms of the oxygenase from a two-component enzyme, p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii. J. Biol. Chem. 281:17044–17053 [DOI] [PubMed] [Google Scholar]
- 41. Sultana A, et al. 2004. Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation. EMBO J. 23:1911–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taguchi T, Okamoto S, Hasegawa K, Ichinose K. 2011. Epoxyquinone formation catalyzed by a two-component flavin-dependent monooxygenase involved in biosynthesis of the antibiotic actinorhodin. Chembiochem 12:2767–2773 [DOI] [PubMed] [Google Scholar]
- 43. Valton J, Fontecave M, Douki T, Kendrew SG, Nivière V. 2006. An aromatic hydroxylation reaction catalyzed by a two-component FMN-dependent monooxygenase. The ActVA-ActVB system from Streptomyces coelicolor. J. Biol. Chem. 281:27–35 [DOI] [PubMed] [Google Scholar]
- 44. Valton J, Mathevon C, Fontecave M, Nivière V, Ballou DP. 2008. Mechanism and regulation of the two-component FMN-dependent monooxygenase ActVA-ActVB from Streptomyces coelicolor. J. Biol. Chem. 283:10287–10296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Berkel WJ, Kamerbeek NM, Fraaije MW. 2006. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124:670–689 [DOI] [PubMed] [Google Scholar]
- 46. Xu Z, Metsä-Ketelä M, Hertweck C. 2009. Ketosynthase III as a gateway to engineering the biosynthesis of antitumoral benastatin derivatives. J. Biotechnol. 140:107–113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.