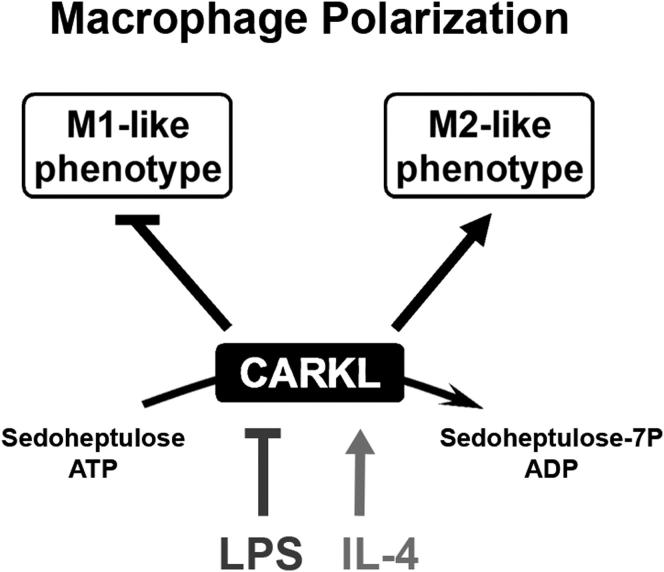
Summary
Immune cells are somewhat unique in that activation responses can alter quantitative phenotypes upwards of 100,000-fold. To date little is known about the metabolic adaptations necessary to mount such dramatic phenotypic shifts. Screening for novel regulators of macrophage activation, we found nonprotein kinases of glucose metabolism among the most enriched classes of candidate immune modulators. We find that one of these, the carbohydrate kinase-like protein CARKL, is rapidly downregulated in vitro and in vivo upon LPS stimulation in both mice and humans. Interestingly, CARKL catalyzes an orphan reaction in the pentose phosphate pathway, refocusing cellular metabolism to a high-redox state upon physiological or artificial downregulation. We find that CARKL-dependent metabolic reprogramming is required for proper M1- and M2-like macrophage polarization and uncover a rate-limiting requirement for appropriate glucose flux in macrophage polarization.
Graphical Abstract
Highlights
► Screened 199 human kinases for their immunoregulatory potential ► CARKL bridges glycolysis, the pentose phosphate pathway, and immune function ► CARKL focuses cellular metabolism toward a “high-redox” state ► CARKL regulation is required for macrophage polarization
Introduction
More than a century after the discovery of macrophages, our understanding of the complexity of innate immune regulation continues to expand (Medzhitov, 2010; Nathan and Ding, 2010). Major recent advances in deciphering macrophage function include the identification of functionally distinct macrophage subtypes, pattern recognition receptors, and macrophages as a functional interface, bridging adaptive and innate immune responses (Gordon and Martinez, 2010; Janeway, 1989; Medzhitov, 2009). Macrophages orchestrate a complex response to external stressors and participate in tissue development, homeostasis, and remodeling. These processes require activation of numerous signaling modules and dramatic alterations in cell morphology and function (Martinon et al., 2009; Rock and Kono, 2008). That said, surprisingly little is known of how energy metabolism is reconfigured to support macrophage activation and effector function.
The concept of cellular metabolic adaptation as a necessary step for functional cellular reprogramming was first described by Otto Warburg in 1924 (Warburg, 1956; Warburg et al., 1924). His observation, that cancer cells primarily generate lactate from glucose even in the presence of ample oxygen, is thought to provide a bioenergetic balance buffering oxygen availability, ATP demand, and carbon building block requirements for cell growth and division (Vander Heiden et al., 2009). In analogy, CD8+ T cells, which also undergo substantial clonal expansion, shift from oxidative metabolism in the resting state toward anaerobic respiration upon activation (Fox et al., 2005; Perl et al., 2002). Very recently, differentiation of Th17 cells and dendritic cell activation was shown to rely on metabolic-reconfiguration (Beurel et al., 2011; Krawczyk et al., 2010; Shi et al., 2011). In macrophages, increased lactate formation and activation of the pentose phosphate pathway (PPP) after phagocytosis have been observed, suggesting potential importance of adapted metabolism on the activation cascade (Drapier and Hibbs, 1988; Schnyder and Baggiolini, 1978). Also, endotoxemia in rats has been shown to influence the PPP in the liver (Kuttner et al., 1982). To what extent proper macrophage activation depends on bioenergetic rearrangement, however, remains poorly understood.
In the current study, we use a screening strategy and identify novel regulators of macrophage activation. We show that specific modulation of glycolytic energy flux is critical to macrophage activation and defines polarity. A number of nonprotein nutrient kinases appear critical in this setting, including CARKL, which we show to be a sedoheptulose kinase of the pentose phosphate pathway. Together, the findings establish a platform for understanding how bioenergetics controls macrophage fate and function.
Results
A Functional Screen for Kinases Involved in Macrophage M1 Polarization
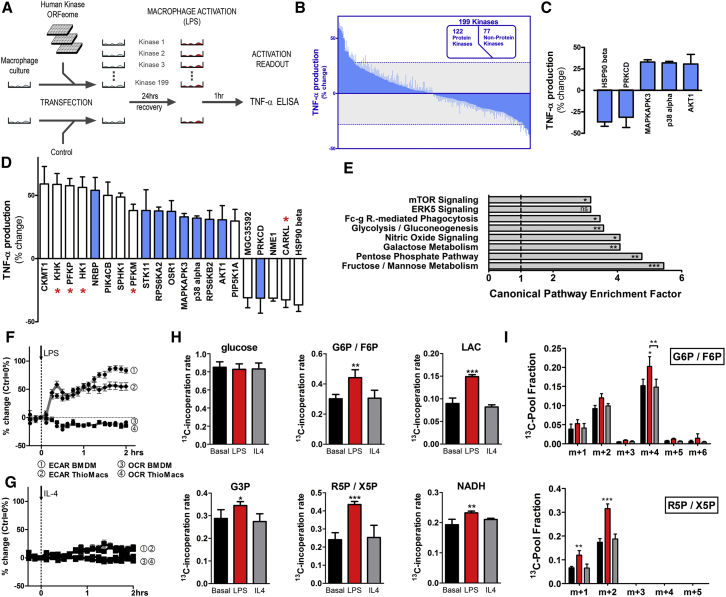
In order to identify novel regulators of macrophage activation, we performed an overexpression screen of 199 known or predicted kinases of the human kinase ORFeome collection (Park et al., 2005). Coding sequences for 122 protein kinases and 77 nonprotein kinases were overexpressed in RAW264.7 macrophages and LPS-induced changes in TNFα secretion monitored by ELISA (Figures 1A and 1B and Table S1). Double-blind analysis of the well-known activation-inducing kinases MAPKAPK3, p38α, and AKT1 revealed prominent potentiation of the LPS-induced TNFα-secretory response (Figure 1C; Kim et al., 2008; Ojaniemi et al., 2003; Schieven, 2009). In contrast, robust inhibitory effects were observed for the known suppressive kinases PKCδ and HSP90-β (Figure 1C; Chen et al., 2005; Kim et al., 2007; Salminen et al., 2008).
Figure 1.
Kinase Screen Reveals Distinct Metabolic Adaptation in Activated Macrophages
(A) Schematic of kinase screen.
(B and C) (B) Kinome modulation of LPS-induced (100 ng/ml; 1 hr) TNFα production (mean change ± SD) in RAW264.7 cells, including (C) kinases with known immune regulatory functions. Dotted lines indicate screen cut-off.
(D) 21 Kinases exceeded the screen cut-off. Blue bars represent protein kinases (PK) and white bars nonprotein kinases (NPK). Kinases involved in primary glucose metabolism are indicated by ∗.
(E) Enrichment analysis for Canonical Pathways.
(F and G) (F) Bone marrow-derived or thioglycollate-elicited macrophages were stimulated with LPS (100 ng/ml) or (G) IL-4 (10 ng/ml), and ECAR and OCR were recorded.
(H and I) Dynamic (nonstationary) metabolic flux anaylsis of LPS or IL-4 stimulated BMDM by incubation with 100% labeled 13C-1-2-glucose. For abbreviations, see text. (H) shows isotope incorporation rate (m+n/total/10 min; m+n is all 13C-labeled molecules irrespective of mass shift, and total is all labeled and unlabeled), and (I) shows isotope distribution; ± SD, n = 5; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Based on the effect size of the double-blind control analysis we set a conservative screen cut-off of ± 4 SDs (∼30% change in TNFα) to determine candidate immunoregulatory kinases. Twenty-one kinases exceeded the cut-off (Figure 1D). Overexpression of 16 kinases potentiated TNFα secretion. The remaining five suppressed the TNFα response (Figure 1D). Interestingly, substantial enrichment of nonprotein kinases over protein kinases was observed in both the activating and suppressive pools of candidate genes (white bars; Figures 1D and S1A). Gene ontology analysis (IPA, Ingenuity) revealed top enrichment scores for “organismal injury,” “inflammatory response,” pathways of cell signaling, and notably, carbohydrate and lipid metabolism (Figure S1B and Tables S2 and S3). Most intriguingly, when analyzed according to classic canonical pathways, 5 of the top 8 pathways comprised core processes of primary metabolism including glycolysis, fructose, mannose, and galactose metabolism, mTOR signaling, and the pentose phosphate pathway (Figure 1E and Table S4). The candidate immunoregulatory kinases responsible for this robust enrichment included hexokinase 1 (HK1), ketohexokinase (KHK), phosphofructokinase platelet-type (PFKP), phosphofructokinase muscle-type (PFKM), and the recently described carbohydrate kinase-like protein, CARKL (EMBL-Bank: AF163573.1). This robust signal overlapped and was in agreement with the enrichment in nonprotein kinases mentioned above (Figures 1D and S1A). Thus, a panel of metabolic nonprotein kinases appears to control macrophage activation.
Defining Cellular Glucose Flux during M1 and M2 Polarization
Considering the robust enrichment for metabolic kinases we hypothesized a substantial reprogramming of metabolism upon macrophage activation. To assess metabolic plasticity upon activation, we recorded extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) in two types of nontransformed primary mouse macrophages (bone marrow-derived macrophages [BMDMs] and thioglycollate-elicited macrophages). LPS stimulation, which drives an M1-like or classical activation pathway, increased ECAR and decreased OCR (Figures 1F and S1C). IL-4, on the other hand, which triggers M2-like activation, induced only a delayed and marginal increase in ECAR but did not affect OCR (Figure 1G). Thus, M1-activation comprises robust metabolic reprogramming toward aerobic glycolysis. To validate these findings, we used nonstationary metabolic flux measurements to trace glucose fate within the cell (Figures 1H and 1I). A 10 min pulse of asymmetrically labeled 13C-1-2-glucose allowed simultaneous monitoring of the pathway flux, origin of labeled metabolites, and thus flux-directionality. In agreement with the data above, LPS-stimulation increased glycolytic-flux (production of glyceraldehyde-3-phosphate [G3P], lactate [LAC] and glucose-6P/fructose-6P [G6P/F6P]) (Figure 1H) and routed a substantial portion of glycolysis-derived carbon into the PPP (m+2 ribose-5P/xylulose5-P [R5P/X5P] in Figure 1I). High m+4 label in G6P/F6P further indicated that glucose is rerouted via glycolysis to the PPP. Sedoheptulose-7-phosphate (S7P), a PPP-derived reaction partner of G3P, displayed relatively high incorporation rates after glucose incubation if compared to, e.g., lactate (Figure S1D). Of note, we observed no significant increase in total S7P flux, but isotopic distribution indicated a minor drop in the m+1 label and a slight increase in m+2. The minor flux of glucose-derived carbon to S7P indicates a small fraction of glucose-derived S7P enters the PPP via G6PD. Increased PPP activity is associated with enhanced reduction of oxidized redox couples. Indeed, LPS stimulation increased the formation of NADH (Figure 1H) and glutathione (GSH), a phenotype sensitive to blockade of glucose-flux by DHEA or oxamate (Figure S1E). The metabolic effect of M2-like activation by IL-4, however, was marginal (Figures 1G–1I and S1C–S1E). Thus, classical macrophage activation is characterized by increased glycolytic and PPP flux.
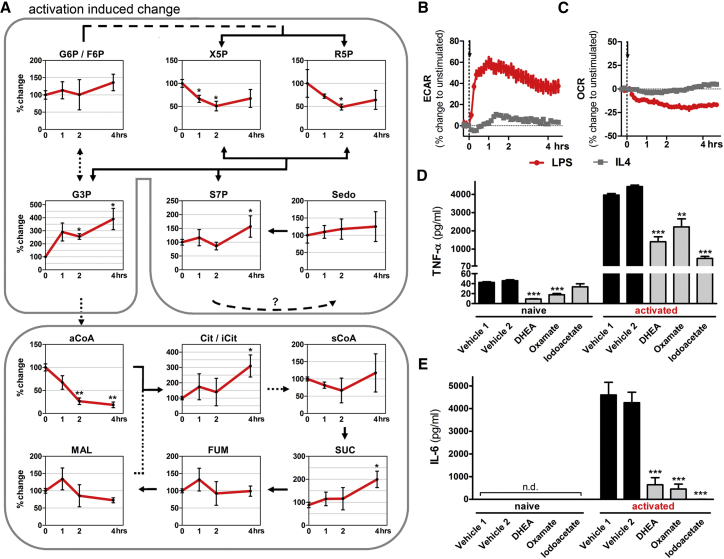
Functional Reconfiguration of Cellular Metabolism during M1 Polarization
A more detailed and time-resolved steady-state metabolomics experiment was used to monitor levels of intermediates in glycolysis, the PPP and tricarboxylic acid cycle (TCA) (Figures 2A and S2A). This approach revealed that LPS-induced polarization induced rapid and significant decreases in X5P and R5P levels, and a slower increase in S7P and G3P with time. These metabolic intermediates constitute a PPP-glycolysis hub. Of note, in the same 4 hr time-frame significant changes were observed in acetyl coenzyme A (aCoA), succinate, and NAD/NADH ratio (Figures 2A and S2B). These findings are supported by LPS-induced ECAR and decreased OCR (Figures 2B and 2C). Again, IL-4 induced only a minimal increase in ECAR but did not affect OCR in both RAW264.7 and primary macrophages (Figures 2B, 2C, and 1G).
Figure 2.
LPS-Induced Polarization in Macrophages Results in a Functional Metabolic Adaptation
(A) Analysis of the initial M1-like activation phase by a steady-state metabolomic time-course in RAW264.7 empty vector control cells (pCtrl = 100%). In the metabolic-pathway illustration, solid lines represent direct metabolite synthesis, whereas dashed lines indicate multiple enzymatic steps.
(B and C) (B) ECAR and (C) OCR recordings of RAW264.7 cells during LPS (100 ng/ml) or IL-4 (10 ng/ml) induced activation.
(D and E) (D) TNF-α and (E) IL-6 cytokine production of RAW264.7 cells pretreated with dehydroepiandrosterone (DHEA, 200 μM), oxamate (40 mM), or iodoacetate (100 μM) 10 min before stimulation with 100 ng/ml LPS for 2 hr (TNF-α) or 6 hr (IL-6). All data represent mean ± SEM of at least three individual experiments; n.d. = not detected, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To test the relevance of metabolic reconfiguration during M1 polarization, we employed inhibitors to interfere with glucose metabolism and measured cytokine production as a functional readout. Importantly, blockade (by DHEA) of glucose-6-phosphate dehydrogenase (G6PD; rate limiting enzyme of the PPP), glyceraldehyde-3-phosphate dehydrogenase (by iodoacetate), and lactate production (by oxamate) significantly reduced activation-induced TNFα and IL-6 production in RAW264.7 (Figures 2D and 2E, respectively) and primary (Figures S2C and S2D, respectively) macrophages. These results confirm a pivotal role for defined metabolic rewiring and select metabolic endpoints in M1 polarization.
CARKL Is a Candidate Immune Regulator In Vitro and In Vivo
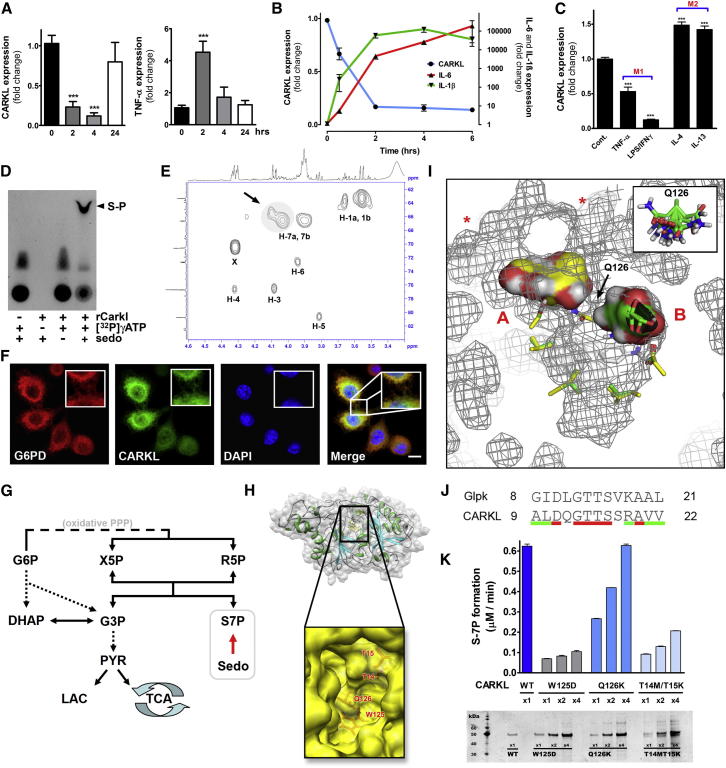
Our kinase screen identified CARKL as possible repressor of LPS-induced macrophage activation. CARKL, in contrast to the two currently known metabolic-kinases (HK, phosphofructokinase), blocked TNFα production (Figure 1D). To begin to define the role of CARKL in immune regulation we measured expression kinetics in peripheral blood mononuclear cells (PBMCs) isolated from volunteers following i.v. injection of LPS (2 ng/kg) (Marsik et al., 2006) (Figure 3A). In this model of human endotoxemia we detected a sharp transient decline of CARKL message followed by normalization at 24 hr. TNFα expression peaked 2 hr post LPS administration. Similar downregulation and kinetics were also observed in cultured human monocytes and, importantly, in nucleated peripheral blood cells in LPS-injected mice (Figure S3A). Thus, rapid CARKL downregulation is a conserved response to acute LPS treatment in mice and humans in vivo and in vitro.
Figure 3.
Regulation, Function, and Structural Evaluation of CARKL
(A) Human CARKL and TNFα mRNA levels in PBMCs from healthy volunteers administered i.v. LPS (2 ng/kg).
(B) CARKL and cytokine mRNA expression in RAW264.7 cells incubated with LPS (100 ng/ml).
(C) CARKL expression in thioglycollate-elicited peritoneal macrophage polarized to either the M1- or M2-like phenotype by stimulation with LPS (100 ng/ml) in combination with IFN (20 ng/ml) or by TNFα (25 ng/ml) for M1 and for M2 with IL-4 (20 ng/ml) or IL-13 (10 ng/ml) for 2 hr. Data represent mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Recombinant CARKL (rCARKL) formed sedoheptulose-phosphate (S-P) as shown by in vitro kinase assay with 32P labeled ATP resolved on thin layer chromatography.
(E) 1H/13C HSQC spectrum of purified reaction product. X = residual HEPES buffer.
(F) Confocal fluorescence imaging of macrophages expressing CARKL_eGFP (green). Nuclei were visualized by DAPI (blue), and for colocalization cells were stained for glucose-6-phosphate dehyrdogenase (G6PD) (red). Scale bar equals 5 μm.
(G) Simplified scheme of glucose metabolism indicating the central position of CARKL functioning as sedoheptulose kinase. Glucose-6-phosphate (G6P), xylulose-5-phosphate (X5P), ribose-5-phosphate (R5P), dihydroxyacetone phosphate (DHAP), glyceraldehyde-3-phosphate (G3P), sedoheptulose-7-phosphate (S7P), sedoheptulose (Sedo), pyruvate (PYR), lactate (LAC), and tricarboxylic acid cycle (TCA).
(H) Three-dimensional model for CARKL protein by comparative modeling. Insert shows opening of the central pocket.
(I) A representative result of computational sedoheptulose docking to CARKL presented in a 3D mesh model where the central cleft is centered and surface accessibility sites are indicated by ∗. AA Q126, defined as flexible AA, is depicted in the inset.
(J) Amino acid (AA) sequence alignment of ATPase domain fragment from glycerol kinase (Glpk) sequence to CARKL (conserved AA red; similar AA green).
(K) Activities of recombinant CARKL point mutants for predicted critical AAs. Silver stain below bar graph indicated equal protein quantities.
In keeping with the marked in vivo regulation, we observed a highly significant reduction in CARKL expression within 30 min post LPS stimulation in RAW264.7 cells (Figure 3B). Messenger RNA levels were reduced by approximately 90% after 2 hr. Interestingly, the timing of CARKL downregulation was synchronous with induction of the prototypical M1 cytokines interleukin IL-6 and IL-1β. To test specificity of the CARKL response to LPS, we also induced macrophage polarization by alternate activation pathways. Whereas primary murine macrophages polarized to an M1-like state showed a significant reduction in CARKL expression, activation with IL-4 or IL-13, agents which drive “M2-like” responses, resulted in mildly increased CARKL expression (Figure 3C). Together, these data strongly suggest a role for CARKL modulation in macrophage polarization in vitro and in vivo in both mouse and man.
CARKL Is a Sedoheptulose Kinase
Murine CARKL is a 476 amino acid (AA) protein containing an FGGY carbohydrate kinase domain. Recent studies have suggested that CARKL is a sedoheptulose kinase (converting sedoheptulose to sedoheptulose-7-phosphate) (Kardon et al., 2008; Wamelink et al., 2008), though direct molecular proof for such a role has been lacking. Using an ADP-accumulation assay, we observed that the introduction of recombinant CARKL and ATP resulted in ADP build-up in the presence but not absence of sedoheptulose, direct evidence that CARKL possesses a sedoheptulose-dependent ATPase activity (data not shown). Further, using thin layer chromatography we found that incubation of CARKL with sedoheptulose and [32P]γATP produced radioactively labeled sedoheptulose-phosphate (Figure 3D). Absence of any single reactant resulted in the complete loss of labeled product. Importantly, 31P-, 1H-, and 13C-NMR analysis of cold reaction product (Figures 3E and S3B) specifically placed CARKL phosphorylation of sedoheptulose at the C-7 carbon, direct evidence of a sedoheptulose kinase activity necessary to link glycolysis and the PPP. For an in-depth technical description of these structural results and enzyme kinetics please refer to Figures S3B and S3C. In keeping with this demonstration, we observed colocalization of CARKL with the first enzyme of the PPP, glucose-6-phosphate dehydrogenase (G6PD), and the metabolic scaffold protein tubulin, by confocal microscopy (Figures 3F and S3D). Thus, CARKL bridges glycolysis and PPP by catalyzing the formation of S7P from sedoheptulose (Figure 3G).
We next used a combination of homology- and comparative-based in silico modeling to predict potential functional domains and substrate interaction sites for CARKL. The resulting model (Figures 3H and S3E), when combined with in silico substrate docking experiments (Figure 3I), predicted several key features for the enzyme-substrate complex: (i) a putative catalytic cleft lies in the center of CARKL with two potential sedoheptulose binding sites; (ii) one face of the cleft comprises a highly conserved ATPase domain similar to that of glycerol kinase, including a homologous pair of threonine residues (Glpk: T13/T14; CARKL: T14/T15) for hydrogen bond formation with ATP (Figure 3J); (iii) a highly flexible glutamine side chain (Q126) separates the central cleft to form two pockets, A and B (Figure 3I); (iv) sedoheptulose docking ranges from an extended linear form in pocket B to a coiled open-chain structure in pocket A (Figure 3I); (v) two additional potential access sites open into pocket A (Figure 3I, asterisks); and (vi) a highly conserved tryptophan (W125) residue limits the internal space of the cavity (Figure 3H, insert). Importantly, this modeling exercise allowed us to predict amino acid residues critical for CARKL activity. In order to test the structural predictions, we generated point mutants of the identified residues (W125D, Q126K, and T14M/T15K), purified the proteins, and tested them for their ability to phosphorylate sedoheptulose (Figure 3K). Notably, all three mutants showed substantially reduced sedoheptulose kinase activity, strongly supporting the structural approach. Specifically, the T14M/T15K mutant, predicted to impair ATP hydrolysis, as well as the W125D substitution, which deforms and adds charge to the catalytic cleft (Figure S3E), exhibited virtually complete abrogation of kinase function. Interestingly, activity of the potential “shuttling” mutant Q126K, while impaired, could be recovered by addition of excess enzyme. All together, these data identify CARKL as a sedoheptulose kinase and provide the first molecular insights into its catalytic mechanism.
CARKL Focuses Glucose Metabolism and Marcrophage Polarization
To investigate the necessity for CARKL in cellular metabolism and ultimately M1 polarization, we generated a stable cell line overexpressing CARKL (pCARKL; Figure 4A). Interestingly, metabolomic profiling of naive pCARKL cells alone revealed significant alterations in intracellular metabolism, including reduced G3P, R-5P, and X-5P levels (Figures 4B and S4A). These findings implicate CARKL in control of metabolic state, and in particular the nonoxidative arm of PPP. In a direct test of this idea, assessment of OCR and ECAR revealed distinct and exclusive reductions in nonmitochondrial respiration (rotenone-resistant oxygen consumption) in pCARKL cells, while ECAR levels were largely unaffected (Figures 4C and S4B).
Figure 4.
CARKL Reconfigures Cellular Metabolism and Represses Macrophage Activation
(A) CARKL protein expression levels in stable overexpressors (pCARKL) and empty vector control cells (pCtrl). Cell lines were derived from RAW264.7 cells.
(B) Metabolic intermediates in pCARKL cells relative to pCtrl cells.
(C) OCR and ECAR of pCARKL cells relative to pCtrl cells.
(D) CARKL expression in pCARKL cells in the presence and absence of LPS.
(E) Relative mRNA expression of (a) CARKL and classical LPS target genes ([b] receptors, [c] cytokines, [d] chemokines) following LPS (100 ng/ml) administration (pCtrl, squares on black; pCARKL, triangles on blue).
(F) MHC class II surface expression of resting (basal, dashed lines) and LPS-activated (solid lines, for 24 hr) macrophages measured by FACS. Histogram is representative of at least three independent experiments.
(G) NF-κB activity measured by a cis-reporting luciferase construct in pCARKL and pCtrl cells before and after activation with LPS (100 ng/ml) for 8 hr.
(H) SOCS3 and SOCS1 mRNA expression in pCARKL (blue) and pCtrl cells (black) relative to unstimulated pCtrl cells.
(I) Nuclear protein fraction from LPS-stimulated (2 hr) pCtrl and pCARKL cells was analyzed by western blot for the presence of phosphorylated STAT3 (STAT3-P) and analyzed by densitometry.
(J) Quantification of intracellular superoxid (O2.-) production rates in macrophages before and after LPS exposure (2 hr) measured by electron spin resonance spectroscopy. Data are means ± SEM of at least three independent experiments; ns = not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
These findings suggest CARKL as a rheostat for cellular metabolism. Using a CMV promoter to express CARKL, we reasoned that pCARKL cells would be largely unaffected by LPS-induced CARKL downregulation, an ideal platform to test the dependency of M1 polarization on CARKL downregulation. Consistent with this hypothesis, pCARKL cells showed no decrease in CARKL gene expression after LPS stimulation (Figure 4D). CARKL overexpression resulted in a severely blunted “proinflammatory” response. Specifically, IL-6, IL-1α, IL-1β, TNFα, TNFsf15, MIP1α (Ccl3), MCP1 (Ccl2), CxCL2, and the IL-13 receptor were repressed upon CARKL overexpression, while TLR4 and CD11b remained unaffected (Figures 4E and S4C). In contrast, the “anti-inflammatory” associated constituents IL-10, macrophage migration inhibitory factor (MIF), and C-C motif cytokine 7 (Ccl7, MCP3), and IL-4R all showed exaggerated responses in the pCARKL cells after LPS. These data, together with reduced MHC class II surface staining upon LPS stimulation (Figure 4F), indicated a blunted M1-like activation profile. Thus CARKL downregulation appears critical for proper M1 polarization.
Next, we explored NFκB/SOCS/STAT3 signaling as well as intracellular superoxide production rates, hallmark mechanistic features of M1-like activation. A luciferase reporter assay revealed a moderately reduced basal and LPS-activated NFκB transcriptional activity in pCARKL cells (Figure 4G), a result confirmed by western blot analysis of nuclear RelA (Figure S4D). IκBα phosphorylation and degradation were not affected (Figure S4E). Further, we observed decreased SOCS3 mRNA expression (Figure 4H) and increased phosphorylation of its target STAT-3 (Figure 4I) in pCARKL cells after LPS stimulation. LPS-induced SOCS3 blocks activation of STAT3, a transcription factor inhibitory to proinflammatory mediator production (Murray, 2005). These findings are in agreement with CARKL-induced repression of proinflammatory gene expression. Similarly, measurement of intracellular superoxide production (O2.-) rates using electron spin resonance (Figure S4F) demonstrated severely blunted radical production (Figure 4J), an effect apparently mediated by sustained S7P formation (Figure S4G). These results indicated “proinflammatory” signal transducers repressed and “anti-inflammatory” signals activated by CARKL.
CARKL Downregulation Is Required for M1-like Metabolic Reprogramming
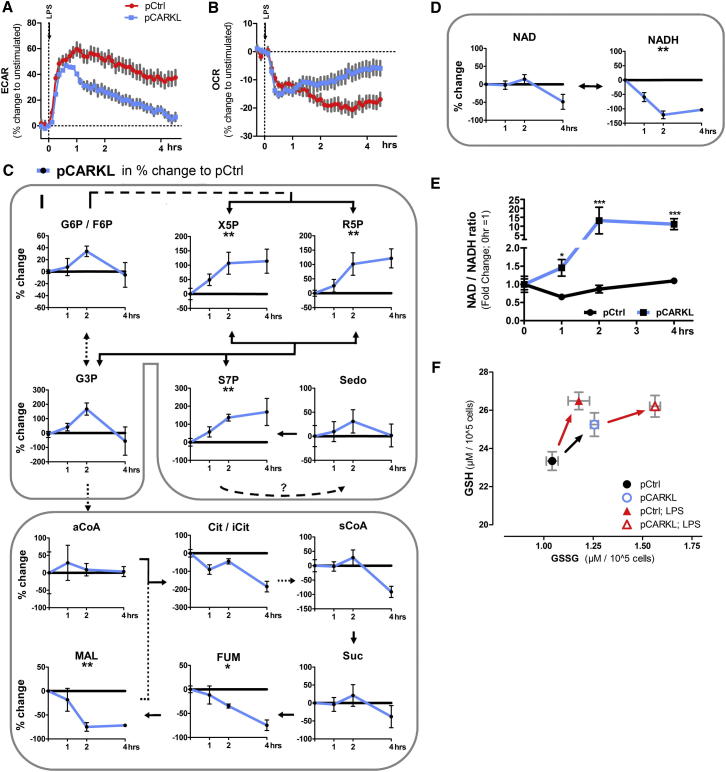
We next compared the ECAR and OCR response of LPS-activated pCARKL and control macrophages. Importantly, in pCARKL cells initial LPS-induced ECAR elevation was unaffected, but extension of this energetic state was blocked (Figure 5A). Again, this finding indicates that a more complex biphasic metabolic reprogramming might be necessary for proper macrophage activation. OCR levels reflected ECAR changes in both control and pCARKL cells (Figure 5B).
Figure 5.
CARKL Integrates Redox State and Glucose Metabolism in Activated Macrophages
(A and B) ECAR and OCR during LPS activation of pCARKL and pCtrl cells.
(C and D) Metabolite profiles of pCtrl and pCARKL cells during initial activation phase (0–4 hrs post 100 ng/ml LPS). Solid lines represent direct metabolite synthesis. Dashed lines indicate multiple enzymatic steps. Data represent delta mean change in % of pCARKL to pCtrl cells (pCtrl = 0%) ± SEM of three independent experiments; to test if a metabolite profile was significantly changed, we used two-way ANOVA as the statistical test (indicated below metabolite name); black line = pCtrl, blue line = pCARKL.
(E) Changes in NAD/NADH peak ratio during activation phase as fold change to unstimulated cells.
(F) GSH and GSSG of resting and LPS-activated (4 hr) pCtrl and pCARKL cells. Data are means ± SEM of three independent experiments; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Metabolomics performed after LPS activation revealed S7P, X-5P, R5P, malate, and fumerate levels linked to CARKL expression (Figures 5C and S5A). An apparent build-up of S7P, X5P, R5P, G3P, and G6P/F6P was observed, concomitant with marked reduction in some TCA cycle intermediates and, importantly, NADH but not NAD+ levels (Figure 5D). In metabolic terms, this equates to a lack of reducing equivalents to drive cellular processes, a prominent increase in NAD/NADH ratio (Figure 5E), and, thus, a marked imbalance in cellular redox state. Additionally, in control cells, LPS activation increased GSH substantially but GSSG only moderately (Figure 5F). IL-4 did not induce such a redox dirft (Figure S5B). In pCARKL cells, LPS induced a more robust increase in GSSG than in GSH, despite a basal increase in the total glutathione pool. These findings support the notion that CARKL acts as a regulator of cellular metabolism and implicate CARKL as a necessary metabolic catalyst for maintaining redox state during activation.
CARKL Directs Macrophage Fate and Is Sufficient for Activation
Given the findings above, we tested CARKL's alleged metabolic function in our macrophage activation model. LPS-induced IL-6 secretion and expression were significantly inhibited in pCARKL cells versus pCtrl control cells (Figures 6A and 4E). Metabolic regulation of IL-6 was also confirmed in primary macrophages by incubation with metformin and rotenone (Figure S6A). Of note, overexpression of transaldolase 1 did not reverse CARKL-mediated cytokine repression, though substitution of galactose as the primary carbon source partly rescued this effect (Figures S6B and S6C). This finding directly implicates metabolic substrate flux as the primary determinant of CARKL's immunomodulatory effects. Most importantly, while overexpression of the catalytically active Q126K mutant recapitulated this marked blunting of IL-6 secretion, the catalytically inactive W125D mutant showed no effect whatsoever (Figure 6A). Thus, the sedoheptulose kinase activity of CARKL is directly related to outcome of macrophage polarization.
Figure 6.
CARKL Regulation Directs the Macrophage Activation Process
(A) Macrophages overexpressing either wild-type or mutant CARKL were stimulated with LPS (100 ng/ml), and subsequent IL-6 secretion was measured in cell-free supernatants 4 hr post activation.
(B) Protein expression levels of miCARKL and control (miCtrl) RAW264.7 cells.
(C) Relative bioavailability of metabolic intermediates in miCARKL and control cells.
(D–F) OCR and ECAR of miCARKL and miCtrl cells and their ECAR (E) and NAD/NADH ratios (F) relative to LPS-activated macrophages (1 hr).
(G–J) TNF-α (G) and IL-6 (H) cytokine secretion before and after LPS activation in miCARKL cells and miCtrl cells. Resting miCARKL and control cells were incubated with Celastrol (1 μg/ml), DHEA (200 μM), oxamate (40 mM), and expression of TNFα (I) and Mrc-1 (J) was measured by RT-PCR.
(K) Mrc-1 expression by IL-4-stimulated miCtrl (black) and miCARKL (white) cells in the absence or presence of buthionine sulphoximine (BSO; 300 μM).
(L and M) TNF-α (L) and Mrc-1 (M) expression of IL-4 (10 ng/ml) activated pCARKL and pCtrl cells in the absence or presence of reduced glutathione ethyl ester (GSH-Et; 5 mM). Incubation time was 4 hrs. Data are mean ± SEM of at least three independent experiments; nd = not detected, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To ultimately test whether CARKL loss was an essential and critical event for macrophage M1 polarization, we generated CARKL knockdown (miCARKL) RAW264.7 cell lines (Figure 6B) and tested for metabolic state and response to LPS (Figures 6C–6H). In agreement with the pCARKL data above, miCARKL cells accumulated intracellular G3P, X5P, and R5P levels (Figures 6C and S4A) and reduced S7P levels. Sedoheptulose remained unaffected. These findings support the idea that CARKL directly controls carbon usage through a simple mass action. CARKL loss promoted rerouting of glucose from aerobic to anaerobic metabolism (Figures 6D and S4B) comparable to LPS induction (Figure 6E). This was accompanied by a similar NAD/NADH ratio (Figure 6F). Notably, LPS stimulation of miCARKL cells caused a marked potentiation of the TNFα, IL-6, and IL-10 response, direct evidence that CARKL downregulation promotes and amplifies M1-like polarization (Figures 6G, 6H, and S6E). Again, this effect was partly rescued by substitution of glucose to galactose as the primary carbon source (Figure S6D). Further, in resting miCARKL cells TNFα (high) and Mrc-1 (low) expression patterns also paralleled LPS activation (Figures 6I, 6J, and S1C), a phenotype resistant to oxamate but sensitive to NFκB and PPP blockade. Together, these data suggested CARKL loss as sufficient to induce a mild M1-like macrophage polarization.
CARKL Sensitizes M2-like Polarization
In contrast to M1-like activation, M2-like activation of macrophages resulted in upregulation of CARKL (Figure 3C and S1C). To investigate possible functional consequences of CARKL level and M2-like polarization, we tested cells with either low or high CARKL levels for basal and IL-4-induced Mrc-1 expression. miCARKL cells exhibited reduced Mrc-1 expression (Figures 6J and 6K). Further, whereas IL-4 increased Mrc-1 expression around 40-fold in control cells, stimulation of miCARKL cells increased Mrc-1 only to untreated control levels (Figure 6K). Depleting GSH pools had no effect on IL-4-induced Mrc-1 in control cells but significantly increased Mrc-1 expression in IL-4 treated miCARKL cells. This suggested altered redox regulation by CARKL loss as one important factor involved in Mrc-1 repression. In BMDM, pharmacologic activation of glucose metabolism by metformin or rotenone totally blunted IL-4 induced Mrc-1 expression (Figure S6G). In both primary and RAW264.7 macrophages, alternative activation resulted in downregulation of TNFα and upregulation of Mrc-1, a phenotype exhibited by pCARKL cells under basal conditions and exaggerated upon IL-4 stimulation (Figures 6L, 6M, and S1C). Importantly, this pCARKL phenotype (TNFαlow/Mrc-1high) was partly sensitive to addition of exogenous GSH (GSH-Et), once again implicating carbon flux and redox control in the functional endpoint. Thus, CARKL sensitizes macrophages to M2 polarization.
Discussion
Macrophage activation screening of ∼200 known and predicted kinases revealed the surprising finding that some of the strongest gene ontology enrichment was observed for pathways regulating carbohydrate metabolism. Indeed, AKT1 and STK11, both identified in our kinase screen, while classically thought of as nutrient sensors, also control a fate switch, from cytotoxic effector to memory CD8+ T cells (Finlay and Cantrell, 2011). More surprising was the robust enrichment of nonprotein kinases potentially capable of contributing directly to metabolic flux through defined pathways. This finding, coupled with our more detailed characterization of CARKL, supports a robust interdependency of metabolism and immune cell function while at the same time highlighting our overall lack of detailed knowledge on the subject.
Indeed, cellular immune activation is a metabolically costly endeavor and cannot operate effectively under energy deficit (Demas et al., 1997). Among the nonprotein kinase candidates observed to modulate TNFα secretion in our screen, HK, the PFK family, and CARKL are strategically positioned to control glucose usage according to cellular requirements. HK regulates intracellular glucose availability. Fructose-1-6-bisphosphate (F-1-6bp) formation by PFK is an irreversible step in glycolysis that determines glycolytic flux and, not surprisingly, is the target of potent antagonistic regulation by key metabolic hormones, such as glucagon and insulin. Our work adds S7P formation by CARKL as a novel rate-limiting lever for the system, balancing the G3P and S7P intermediates of nonoxidative PPP and glycolysis (Figure S7). Interestingly, a novel sedhoheptulose-1-7-bisphosphatase, which forms S7P from S1-7bp, was very recently reported as a crucial enzyme in yeast ribosome biogenesis with obvious consequences (Clasquin et al., 2011).
One particularly interesting observation was the highly significant drop of NADH levels in pCARKL cells during macrophage activation, which resulted in substantial redox shift (Figure 5D). PPP activity contributes to reduction of redox couples via NADPH. We therefore expected and observed increased GSH and NADH generation during M1 activation, a phenotype that could be reversed either by administration of DHEA to block the PPP directly or by counterbalancing CARKL loss. M2-like activation, on the other hand, resulted in an upregulation of CARKL which was not followed by increased GSH or NADH formation. These data represent a functional distinction between the two polarization states that is, importantly, CARKL dependent. Further, the pattern of baseline changes observed in the pyridine pool in miCARKL and pCARKL cells (Figure S4A) provides an interesting potential link to poly ADP-ribose polymerase (PARP) protein family involvement. PARPs are responsible for NAD degradation and important for immune cell function, e.g., activation of NFκB (Oliver et al., 1999; Pollak et al., 2007). In keeping with this idea, NADP pools were much less affected than NAD in our system, a common feature of PARP-mediated pyridine regulation (Pellny et al., 2009).
It has been reported that several master regulators of inflammation are sensitive to ROS and that their function is dependent on cellular redox states (Kazama et al., 2008; Kumar et al., 1992; Toledano and Leonard, 1991; Wang et al., 1999). For instance, NFκB is sensitive to cellular redox equilibrium (Perl et al., 2002). This confirms the recent findings of Kawauchi and colleagues reporting the bidirectional influence of NFκB and glucose metabolism in cancer cell physiology (Kawauchi et al., 2008). Interestingly, although CARKL reduced total active NFκB, relative induction by LPS was only slightly affected. It remains to be established how exactly CARKL and the NFκB system rely on each other. We further report CARKL as negative regulator of LPS-induced SOCS3 expression, without influencing SOCS1 levels, but resulting in enhanced STAT3 phosphorylation. SOCS3 is commonly known as repressor of STAT3. It will be interesting to see whether CARKL directly impacts the SOCS/STAT system (Figure 7). Specific SOCS isoforms appear critical to eliciting pro- versus anti-inflammatory macrophage states (O'Shea and Murray, 2008; Whyte et al., 2011; Yoshimura et al., 2007).
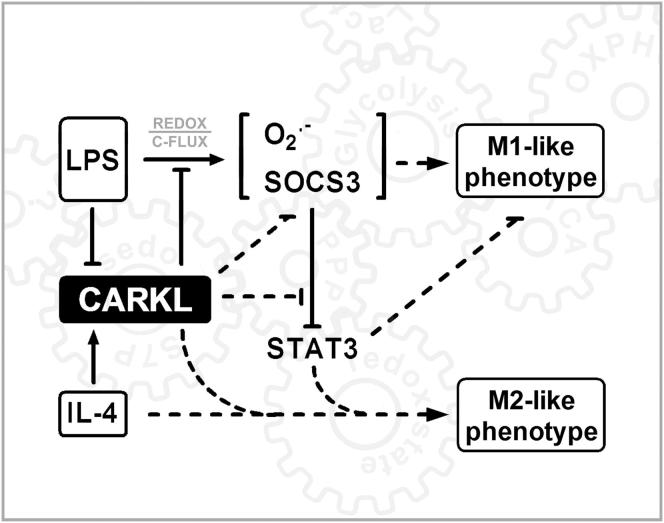
Figure 7.
CARKL Regulation of M1/M2 Activation
Schematic shows how CARKL may direct activation phases/fates of macrophages. CARKL directs carbon reshuffling (C-Flux) between glycolysis and PPP (Figure S7). The resulting alteration in bioenergetics likely functions as a regulatory system analogous to classical protein-based activation schemes (as, for example, for inositol-phosphates). Dashed lines represent effects for which direct evidence exists but where exact molecular mechanisms remain to be defined.
And what of clinical relevance? Metabolic reconfiguration and mild M1-like polarization are triggered by CARKL loss even in the absence of additional stimulus. While the phenomenon is not fully understood, cystinosis patients, who harbor a 57 kb deletion affecting CARKL and CTNS, despite showing no strong signs of inflammation, respond positively to continuous anti-inflammatory treatment (Haycock et al., 1982). Further, our data may provide insight into potential new modes of action for drugs which clearly bridge both immune and metabolic signaling, such as glucocorticoids and rapamycin.
In summary, this study identifies CARKL as a sedoheptulose kinase orchestrating pro- and anti-inflammatory immune responses through metabolic control. Our results highlight the fundamental nature of metabolic reconfiguration in macrophage activation and define unique metabolic demands of distinct activation states.
Experimental Procedures
Reagents
Unless otherwise stated all reagents were purchased from Sigma, all cytokines from R&D Systems, and Celastrol from InvivoGen. [32P]γ-ATP was a generous gift of O. Hantschel (CeMM, Austria).
Cell Culture and Animals
RAW264.7 (ATCC) and all other cells were cultured in DMEM high glucose supplemented with 10% heat-inactivated FBS and 50 μg/ml gentamycin. Thioglycollate-elicited mouse peritoneal macrophages were isolated as described by Wassenberg et al. (1999). Bone marrow-derived macrophages were differentiated by 10% L929 conditioned media for 5 days or by recombinant murine M-CSF (2ng/ml) (Figure S3A, lower left graph) as previously described (Haschemi et al., 2007). Male C57BL/6J mice were purchased from JAX.
Kinase Screen
RAW264.7 macrophages were transfected using Lipofectamine 2000 (Invitrogen). Each individual human kinase ORF (Park et al., 2005) was transfected to three wells. Twenty-four hours after transfection, cells were incubated with 100 ng/ml LPS for 1 hr. Cell-free supernatant was assayed for TNFα production. For further information please refer to Supplemental Experimental Procedures.
Q-PCR
For Q-RT-PCR analysis, total RNA was extracted and reverse-transcribed using commercial kits (QIAGEN, Applied Biosystems). All subsequent Q-RT-PCRs were performed on an AbiPRISM 7900HT real-time cycler using iTaq SYBR Green Supermix with ROX (BioRad). For further information and primers please refer to Supplemental Experimental Procedures and Table S2.
Cloning and Generation of Stable Cell Lines
A sequence-verified murine CARKL cDNA (clone MMM1013 OpenBiosystems) lacking 5′ and 3′ UTRs was cloned into pcDNA6 (Invitrogen). miRNA-adapted short hairpin RNA targeting mouse CARKL generated in pSM2 vector (Open Biosystems, clone ID: V2MM_41673 F4) was subcloned into the LMP vector. The pFUSE_eGFP plasmid was used to generate pCARKL_eGFP fusion. All stable cell lines were derived from the RAW264.7 cell line. Refer to Supplemental Experimental Procedures for further information.
Generation of Recombinant Mutant and Wild-Type CARKL
To generate recombinant wild-type or mutant CARKL, we employed in vivo site-directed biotinylation as a purification strategy. The full-length open-reading-frame of mouse CARKL or the mutant (W125D, Q126K, and T14M/T15K; see below) was cloned into PinPoint Xa-1 vector (Promega) and purification from E.coli was performed according to the PinPoint manual (Promega) with final removal of the purification tag. For detailed information see Supplemental Experimental Procedures.
Sedoheptulose Isolation from Sedum spectabile
Sedoheptulose was isolated according to a modified protocol of Schmidt et al. (1998). The final sedoheptulose preparation was ∼90% pure as measured by NMR analysis (data not shown). For further information see Supplemental Experimental Procedures.
Biochemical Characterization of CARKL Activity
We used the ADP Quest Assay (DiscoveRx) to indirectly measure S7P formation by ADP accumulation over time. Radioactive kinase assays were performed by incubating recombinant CARKL with [32P]γ-ATP in kinase reaction buffer (pH 7.6) containing 25 mM HEPES, 20 mM KCl, 10 mM MgCl2, 10 mM ATP, and 10 mM sedoheptulose for 15 min at 30°C. The reaction mixture was resolved on thin layer chromatography plates (Merck), developed with MeOH:Chloroform:H20 (5:5:1) and subsequently exposed to film. For purification of S7P please refer to Supplemental Experimental Procedures.
Nuclear Magnetic Resonance
Nuclear magnetic resonance (NMR) spectra were recorded at 297 K on a Bruker DPX 400 instrument at 400.13 MHz for 1H, 100.62 MHz for 13C, and 161.97 MHz for 31P using a 5 mm broad band probe with Z gradients. For detailed information see Supplemental Experimental Procedures.
3D Modeling and Docking Experiments of CARKL
The model of CARKL was built based on the 3D structure of xylulose kinase (PDB code: 2ITM) by using MODWEB for homology and comparative modeling of protein 3D structures (Eswar et al., 2003; Pieper et al., 2009) with subsequent optimization. A detailed workflow for modeling and sedoheptulose docking is accessible in Supplemental Experimental Procedures.
Western Blot
Total-cell lysates were prepared by harvesting cells in cell lysis buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). Proteins were resolved on 4%–12% polyacrylamide gels (Invitrogen) and transferred to PVDF membranes (Bio-Rad). Antibodies: anti-CARKL (ab69920, Abcam Inc.), anti-Stat-P (Tyr705, clone D3A7), and anti-histone H3 (clone D1H2) (Cell Signaling Technology), and anti-β-Actin (A5316, Sigma). Nuclear fractions were prepared using Nuclear Extraction Kit (Panomics).
Site-Directed Mutagenesis
Expression plasmids PinPoint Xa_CARKL (bacterial expression) and pCARKL (eukaryotic expression), containing the mouse CARKL cDNA were mutated using the QuickChange site-directed mutagenesis kit II (Stratagene).
FACS
Cells were incubated with FITC-labeled mAb MHC class II (Invitrogen). More than 0.5 × 105 cells were analyzed by flow cytometry (FACSCalibur; BD). Surface marker analysis was performed using FlowJo software (Tree Star, Inc.).
Confocal Laser Scanning Microscopy
Stable CARKL_eGFP-expressing macrophages were cultured in tissue culture-treated glass slides (BD Falcon), and specimens were then examined after staining using a Zeiss LSM5 laser scanning Microscope (Carl Zeiss Optics). A detailed protocol is accessible in Supplemental Experimental Procedures.
Luciferase Reporter Assay
PathDetect NFκB cis-reporting luciferase plasmid for measuring transcription factor activation contained a basic TATA box in addition to five repetitive NFκB enhancer element binding sites (Stratagene). RAW264.7 cells were seeded into 48-well plates and transfected 1 day thereafter at 1.0 μg/well of reporter construct using Superfect reagent (QIAGEN) according to the manufacturer's instructions. Twenty-four hours later, cells were treated with or without LPS (100 ng/ml). After 8 hr, protein extracts were quantified for normalization and assessed for luciferase activity (Promega) according to the manufacturer's instructions.
GSH and GSSG Measurements
Oxidized and reduced glutathione were measured using GSH/GSSG-Glo assay provided by Promega according to provided protocol.
Cytokine Measurements
TNF-α and IL-6 levels in cell culture supernatants were measured using Mouse TNF-α/Tnfsf1a and IL-6 Quantikine ELISA Kits purchased from R&D Biosystems, according to the protocol provided by the manufacturer.
Electron Spin Resonance
We employed CMH (1-Hydroxy-2,2,5,5-tetramethyl-pyrrolidine-3-carboxylic acid methyl ester) spin probe (Noxygen) to measure absolute superoxide generation rates in real time (Figure S4F) using a Bruker EMX spectrometer. For detailed protocol see Supplemental Experimental Procedures.
Cellular Respiration and Extracellular Acidification
XF24 Extracellular Flux Analyzer (Seahorse Biosciences) was used to determine the bioenergetic profile of intact cells (Spolarics and Wu, 1997). Briefly, cells were seeded (70,000 cells/well) in XF24 plates and allowed to recover for 24 hr before the OCR and ECAR were assessed in glucose-containing media (Seahorse Biosciences). Results were normalized to the actual cell count immediately after OCR and ECAR recordings.
Metabolomic Analysis
Metabolites were extracted from cells by 80/20 methanol/water solvent, separated via hydrophilic interaction liquid chromatography (HILIC), and detected online using an Agilent 6410 Triple Quadrupole mass spectrometer (MS/MS) with an electrospray ionization (ESI) source, as detailed in Supplemental Experimental Procedures.
Flux Analysis
Control, LPS (100 ng/ml, 1 hr), or IL-4 (10 μg/ml, 2 hr) stimulated primary macrophages (BMDM) were incubated with 100% 13C-1-2-glucose-containing media for exactly 10 min and subsequently quenched with liquid nitrogen. Carbon-13 mass isotopomer analysis of metabolites was carried out using liquid chromatography-mass spectrometry (LC-MS), according to the protocol of Lorenz et al. (2011), as detailed in Supplemental Experimental Procedures.
Human and Murine In Vivo Models of Endotoxemia
We employed banked human PBMC mRNA samples (n = 6) from a previously published study where participants received an intravenous bolus containing 2 ng/kg LPS (Marsik et al., 2006). For sublethal murine in vivo endotoxemia, we injected 50 μg/kg LPS (Sigma) into the tail vein of male C57BL/6J mice and isolated PBMCs from mice before and after LPS injection (n = 3–4). All animal experiments were carried out according to an ethical animal license protocol and contract approved by the Medical University Vienna (BMWF-66.009/0140-II/10b/2010).
Statistical Analyses
All data unless otherwise indicated are shown as mean ± SEM and were tested using two-tailed Student's t test or ANOVA (p < 0.05). All figures and statistical analyses were generated using GraphPad Prism 4.
Acknowledgments
The authors are indebted to D.W. Pettigrew, K. Hagenbichler, M. Ozsvar, A. Jais, D. Baumgartner, M. Mayerhofer, M. Kubicek, M. Raith, B. Jilma, O. Hantschel, A. Goncalves, and C.J. Binder for their support. Further, the authors would like to thank A. Hofinger for NMR spectra and the Cell Sorting Core Unit of MUW. This work was supported in part by the CCHD Ph.D. Program funded by the Austrian Science Fund (FWF) (D.K. W1205); by R01 (GM088666 to L.E.O.); FWF (P22258-B12 to B.K.); by DoD Center for Integration of Medicine and Innovative Technology; and by NIH grants 1K25DK092558-01 (C.R.E.) and the Molecular Phenotyping Core of the Michigan Nutrition Obesity Research Center (DK089503). We also thank the Julie Henry Fund of the Transplant Institute at the Beth Israel Deaconess Medical Center, Boston, MA.
Published online: June 5, 2012
Footnotes
Supplemental Information includes seven figures, five tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at doi:10.1016/j.cmet.2012.04.023.
Contributor Information
Arvand Haschemi, Email: arvand.haschemi@meduniwien.ac.at.
J. Andrew Pospisilik, Email: pospisilik@immunbio.mpg.de.
Supplemental Information
References
- Beurel E., Yeh W.I., Michalek S.M., Harrington L.E., Jope R.S. Glycogen synthase kinase-3 is an early determinant in the differentiation of pathogenic Th17 cells. J. Immunol. 2011;186:1391–1398. doi: 10.4049/jimmunol.1003511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.J., Hsu K.W., Tsai J.N., Hung C.H., Kuo T.C., Chen Y.L. Involvement of protein kinase C in the inhibition of lipopolysaccharide-induced nitric oxide production by thapsigargin in RAW 264.7 macrophages. Int. J. Biochem. Cell Biol. 2005;37:2574–2585. doi: 10.1016/j.biocel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Clasquin M.F., Melamud E., Singer A., Gooding J.R., Xu X., Dong A., Cui H., Campagna S.R., Savchenko A., Yakunin A.F. Riboneogenesis in yeast. Cell. 2011;145:969–980. doi: 10.1016/j.cell.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demas G.E., Chefer V., Talan M.I., Nelson R.J. Metabolic costs of mounting an antigen-stimulated immune response in adult and aged C57BL/6J mice. Am. J. Physiol. 1997;273:R1631–R1637. doi: 10.1152/ajpregu.1997.273.5.R1631. [DOI] [PubMed] [Google Scholar]
- Drapier J.C., Hibbs J.B., Jr. Differentiation of murine macrophages to express nonspecific cytotoxicity for tumor cells results in L-arginine-dependent inhibition of mitochondrial iron-sulfur enzymes in the macrophage effector cells. J. Immunol. 1988;140:2829–2838. [PubMed] [Google Scholar]
- Eswar N., John B., Mirkovic N., Fiser A., Ilyin V.A., Pieper U., Stuart A.C., Marti-Renom M.A., Madhusudhan M.S., Yerkovich B., Sali A. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003;31:3375–3380. doi: 10.1093/nar/gkg543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay D., Cantrell D.A. Metabolism, migration and memory in cytotoxic T cells. Nat. Rev. Immunol. 2011;11:109–117. doi: 10.1038/nri2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C.J., Hammerman P.S., Thompson C.B. Fuel feeds function: energy metabolism and the T-cell response. Nat. Rev. Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- Gordon S., Martinez F.O. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Haschemi A., Wagner O., Marculescu R., Wegiel B., Robson S.C., Gagliani N., Gallo D., Chen J.F., Bach F.H., Otterbein L.E. Cross-regulation of carbon monoxide and the adenosine A2a receptor in macrophages. J. Immunol. 2007;178:5921–5929. doi: 10.4049/jimmunol.178.9.5921. [DOI] [PubMed] [Google Scholar]
- Haycock G.B., Al-Dahhan J., Mak R.H., Chantler C. Effect of indomethacin on clinical progress and renal function in cystinosis. Arch. Dis. Child. 1982;57:934–939. doi: 10.1136/adc.57.12.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway C.A., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Kardon T., Stroobant V., Veiga-da-Cunha M., Schaftingen E.V. Characterization of mammalian sedoheptulokinase and mechanism of formation of erythritol in sedoheptulokinase deficiency. FEBS Lett. 2008;582:3330–3334. doi: 10.1016/j.febslet.2008.08.024. [DOI] [PubMed] [Google Scholar]
- Kawauchi K., Araki K., Tobiume K., Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- Kazama H., Ricci J.E., Herndon J.M., Hoppe G., Green D.R., Ferguson T.A. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity. 2008;29:21–32. doi: 10.1016/j.immuni.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.C., Jeon W.K., Hong H.Y., Jeon K.B., Hahn J.H., Kim Y.M., Numazawa S., Yosida T., Park E.H., Lim C.J. The anti-inflammatory activity of Phellinus linteus (Berk. & M.A. Curt.) is mediated through the PKCdelta/Nrf2/ARE signaling to up-regulation of heme oxygenase-1. J. Ethnopharmacol. 2007;113:240–247. doi: 10.1016/j.jep.2007.05.032. [DOI] [PubMed] [Google Scholar]
- Kim C., Sano Y., Todorova K., Carlson B.A., Arpa L., Celada A., Lawrence T., Otsu K., Brissette J.L., Arthur J.S., Park J.M. The kinase p38 alpha serves cell type-specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat. Immunol. 2008;9:1019–1027. doi: 10.1038/ni.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk C.M., Holowka T., Sun J., Blagih J., Amiel E., DeBerardinis R.J., Cross J.R., Jung E., Thompson C.B., Jones R.G., Pearce E.J. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115:4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Rabson A.B., Gélinas C. The RxxRxRxxC motif conserved in all Rel/kappa B proteins is essential for the DNA-binding activity and redox regulation of the v-Rel oncoprotein. Mol. Cell. Biol. 1992;12:3094–3106. doi: 10.1128/mcb.12.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttner R.E., Schumer W., Apantaku F.O. Effect of endotoxin and glucocorticoid pretreatment on hexose monophosphate shunt activity in rat liver. Circ. Shock. 1982;9:37–45. [PubMed] [Google Scholar]
- Lorenz M.A., Burant C.F., Kennedy R.T. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal. Chem. 2011;83:3406–3414. doi: 10.1021/ac103313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsik C., Sunder-Plassmann R., Jilma B., Kovar F.M., Mannhalter C., Wagner O., Rumpold H., Endler G. The C-reactive protein (+)1444C/T alteration modulates the inflammation and coagulation response in human endotoxemia. Clin. Chem. 2006;52:1952–1957. doi: 10.1373/clinchem.2006.069823. [DOI] [PubMed] [Google Scholar]
- Martinon F., Mayor A., Tschopp J. The inflammasomes: guardians of the body. Annu. Rev. Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30:766–775. doi: 10.1016/j.immuni.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140:771–776. doi: 10.1016/j.cell.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Murray P.J. The primary mechanism of the IL-10-regulated antiinflammatory response is to selectively inhibit transcription. Proc. Natl. Acad. Sci. USA. 2005;102:8686–8691. doi: 10.1073/pnas.0500419102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C., Ding A. Nonresolving inflammation. Cell. 2010;140:871–882. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- O'Shea J.J., Murray P.J. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaniemi M., Glumoff V., Harju K., Liljeroos M., Vuori K., Hallman M. Phosphatidylinositol 3-kinase is involved in Toll-like receptor 4-mediated cytokine expression in mouse macrophages. Eur. J. Immunol. 2003;33:597–605. doi: 10.1002/eji.200323376. [DOI] [PubMed] [Google Scholar]
- Oliver F.J., Ménissier-de Murcia J., Nacci C., Decker P., Andriantsitohaina R., Muller S., de la Rubia G., Stoclet J.C., de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Hu Y., Murthy T.V., Vannberg F., Shen B., Rolfs A., Hutti J.E., Cantley L.C., Labaer J., Harlow E., Brizuela L. Building a human kinase gene repository: bioinformatics, molecular cloning, and functional validation. Proc. Natl. Acad. Sci. USA. 2005;102:8114–8119. doi: 10.1073/pnas.0503141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellny T.K., Locato V., Vivancos P.D., Markovic J., De Gara L., Pallardó F.V., Foyer C.H. Pyridine nucleotide cycling and control of intracellular redox state in relation to poly (ADP-ribose) polymerase activity and nuclear localization of glutathione during exponential growth of Arabidopsis cells in culture. Mol Plant. 2009;2:442–456. doi: 10.1093/mp/ssp008. [DOI] [PubMed] [Google Scholar]
- Perl A., Gergely P., Jr., Puskas F., Banki K. Metabolic switches of T-cell activation and apoptosis. Antioxid. Redox Signal. 2002;4:427–443. doi: 10.1089/15230860260196227. [DOI] [PubMed] [Google Scholar]
- Pieper U., Eswar N., Webb B.M., Eramian D., Kelly L., Barkan D.T., Carter H., Mankoo P., Karchin R., Marti-Renom M.A. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2009;37(Database issue):D347–D354. doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak N., Dölle C., Ziegler M. The power to reduce: pyridine nucleotides—small molecules with a multitude of functions. Biochem. J. 2007;402:205–218. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock K.L., Kono H. The inflammatory response to cell death. Annu. Rev. Pathol. 2008;3:99–126. doi: 10.1146/annurev.pathmechdis.3.121806.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A., Paimela T., Suuronen T., Kaarniranta K. Innate immunity meets with cellular stress at the IKK complex: regulation of the IKK complex by HSP70 and HSP90. Immunol. Lett. 2008;117:9–15. doi: 10.1016/j.imlet.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Schieven G.L. The p38alpha kinase plays a central role in inflammation. Curr. Top. Med. Chem. 2009;9:1038–1048. doi: 10.2174/156802609789630974. [DOI] [PubMed] [Google Scholar]
- Schmidt U., Stiller R., Brade H., Thiem J. Unusual Phosphorylation of Sedoheptulose by Means of Hexokinase. Synlett. 1998;1998:125–126. [Google Scholar]
- Schnyder J., Baggiolini M. Role of phagocytosis in the activation of macrophages. J. Exp. Med. 1978;148:1449–1457. doi: 10.1084/jem.148.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.Z., Wang R., Huang G., Vogel P., Neale G., Green D.R., Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolarics Z., Wu J.X. Role of glutathione and catalase in H2O2 detoxification in LPS-activated hepatic endothelial and Kupffer cells. Am. J. Physiol. 1997;273:G1304–G1311. doi: 10.1152/ajpgi.1997.273.6.G1304. [DOI] [PubMed] [Google Scholar]
- Toledano M.B., Leonard W.J. Modulation of transcription factor NF-kappa B binding activity by oxidation-reduction in vitro. Proc. Natl. Acad. Sci. USA. 1991;88:4328–4332. doi: 10.1073/pnas.88.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamelink M.M., Struys E.A., Jansen E.E., Levtchenko E.N., Zijlstra F.S., Engelke U., Blom H.J., Jakobs C., Wevers R.A. Sedoheptulokinase deficiency due to a 57-kb deletion in cystinosis patients causes urinary accumulation of sedoheptulose: elucidation of the CARKL gene. Hum. Mutat. 2008;29:532–536. doi: 10.1002/humu.20685. [DOI] [PubMed] [Google Scholar]
- Wang F., Wang L.Y., Wright D., Parmely M.J. Redox imbalance differentially inhibits lipopolysaccharide-induced macrophage activation in the mouse liver. Infect. Immun. 1999;67:5409–5416. doi: 10.1128/iai.67.10.5409-5416.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Warburg O., Posener K., Negelein E. Ueber den Stoffwechsel der Tumoren. Biochem. Z. 1924;152:319–344. [Google Scholar]
- Wassenberg J.J., Dezfulian C., Nicchitta C.V. Receptor mediated and fluid phase pathways for internalization of the ER Hsp90 chaperone GRP94 in murine macrophages. J. Cell Sci. 1999;112:2167–2175. doi: 10.1242/jcs.112.13.2167. [DOI] [PubMed] [Google Scholar]
- Whyte C.S., Bishop E.T., Rückerl D., Gaspar-Pereira S., Barker R.N., Allen J.E., Rees A.J., Wilson H.M. Suppressor of cytokine signaling (SOCS)1 is a key determinant of differential macrophage activation and function. J. Leukoc. Biol. 2011;90:845–854. doi: 10.1189/jlb.1110644. [DOI] [PubMed] [Google Scholar]
- Yoshimura A., Naka T., Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.