Abstract
Cis-acting regulatory elements of the wheat acetyl-CoA carboxylase (ACC) gene family were identified by comparing the promoter activity of 5′ end gene fragments fused to a reporter gene in two transient expression systems: wheat protoplasts and epidermal cells of mature embryos. Expression of the plastid and the cytosolic ACC genes is each driven by two nested promoters responsible for the synthesis of two transcript types. The internal promoter is located in an intron removed from transcripts originating at the first promoter. These complex promoters, which are different for the cytosolic and plastid ACC genes, control tissue-specific expression of the enzymatic activity supplying cytosolic, plastid, and mitochondrial pools of malonyl-CoA. The activity of one such complex promoter, driving expression of one of the cytosolic ACC genes, was studied throughout development of transgenic wheat plants carrying a full-length promoter–reporter gene fusion. High activity of the promoter was detected in the coleoptile, in the upper sheath section of the leaf, on the top surface of the ovary, in some sections of the main veins in the lemma and glume, and in abaxial epidermis hair cells of the lemma, glume, and rachis. The findings are consistent with the developmental and environmental requirements for very-long-chain fatty acids and flavonoids, whose synthesis begins with the ACC reaction in the cytosol of these specific cell types.
Modern bread wheat has a large hexaploid genome, making molecular genetic studies difficult. We have been studying the structure, evolution, and function of the family of genes encoding the enzyme acetyl-CoA carboxylase (ACC) during wheat development (1–7).
In plants, separate ACC isozymes supply the malonyl-CoA pools used for de novo fatty acid (FA) biosynthesis in plastids and mitochondria, and for FA elongation and flavonoid (FL) and stilbene biosynthesis in the cytosol (8, 9). FAs are essential in membrane biogenesis, lipoic acid synthesis, production of oil as storage material, cuticular wax (CW) synthesis, signaling, and pollen–stigma interaction. FLs play multiple roles in plants as pigments and UV protectants, defense compounds, and as signals (10). Malonyl-CoA is also used for protein and small-molecule malonylation.
Our previous study (5) showed that significant regulation of cytosolic and plastid ACC genes in young wheat plants is accomplished at the transcript level. The number and identity of transcribed genes were established by cDNA and genomic DNA sequence comparisons. Transcription start sites and splicing patterns of leader introns were identified for multiple genes, including homoeologs and paralogs. We found that in wheat seedlings, the plastid ACC mRNA level is high in the middle part of the plant and low in roots and leaf blades. The three plastid ACC-homoeologous genes are equivalent in their level of expression. Cytosolic ACC mRNA accumulates to a high level in the lower sheath section of the plant. For the cytosolic ACC genes, we found that transcripts of the three homoeologs had similar tissue distribution and abundance, whereas a fourth gene, a paralog, was hardly transcribed. The differences between expression of plastid and cytosolic ACC genes were suggested to reflect, respectively, the demand for long-chain FAs needed for membrane lipids in dividing meristematic cells and during chloroplast biogenesis, and for CW [derived from very-long-chain FAs (VLCFAs)] and FL biosynthesis to provide protection against biotic and abiotic stresses in newly emerging leaves.
To more effectively address the questions of structure and function for multiple paralogs and homoeologs, we describe the complex nature of the promoters and putative regulatory elements of the hexaploid wheat (Triticum aestivum) ACC genes. We examined promoter-reporter fusions in three systems: (i) transient expression in protoplasts prepared from cells grown in culture; (ii) transient expression in bombarded epidermal cells of mature wheat embryos; and (iii) organ-specific expression during a complete cycle of development in transgenic wheat plants.
Materials and Methods
A series of fusions of the Acc promoters (GenBank accession nos. AF029897, AF305204, AF305205, and AF305207) and the β-glucuronidase (GUS) reporter gene (Figs. 1 and 2) was prepared by using native and PCR-engineered restriction sites. Transient expression of the GUS reporter gene in wheat protoplasts and in wheat embryos was measured by delivering plasmid DNA by electroporation (11) and particle bombardment (12), respectively. Transgenic wheat lines were generated by the particle-bombardment method (12). GUS activity was determined fluorometrically (transient expression; ref. 13) or by histochemical staining (transgenic plants; ref. 14). RT-PCR experiments were performed as described (5). Additional information about materials and methods used in this study is provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Fig. 1.
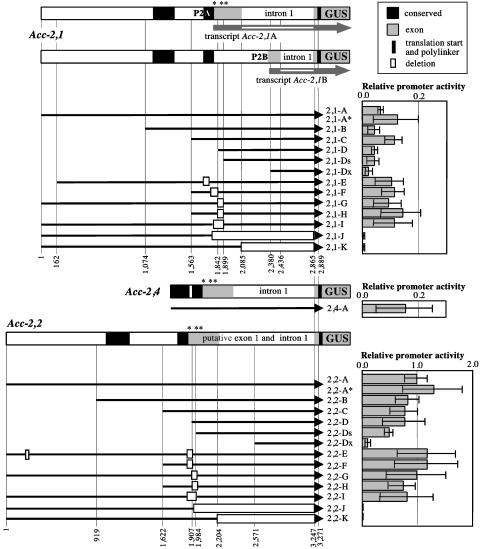
Structure of the Acc-2,1, Acc-2,2, and Acc-2,4 promoter–GUS fusions and relative transient expression of GUS in wheat protoplasts. Numbers identifying key features of the constructs correspond to nucleotide positions in cloned gene fragments (Supporting Materials and Methods). The approximate position of leader AUG codons is indicated by asterisks. These triplets were changed to non-AUG codons in constructs 2,1-A* and 2,2-A*. P2A and P2B, alternative promoters. 5′ terminal deletions are indicated by shortened black arrows, and internal deletions are marked with rectangles. The structure of the two transcript types (Acc-2A and Acc-2B) is illustrated for gene Acc-2,1. Both transcript types were identified for gene Acc-2,4 as well. Only the most upstream 5′ splice site found in transcript Acc-2,1B is shown. Other sites used for splicing of the intron in Acc-2 genes are located 23 and 30 nucleotides further downstream. Activity of construct 2,2-A was taken as 1.
Fig. 2.
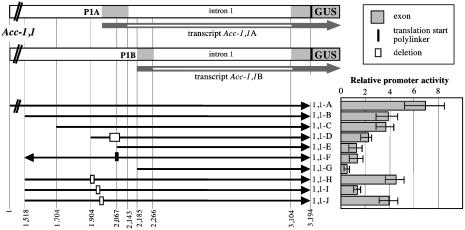
Structure of the Acc-1,1 promoter–GUS fusions and relative transient expression of GUS in wheat protoplasts. Numbers identifying key features of the constructs correspond to nucleotide positions in cloned gene fragments (Supporting Materials and Methods). P1A and P1B, alternative promoters. 5′ terminal deletions are indicated by shortened black arrows, and internal deletions are marked with rectangles. Fragment inversion in construct 1,1-F is marked with an arrow facing left. The structure of the two transcript types (Acc-1,1A and Acc-1,1B) was determined (5). Activity of the construct 2,2-A construct (Fig. 1) was taken as 1.
Results
The three homoeologous chromosomes of hexaploid wheat diverged only recently (4) and we assumed that a detailed promoter analysis of one of the three homoeologs should be sufficient to draw conclusions about all of them. Our previous study showed that for each gene, Acc-2 and Acc-1, all three homoeologs are active, have the same mRNA expression pattern during seedling development, and accumulate mRNAs to a very similar level. Acc-2,2 arose by gene duplication earlier in wheat evolution and may have already acquired new functions. Therefore, we analyzed the promoter of only one of the Acc-1 homoeologs (Acc-1,1) and promoters of two Acc-2 paralogs: Acc-2,1 and Acc-2,2. We also measured activity of the promoter of gene Acc-2,4, an Acc-2,1 homoeolog.
The promoter analysis focused on the sequences upstream of the ACC translation start and extended ≈2 kb upstream of the transcription start sites. The constructs consisting of a series of deletions and mutations are diagrammed in Figs. 1 and 2. Promoter activity was assessed by monitoring GUS. We began the analysis by testing the entire set of constructs in a transient expression system using protoplasts derived from undifferentiated cells grown in suspension culture.
Cytosolic ACC Genes Are Transcribed from Two Promoters. The activity of promoters of two homoeologs, Acc-2,1 and Acc-2,4, was low but was easily detectable above a no-promoter control, whereas the activity of the promoter of gene Acc-2,2, their paralog, was significantly higher (Fig. 1). The promoter activity of construct 2,2-A confirms that the Acc-2,2 gene can be transcriptionally active, at least in some cell types (represented here by protoplasts), although it was not detected in wheat leaves by using RT-PCR (5). Deletion of large 5′ end fragments (constructs 2,1-B, 2,1-C, 2,1-D, and 2,1-Ds, and 2,2-B, 2,2-C, 2,2-D, and 2,2-Ds) had little or no effect on the activity of the Acc-2 promoters in protoplasts (Fig. 1), even though constructs 2,1-Ds and 2,2-Ds had deletions extending into the leader of the previously identified transcripts. Mutagenesis of all three AUG codons of the leader of transcript Acc-2A (constructs 2,1-A* and 2,2-A*) or deletion of conserved fragments encoding this part of the leader (constructs 2,1-E to 2,1-I and 2,2-E to 2,2-I) had no significant effect on GUS expression (Fig. 1). These results show that neither the upstream AUG codons nor the conserved sequence elements around them are involved in the regulation of gene expression in protoplasts.
We postulated that two promoters are used to transcribe Acc-2 genes to produce transcript types Acc-2A and Acc-2B (5). We have now determined the structure and approximate start site of the second transcript type (Fig. 1) by using the primer-walking RT-PCR approach. The promoter used for this transcript (P2B) must be located in the 5′ half of the leader intron of transcript Acc-2A transcribed from promoter P2A. Comparisons of several Acc-2 genes from different wheats revealed sequence conservation of the region containing the P2B promoter and the 5′ splice sites of the first intron of transcript Acc-2B (data not shown). The location of the P2B promoter is supported by the results of the deletion analysis: constructs 2,1-Dx and 2,2-Dx had significantly lower activity than did constructs 2,1-Ds and 2,2-Ds, respectively. This decrease was much more pronounced for the Acc-2,2 gene. Deletion of the downstream promoter P2B in constructs 2,1-K and 2,2-K, and 2,1-J and 2,2-J eliminated the activity entirely (Fig. 1). We therefore conclude that in protoplasts the activity of constructs 2,1-D and 2,2-D represents the activity of the downstream promoter P2B. Thus, the ACC2 genes contain nested promoters, of which the internal one lies within an intron transcribed normally from the upstream promoter.
Two Promoters also Drive Expression of the Plastid ACC Genes. Construct 1,1-A, derived from gene Acc-1,1, had the highest activity of all of the constructs tested in protoplasts (Fig. 1 and 2). It was 7-fold higher than construct 2,2-A and 70- to 100-fold higher than activity of constructs 2,1-A and 2,4-A. Deletion of 5′ end fragments lowered this activity gradually (Fig. 2, constructs 1,1-B, 1,1-C, 1,1-E, and 1,1-G). Construct 1,1-D, with both a 5′ end deletion and an internal deletion, showed activity intermediate between 1,1-C and 1,1-G. Two deletions within the putative promoter P1A and leader region in constructs 1,1-H and 1,1-J had no effect on the activity compared with construct 1,1-B, from which they were derived. In contrast, a small deletion in construct 1,1-I lowers the activity to the same level found for either construct 1,1-E, in which the entire promoter region was deleted, or construct 1,1-F, in which the putative promoter was inserted in reverse orientation. These results indicate that the core promoter P1A is located within a 100-bp fragment at the 5′ end of construct 1,1-D, which is immediately upstream of the upstream transcription start site and that its essential portion is located within a 40-bp fragment deleted in construct 1,1-I (Fig. 2). A sequence motif similar to the “TATA box” found in well expressed plant genes (15) is present within an AT-rich sequence found in this region (AATTTATTATTTTA). However, full activity of the upstream promoter P1A requires additional sequence elements located further upstream, some >500 base pairs upstream, as suggested by the higher activity of construct 1,1-A.
Construct 1,1-E retained a significant level of activity despite deletion of most of the sequences encoding the leader of transcript Acc-1,1A (Fig. 3; ref. 5), as well as the core promoter P1A described above. This result suggested that a second promoter (P1B) located further downstream was used for the Acc-1,1B transcript (Fig. 2) detected by the RT-PCR experiments. Some sequence elements important for the activity of this downstream promoter are located within a 100-bp fragment present at the 5′ end of the DNA fragment in construct 1,1-E immediately upstream of the downstream transcription start site. Its deletion (construct 1,1-G) reduces the promoter activity significantly, although not completely. A second sequence motif similar to the TATA box is present within an AT-rich sequence found in this region (TCTATTTATCTCTTT). The spatial arrangement of the two promoters suggests that utilization of the two transcription start sites is coupled with alternative splicing of the first intron. The RT-PCR cloning experiments showed that the upstream splicing site was used exclusively for transcripts initiated at the upstream transcription start site and vice versa. The differential accumulation of the two types of transcripts established that in young wheat plants the upstream promoter was stronger than the downstream one. This finding appears to be the case in wheat protoplasts as well.
Fig. 3.
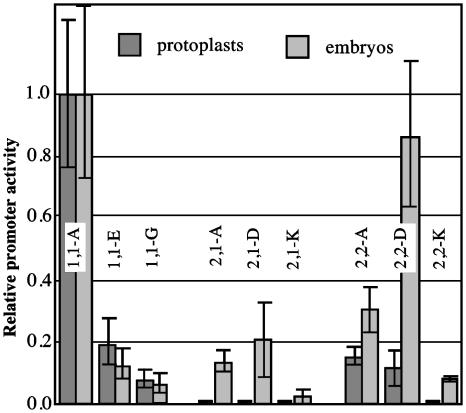
Comparison of transient expression of GUS for selected Acc promoter fusions in wheat protoplasts and in mature embryos. Activity of the construct 1,1-A was taken as 1 for each series. The protoplast results were calculated from data shown in Figs. 1 and 2.
Relative Strength of Acc Promoters Is Different in Protoplasts and Mature Wheat Embryos. In the next step, a group of constructs was tested in imbibed mature wheat embryos at a very early germination stage (Fig. 3). These embryos included multiple cell types, but the majority of the GUS activity measured in the particle bombardment transient expression assay comes from the epidermal cells. The purpose of this experiment was two-fold: (i) to confirm conclusions of the promoter activity of the gene fragments and the alternative promoters as defined in the protoplast experiments, and (ii) to reveal new information on promoter tissue specificity by contrasting dedifferentiated protoplasts with embryo epidermal cells. All full-length promoter constructs showed significant transient activity in embryos. The relative promoter activity of constructs 1,1-A, 1,1-E, and 1,1-G was the same in both transient expression systems. With regard to the cytosolic gene promoters, the relative expression driven by construct 2,1-A was significantly higher, and by construct 2,2-A, was slightly higher in embryos than in protoplasts. The Acc-2,1 promoter (construct 2,1-A) showed relatively little activity in protoplasts but in embryos the activity reached a level of 50% of construct 2,2-A (Figs. 1 and 3). Deletion 2,1-D has no effect, but deletion 2,1-K significantly reduces activity of the Acc-2,1 promoter in embryos. A similar relationship was observed in protoplasts. For the Acc-2,2 promoter, deletion in construct 2,2-D, removing the upstream promoter P2A and any upstream regulatory sequence, increases GUS activity, suggesting that this part of the gene contains an element repressing its expression in the epidermis of imbibed mature embryos. This deletion has no effect on the activity of the Acc-2,2 promoter in protoplasts. Deletion in construct 2,2-K decreases the promoter activity in embryos significantly, but not as much as in protoplasts (Figs. 1 and 3).
These results confirm the arrangement and relative activity of the alternative promoters of each of the Acc genes. The activity of the Acc-2,1 promoter increases >10-fold, relative to the Acc-1,1 gene, when transient expression in embryos is compared with that in protoplasts, suggesting an increased demand for cytosolic malonyl-CoA for either FA elongation or FL biosynthesis, or both, in epidermal cells. Protoplasts are not expected to synthesize large amounts of VLCFAs, in contrast with epidermal cells, where they are needed for biosynthesis of the wax deposited on the surface of various plant organs. Furthermore, it was reported previously that cells of a maize suspension culture, which is similar to our wheat culture used for protoplast preparation, do not accumulate FLs (16).
Activity of the Promoter(s) of the Acc-2,1 Gene in Transgenic Wheat: Young Plant Development. Tissue specificity of the Acc-2,1 promoter revealed by the transient expression experiments prompted its analysis during a complete wheat developmental cycle using transgenic plants carrying full-length promoter construct 2,1-A. These results were reproducible in multiple lines of transgenic plants and dependent on the presence of the transgene.
Activity of the Acc-2,1 promoter was detected initially in the coleoptile (marker 1, Fig. 6, which is published as supporting information on the PNAS web site) when the seedlings were 5 days old (one-leaf stage). The highest expression level was observed in coleoptiles of 8-day-old seedlings when the plant was at the two-leaf stage and before formation of the collar on the first full leaf. No expression was detected in the two leaves developing inside of the coleoptile. GUS staining in the coleoptile decreased by day 12, when the plant was at the two-leaf stage and after a collar was formed on the first full leaf, where GUS activity was detected in the collar of the first leaf and immediately below it (marker 2, Fig. 6). No expression was observed in the inner (second) leaf. By day 21, when the plant was at the four-leaf stage with collars formed on the first, second, and third leaf, GUS staining in the coleoptile, already undergoing senescence, disappeared completely (Inset I, Fig. 6). Staining of the upper sheath section was observed for all consecutive leaves with the oldest leaves showing less intense staining covering a larger section of the sheath. At the four-leaf stage, the highest expression level was observed in and below the collar of the second leaf. Lower expression was detected in the collars of the first and third leaf, and no expression was detected on the fourth leaf, which has not yet formed its collar. A cross section revealed GUS staining preferentially in and around the veins of the coleoptile (Inset II, Fig. 6). Staining of the upper sheath section was diffuse and did not allow identification of individual stained cells. No GUS expression from construct 2,1-A was detected in any other part of the young plant leaves or in roots.
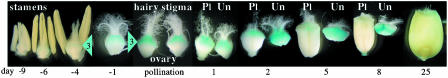
Activity of the Promoter(s) of the Acc-2,1 Gene in Transgenic Wheat: Flower and Seed Development. GUS expression driven by the Acc-2,1 promoter can first be detected in the ovary 4 days before pollination. The expression increases gradually and reaches a maximum at pollination. Expression decreases as the seed develops, with only weak GUS staining detectable 8 days after pollination (marker 3, Fig. 4). The strong GUS expression is confined to the stigma surface of the ovary (see cross sections, Fig. 7, which is published as supporting information on the PNAS web site). In contrast, GUS expression in the unpollinated ovary (anthers removed before pollination) remains high for several days and then decreases slowly as the unpollinated ovary shrinks and dies (Fig. 4). No expression was observed in anthers, pollen, or embryos.
Fig. 4.
Activity of the Acc-2,1 promoter during wheat flower and seed development. GUS staining of transgenic wheat plants transformed with construct 2,1-A. Stamens showed no GUS staining and were removed from some flowers shown. Pl, pollinated; Un, unpollinated; pollination, day 0. Major site of GUS activity is marked with a blue triangle.
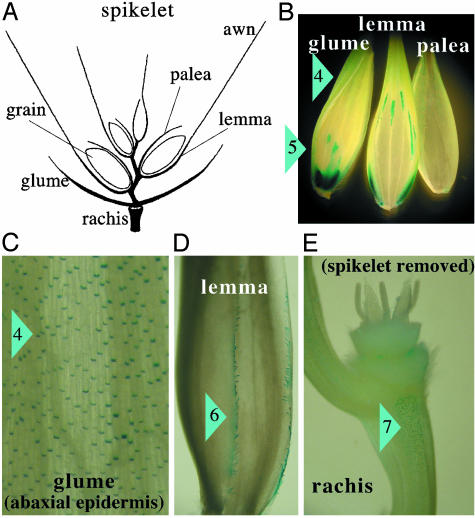
Strong GUS expression was detected in different organs of the developing spikelet (Fig. 5A) in the basal parts of epidermal hair cells (small cells between long epidermal cells arranged in rows) of the outer (abaxial) epidermis of the upper part of the glume and lemma (marker 4, Fig. 5 B and C). Most of the hairs themselves were not visible in our stained preparations, except for a single row of large hairs along the edges of the glume and lemma, which showed strong GUS staining (marker 6, Fig. 5D). The highest number of stained hair cells was observed in areas exposed to the environment; e.g., in areas of the glume not covered by the lemma. Few stained cells were seen on the internal surfaces (adaxial epidermis) and then only near the edges of the organs. Compared with the glumes and lemma, relatively few stained cells were seen in the outer epidermis of the palea. A similar staining pattern of individual cells was seen on the rachis, with the highest number of stained cells in those parts of the epidermis fully exposed to the environment (marker 7, Fig. 5E). Stronger expression was detected in and around some sections of the major veins of glumes and lemma (marker 5, Fig. 5B), which was similar to the staining of the veins in the upper part of the coleoptile (Fig. 6). These stained sections of the veins were often concentrated at the bottom of the glumes and lemma. Expression in both locations of the glumes and lemma was maximal at pollination (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 5.
Activity of the Acc-2,1 promoter in developing spikelets. GUS staining of transgenic wheat plants transformed with construct 2,1-A. (A) Main structures of a mature spikelet. (B) Glume, lemma, and palea. (C) Enlargement of the upper part of the glume. (D) Hairs at the edge of the lemma. (E) The rachis. Major sites of GUS activity are marked with blue triangles and numbers: 4, hair cells of the outer (abaxial) epidermis of the upper part of glume and lemma; 5, section of major veins of the glume and lemma; 6, large hairs at the edge of the lemma; and 7, epidermis of the rachis.
Correlation Between Acc Transcript Levels and Activity of the Acc-2,1 Promoter Revealed by GUS Staining in Transgenic Plants. We observed previously that Acc-2 transcripts reach their maximum level in the lower sheath section of young wheat plants (5). For the Acc-2,1 gene, this pattern correlates well with the elevated promoter activity detected by GUS staining in leaves of transgenic wheat plants carrying construct 2,1-A (Fig. 6).
We measured levels of both plastid and cytosolic ACC mRNAs in three leaves of a 14-day-old plant (Fig. 9, which is published as supporting information on the PNAS web site). The overall pattern of expression of Acc genes is very similar in all three leaves, and is similar to the patterns described (5). The highest level of the plastid ACC mRNA is found at the base of the third (youngest) leaf (Fig. 9A). The maximum level of the cytosolic ACC mRNA is highest in the midsection of the third leaf (Fig. 9B). This section also shows a relatively high level of the plastid ACC mRNA. All three sections of the first (oldest) and the second leaf show decreased levels of both ACC mRNAs.
There is a clear delay between the accumulation of Acc-2 mRNA (midsection of the youngest leaf) and GUS staining (midsection of older leaves that already formed collar), presumably caused by the time needed for GUS to accumulate. It is likely that the promoter activity is high in cells 1–3 days before GUS staining is actually detected. Cells of the middle section of the third leaf in which Acc-2 mRNA level is the highest (Fig. 9B) will be positioned in the upper sheath section of the third leaf a few days later, which is where the GUS staining is the strongest (Fig. 6). By contrast, no staining was observed in any part of the leaf blade, where there was insufficient mRNA made earlier for GUS to accumulate to a level detectable by staining.
GUS staining experiments reveal, with the delay discussed above, specific Acc-2 gene expression in a small subset of cells of each organ that transcribe this gene at a high level. These properties are not reflected properly in the direct mRNA level measurements by real-time PCR, which include all cells in each section and therefore cannot reveal differences between cell types, such as mesophyll and epidermal cells, and different times in their development. For example, there is no difference in the Acc-2 mRNA level in ovaries 9 days before and at the time of pollination (Fig. 9B). GUS staining, however, indicates increasing expression of the genes during this time, but only in the epidermal cells on the top surface of the ovary (Fig. 4).
Discussion
Our transcription and promoter analysis revealed four levels of structural and functional divergence among members of the Acc gene family. First, the promoters of the Acc-1 and Acc-2 genes are completely different. Acc-1 was created by duplication of an ancestral Acc-2 gene shortly after divergence of the grass family (6), a fact reflected in significant coding sequence similarity between these paralogs. This gene acquired the plastid function performed originally by the multisubunit ACC of endosymbiont origin still found in dicots (8). Second, promoters of paralogous Acc-2 genes (e.g., Acc-2,1 and Acc-2,2) share blocks of conserved sequences, but in other parts, differ sufficiently to have an altered specificity acquired during their extended divergent evolution (1, 6). Third, both Acc-1 and Acc-2 genes have nested alternative promoters with different tissue specificity. Fourth, promoters of all three homoeologs of the Acc-1 and Acc-2 genes retained their activity and specificity. Sequence differences between them can be attributed primarily to the divergent evolution of the wheat diploid ancestors, which did not affect gene function. Fewer changes were introduced during the evolution of tetraploid and hexaploid wheat. These polyploidization events occurred <1 million and 10,000 years ago, respectively (4).
What is the purpose of such divergent complex promoters? We propose that a significant level of cell-specific expression of ACC isozymes to supply the plastid, mitochondrial, and cytosolic pools of malonyl-CoA is accomplished at the transcriptional level. The composite nested promoters found in the Acc genes may be commonly used in grasses; for the Acc genes, the core of the downstream promoter is located in the first intron of transcripts made from the upstream promoter. This arrangement is also found in the well studied promoters of the rice β-tubulin gene (17) and the maize ubiquitin gene (18), where the core promoters and their associated cis-acting regulatory elements are intertwined with translation signals encoded in the transcript leaders and the splicing signals needed to remove the leader introns.
Malonyl-CoA, which is derived from acetyl-CoA by the action of ACC, is a key metabolite, a source of activated carbon units for primary and secondary metabolism. This enzymatic activity is essential for different subcellular compartments. Selective herbicides kill grasses such as wheat by specific binding and inhibition of the enzymatic activity in plastids (19), and possibly also in mitochondria (9). Arabidopsis mutants in which the cytosolic ACC gene is disrupted are embryo-lethal and deposit triacylglycerides lacking VLCFAs (20). Finally, all current evidence (8) implicates the ACC reaction as an important regulatory step, controlling metabolite flow during plant development and in response to stress.
The demand for ACC activity in wheat is met by expression of a small gene family encoding multidomain enzymes. Expression of the plastid isozyme supplying malonyl-CoA for FA synthesis for membrane lipids in dividing and fast growing cells, and during chloroplast development in mesophyll cells, is a part of one developmental program. In some cells (e.g., some epidermal cells), a significant fraction of FAs made in plastids is further elongated and used for CW synthesis in these cells. In this case, expression of both plastid and cytosolic ACCs is likely to be coordinated and under control of the regulatory circuits of VLCFA synthesis. On the other hand, FL synthesis requiring activity of the cytosolic ACC is likely to be regulated by different developmental and environmental cues. Finally, the mitochondrial pool of malonyl-CoA in plants is either derived from the cytosol, or, as recently suggested for grasses, is supplied by ACC localized in the organelle (9). In grasses, the product of the same Acc gene appears to be imported to both plastids and mitochondria. We believe that much of the differential and coordinated expression required to regulate ACC is provided by its nested alternative promoters.
We noted the following correlations between the strong activity of the cytosolic ACC promoter in specific cells of some organs detected in the transgenic-wheat experiments and plant developmental processes.
(i) The sheath section of the leaf. CW deposited on the epidermal cells acts as a component of the protective barrier against biotic and abiotic stresses, such as bacterial and fungal pathogens, and water loss during drought. FLs add another layer of protection to the plant, against UV radiation and possibly against pathogen attack. Our observations are consistent with earlier reports (21) from various studies of monocots; e.g., on accumulation of CW on emerging leak leaves, secretion of CW by cork cells of the leaf abaxial epidermis in sorghum, greatly affected in bloomless mutants (22, 23), and on rapid accumulation of FLs in young leaves of barley as they emerge from the coleoptile (24).
(ii) The top surface of the ovary. VLCFA derivatives secreted on the surface of pollen and stigma have been shown to participate in pollen–stigma interaction in various plants (25–27), including wheat (28) with the lipid-rich pollen coat playing a role in plants with “dry stigmas” and lipid-rich secretions on “wet stigmas” playing the role in other plants. Our results show a high level of expression of the cytosolic ACC gene on the top surface of the ovary, suggesting an elevated rate of CW secretion by epidermal cells in this part of the organ.
(iii) Hair cells in the abaxial epidermis of the lemma and glume. Glumes and lemma are characterized by a very dense cuticle layer on their epidermal cells. The accumulation of CW on these organs adds to the protection of the developing seed in a manner similar to that described above for the developing leaves. A recent study on barley (29) suggested an additional role for CW deposited on these organs: regulation of ear wetting and prevention of “in-ear” sprouting. Our study extends these observations by implicating hair cells as important producers of CW. Another observation is also revealing: the hair cells in the abaxial epidermis of glumes and lemmas are active in this process and mostly in those areas that are exposed to the environment. A similar role could be assigned to specific cells in the exposed parts of the rachis. Thus, all of our observations on the expression of the Acc gene family are consistent with the known physiology of parts of developing wheat plants.
Supplementary Material
Acknowledgments
We thank J. E. Fry and N. V. Sidorova (Monsanto, St. Louis) for providing wheat cell cultures, training, and valuable advice. This work was supported in part by grants from the Consortium for Plant Biotechnology Research, Monsanto, and a fellowship from the German Academic Exchange Service (to E.Z.).
Abbreviations: ACC, acetyl-CoA carboxylase; FA, fatty acid; VLCFA, very-long-chain FA; CW, cuticular wax; FL, flavonoid; GUS, β-glucuronidase.
References
- 1.Faris, J., Sirikhachornkit, A., Haselkorn, R., Gill, B. & Gornicki, P. (2001) Mol. Biol. Evol. 18, 1720–1733. [DOI] [PubMed] [Google Scholar]
- 2.Gornicki, P., Podkowinski, J., Scappino, L. A., DiMaio, J., Ward, E. & Haselkorn, R. (1994) Proc. Natl. Acad. Sci. USA 91, 6860–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gornicki, P., Faris, J., King, I., Podkowinski, J., Gill, B. & Haselkorn, R. (1997) Proc. Natl. Acad. Sci. USA 94, 14179–14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang, S., Sirikhachornkit, A., Su, X. J., Faris, J., Gill, B., Haselkorn, R. & Gornicki, P. (2002) Proc. Natl. Acad. Sci. USA 99, 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Podkowinski, J., Jelenska, J., Sirikhachornkit, A., Zuther, E., Haselkorn, R. & Gornicki, P. (2003) Plant Physiol. 131, 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang, S. X., Sirikhachornkit, A., Faris, J. D., Su, X. J., Gill, B. S., Haselkorn, R. & Gornicki, P. (2002) Plant Mol. Biol. 48, 805–820. [DOI] [PubMed] [Google Scholar]
- 7.Podkowinski, J., Sroga, G. E., Haselkorn, R. & Gornicki, P. (1996) Proc. Natl. Acad. Sci. USA 93, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikolau, B. J., Ohlrogge, J. B. & Wurtele, E. S. (2003) Arch. Biochem. Biophys. 414, 211–222. [DOI] [PubMed] [Google Scholar]
- 9.Focke, M., Gieringer, E., Schwan, S., Jansch, L., Binder, S. & Braun, H.-P. (2003) Plant Physiol. 133, 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkel-Shirley, B. (2001) Plant Physiol. 127, 1399–1404. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou, H., Stiff, C. M. & Konzak, C. F. (1993) Plant Cell Rep. 12, 612–616. [DOI] [PubMed] [Google Scholar]
- 12.Becker, D., Brettschneider, R. & Lorz, H. (1994) Plant J. 5, 299–307. [DOI] [PubMed] [Google Scholar]
- 13.Jefferson, R. A. (1987) Plant Mol. Biol. Rep. 5, 385–403. [Google Scholar]
- 14.McCabe, D. E., Swain, W. F., Martinelli, B. J. & Christou, P. (1988) Bio/Technology 6, 923–926. [Google Scholar]
- 15.Sawant, S. V., Singh, P. K., Gupta, S. K., Madnala, R. & Tuli, R. (1999) J. Genet. 78, 123–131. [Google Scholar]
- 16.Grotewold, E., Chamberlin, M., Snook, M., Siame, B., Butler, L., Swenson, J., Maddock, S., Clair, G. S. & Bowen, B. (1998) Plant Cell 10, 721–740. [PMC free article] [PubMed] [Google Scholar]
- 17.Morello, L., Bardini, M., Sala, F. & Breviario, D. (2002) Plant J. 29, 33–44. [DOI] [PubMed] [Google Scholar]
- 18.Salgueiro, S., Pignocchi, C. & Parry, M. A. J. (2000) Plant Mol. Biol. 42, 615–622. [DOI] [PubMed] [Google Scholar]
- 19.Zagnitko, O., Jelenska, J., Tevzadze, G., Haselkorn, R. & Gornicki, P. (2001) Proc. Natl. Acad. Sci. USA 98, 6617–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baud, S., Guyon, V., Kronenberg, J., Wuilleme, S., Miquel, M., Caboche, M., Lepiniec, L. & Rochat, C. (2003) Plant J. 33, 75–86. [DOI] [PubMed] [Google Scholar]
- 21.Rhee, Y., Hlousek-Radojcic, A., Ponsamuel, J., Liu, D. & Post-Beittenmiller, D. (1998) Plant Physiol. 116, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenks, M. A., Rich, P. J. & Ashworth, E. N. (1994) Int. J. Plant Sci. 155, 506–518. [Google Scholar]
- 23.Jenks, M. A., Rich, P. J., Rhodes, D., Ashworth, E. N., Axtell, J. D. & Ding, C.-K. (2000) Phytochemistry 54, 577–584. [DOI] [PubMed] [Google Scholar]
- 24.Liu, L., Gitz, D. C. I. & McClure, J. W. (1995) Physiol. Plant. 93, 725–733. [Google Scholar]
- 25.Hiscock, S. J., Hoedemaekers, K., Friedman, W. E. & Dickinson, H. G. (2002) Int. J. Plant Sci. 163, 1–16. [Google Scholar]
- 26.Wolters-Arts, M., Van der Weerd, L., Van Aelst, A. C., Van der Weerd, J., Van As, H. & Mariani, C. (2002) Plant Cell Environ. 25, 513–519. [Google Scholar]
- 27.Chen, X., Goodwin, S. M., Boroff, V. L., Liu, X. & Jenks, M. A. (2003) Plant Cell 15, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, A. M., Xia, Q., Xie, W. S., Dumonceaux, T., Zou, J. T., Datla, R. & Selvaraj, G. (2002) Plant J. 30, 613–623. [DOI] [PubMed] [Google Scholar]
- 29.King, R. W. & von Wettstein-Knowles, P. (2000) Euphytica 112, 157–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.