Abstract
The genome of tobacco mosaic virus (TMV) encodes replicase protein(s), movement protein (MP), and capsid protein (CP). On infection, one or more viral proteins direct the assembly of virus replication complexes (VRCs), in association with host-derived membranes. The impact of CP-mediated resistance on the structures of the replication complexes was examined in nontransgenic and transgenic BY-2 cell lines that produce wild-type CP, mutant CPT42W, and Ds-Red, which was targeted to endoplasmic reticulum by using immunofluorescence and 3D microscopy. We developed a model of VRCs that shows a clear association of MP with and surrounding the endoplasmic reticulum. Replicase is located within the MP bodies, as well as isolated sites throughout the cell. CP surrounds the VRCs. CP enhances the production of MP and increases the size of the VRC; however, the mutant CPT42W reduces the amount of MP and interferes with the formation of VRCs. We propose a regulatory role of the CP in the establishment of the VRC. We suggest that the lack of formation of VRCs restricts the efficiency of virus replication and the formation of virus movement complexes, resulting in restriction of cell–cell spread of infection. This results in higher levels of plant CP-mediated protection provided by CPT42W.
Positive-strand RNA viruses share fundamental similarities in RNA replication despite differences in genome organization, virion morphology, and host range. On entering the cell, the messenger-sense genome is translated to yield a variety of proteins, some of which direct the assembly of virus replication complexes (VRCs) that are invariably associated with host membrane (1–4).
The 6.3-kb (+) sense RNA genome of tobacco mosaic virus (TMV) encodes two proteins essential for replication of the genome (replicase, 126/183 kDa), a 30-kDa movement protein (MP) that is essential for cell–cell spread of infection, and a 17.5-kDa capsid protein (CP). Cytological studies of TMV-infected cells showed VRCs associated with cytoplasmatic inclusions bodies, called viroplasm [also called X-bodies (5)], that enlarge throughout infection. Electron microscopy revealed that these inclusion bodies contained ribosomes, tubules, and 126/183-kDa replication proteins (6–9).
Recent studies show that the 126-kDa protein associates with the endoplasmic reticulum (ER) in the absence of other viral proteins, and it was suggested that association may occur via either membrane-bound host proteins or a membrane insertion of an amphipathic helix detected in the protein (10). The MP behaves as an intrinsic membrane protein, promotes the formation of ER aggregates, and probably facilitates the establishment of TMV replication complexes that contain viral RNA, replicase, and MP (11–15).
Transgenic plants that produce TMV CP interfere with disassembly of TMV and thereby confer resistance to TMV infection (16). Mutant CP that is changed at amino acid 42 [Thr-42 → Trp (T42W)] interferes with virus disassembly. In addition, it reduces production of MP on infection, thereby reducing cell–cell spread of infection. The mutant, CPT42W, also reduces the formation of the MP inclusion bodies (17).
To gain a more complete understanding of the structure of TMV replication complexes and the relationship of these structures with the mechanisms of CP-mediated resistance, each virus protein was localized by using immunofluorescence and 3D microscopy (confocal optical sections and digital image reconstruction). We analyzed the spatial and temporal changes in distribution of each viral protein in infected protoplasts of nontransgenic and transgenic BY-2 cell lines that produce wild-type CP (BY-CP), mutant CP (BY-CPT42W), and Ds-Red that was targeted to ER (ER-DsRed). Our data provides highly detailed information of the structure and localization of the VRCs during infection. CP enhances the production of MP and increases the size of the VRC; CPT42W reduces MP production and interferes with the formation of VRCs. We propose a regulatory role of the CP in establishing the VRC. We suggest that the lack of formation of VRCs restricts the efficiency of virus replication and the formation of virus movements complexes (S. Kawakami and R.N.B., unpublished work) that restricts cell–cell spread of infection. This results in higher levels of plant protection provided by the CPT42W (18), suggesting that several different resistance mechanism play a role in CP-mediated resistance.
Materials and Methods
BY-2 Transgenic Lines. Transgenic BY-2 cell lines that produce wild-type CP or mutant CP were generated via Agrobacterium tumefaciens-mediated transformation by using the clones pMONCP-T42W (18) and pCGN-CPU1 (17).
Protoplast Isolation and Transfection. To obtain infectious viral RNA from the full-length cDNA clone of TMV RNA, pU3/4–12 (19), the MEGAscript Transcription kit (Ambion, Austin, TX) was used, following the manufacturer's recommendations. Reactions were supplemented with m7G(5′)ppp(5′)G Cap Analog (Ambion). Tobacco BY-2 protoplasts were prepared from suspension cell cultures essentially as described by Watanabe et al. (20), and 1.5 × 106 cells were inoculated by electroporation and incubated at 28°C in the dark.
Immunocytochemistry. Infected protoplasts were harvested and fixed in freshly prepared 3% paraformaldehyde buffer (50 mM potassium phosphate, pH 6.7/10 mM EGTA) for 1 h and then washed twice with PBS. An aliquot of cells was allowed to settle on polyethyleneimine (PEI)-coated (1%) coverslips and then treated with blocking buffer (PBS/1% BSA) containing 0.1% Triton X-100 for 8 min, followed by three rinses with blocking buffer for 20 min. The protoplasts were incubated for 2 h at room temperature with primary antibodies [anti-CP (produced in mice) and anti-replicase (produced in rabbit)], followed by three washes with PBS. The last incubation was done with the Alexa 488-conjugated anti-MP antibody for 2 h and washing again as above. Finally, the coverslip was inverted and mounted in PBS on a slide with a chamber to avoid damaging the cell. Anti-rabbit and anti-mouse antibodies conjugated with Cy3 and with Cy5, respectively, were used as secondary antibodies (Jackson ImmunoResearch, diluted 1/50).
Confocal Microscopy. Confocal microscopy was conducted with a Leica SP2 inverted confocal microscope. The scanning was done at ×63 magnification (HCX PL APO 63.0 × 1.32 oil immersion lens) by using a 488 Argon laser, a Kripton 568 laser, and a HeNe 634 laser as excitation light sources. Optical sections were taken at 0.4-μm intervals. The wavelength settings of the acquisition detectors were adjusted to minimize crossover between the emission wavelengths of each fluorophore, and scanning was conducted in a sequential mode to further minimize crossover.
Three-Dimensional Reconstruction. The 3D reconstruction of the confocal image stacks and background subtraction were accomplished by using imaris v.3.3 software (Bitplane, St. Paul, MN). The imaris Surpass module was used to calculate the blending 3D reconstructions to generate the cutaway visualizations. Blending calculations allow one to adjust the transparency of each channel (one channel per fluorophore) making it possible to view the composition of the structures. The imaris Coloc module was used to calculate the colocalized voxels (volume unit, analogous to a pixel in two dimension images) between the different channels. We used a polygon manual selection mode to select the colocalized voxels with correlation coefficients ≥0.99. The colocalization channels in some figures were adjusted to increase the detection of colocalization; therefore, these values do not reflect the intensity of colocalization but localization per se.
Results
To gain a more complete understanding of the mechanism of TMV replication, we analyzed the spatial and temporal changes in distribution of each viral protein (i.e., replicase, MP, and CP) in BY-2 protoplasts infected with TMV-RNA. We localized all viral proteins simultaneously by using immunofluorescence and 3D microscopy (confocal optical sections and 3D digital reconstructions) in protoplasts harvested at 7, 10, 16, and 24 h postinfection (hpi). Three-dimensional reconstructions and background subtraction were performed by using imaris.
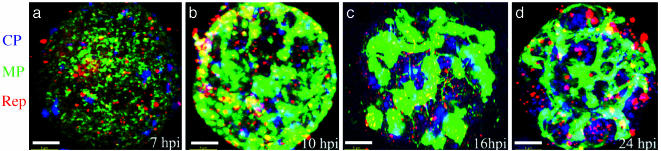
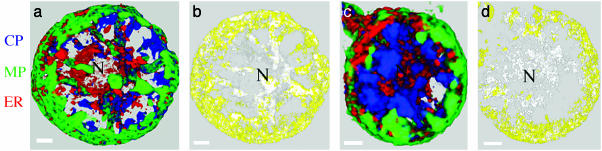
A representative sequence of the distribution of each protein during the virus infection is shown in Fig. 1. Rotation movies of the 3D reconstructions provided in Fig. 1 are available as Movies 1–4, which are published as supporting information on the PNAS web site. The latter enhances greatly the visualization of distribution and localization of antibodies. The MP (green in the visualization) formed punctate structures at early stages of the infection, small bodies at mid-stage, and interconnected large bodies during late stages of the infection. These data are in agreement with previous work using a TMV construct in which the GFP was fused with MP (13). The replicase (red in the visualization) is in small puncta scattered throughout the cell that increase in size as the infection progresses, whereas localization per se remains unchanged. The CP (blue in the visualization) forms irregular bodies that also grow in size during the infection: at late stages, CP is most abundant in bodies around the nucleus, as well as in cytoplasmic strands that connect the nucleus with the cortex of the cell.
Fig. 1.
Three-dimensional maximum-intensity projection reconstruction of BY-2 protoplasts infected with TMV through an infection cycle. Protoplasts were collected at 7 hpi (a), 10 hpi (b), 16 hpi (c), and 24 hpi (d), stained with antibodies, and observed by confocal microscopy: images were interpreted by imaris. Immunostaining color code: green, MP; blue, CP; red, replicase. Three-dimensional rotation reconstruction can be observed in Movies 1–4. (Scale bars, 5 μm.)
Viral Replication Complexes. During early stages of infection, MP and replicase are primarily found in the large bodies throughout the cell, and at later stages the CP becomes associated with these structures. Therefore, we refer to these structures as virus replication complexes (VRCs); it is proposed that replication of viral RNA (vRNA) and some or most of the translation of viral proteins take place in VRC (14).
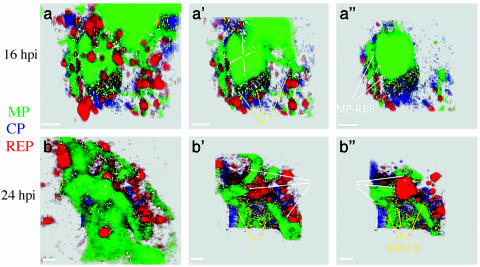
To further analyze the composition of the VRCs, we determined the frequency of colocalization of MP with CP and replicase. The colocalization channels are shown in a close-up of a small section of a protoplast that contains several VRCs from cells at 16 and 24 hpi (Fig. 2). The colocalized voxels (volume unit) with correlation coefficients ≥0.99 were selected with software assistance. This is a more accurate way to visualize the colocalization of fluorophores than simply superimposing the images because it considers three dimensions in selecting the colocalized volumes. We generated two colocalization channels: one channel is visualized as white and contains the colocalization between replicase (red) and MP (green), and the other is visualized as yellow and shows colocalization between CP (blue) and MP.
Fig. 2.
Detailed view of VRCs in BY-2 protoplasts infected by TMV. Protoplasts were immunostained and viewed as in Fig. 1, and data were analyzed by focusing on the structure and composition of VRCs. Three cutaway sections of rendered volumes (a, a′, a″, b, b′, and b″) were made through the VRCs along the z axis. Immunostaining color code: green, MP; blue, CP; red, replicase. Colocalization color code: yellow, MP-CP (yellow arrows); white, MP-replicase (white arrows). (a) At 16 hpi. (b) At 24 hpi. (Scale bars, 1 μm.)
During the period of the experiments, the primary component of the VRC is MP, and replicase is in close association with the MP bodies. We observed two types of bodies that contain replicase. The larger bodies are usually located near the periphery of the MP bodies and exhibit colocalization of proteins at the interface between MP and replicase. Replicase was also observed without an apparent interaction with the VRCs. A second smaller type of replicase body was observed within the MP body; these bodies are readily seen in cutaway sections on the colocalization channel (enhanced for increased visualization). At 16 hpi, the white channel shows small points colocalization of MP and replicase within the MP bodies (Fig. 2, e.g., see white arrows in a′ and a″). At 24 hpi, the small white puncta are evenly distributed within the MP bodies (Fig. 2, e.g., see white arrows in b′ and b″).
The CP is also associated with the VRC, primarily at the periphery of the VRCs. At 16 hpi, the colocalization of MP and CP (yellow) shows that the small CP bodies are in close association with the MP mass but only display colocalization near the interface of each body (Fig. 2, e.g., see yellow arrows in a′). At 24 hpi, the MP bodies are larger and show more colocalization of CP and MP at surfaces (Fig. 2 b, b′, and b″; see also Fig. 6 a and a′; the colocalization is shown in the white cannel).
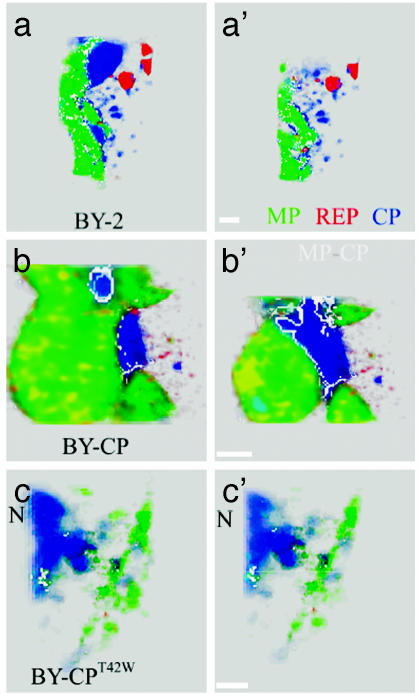
Fig. 6.
Detailed cutaway view of the VRCs in nontransgenic and transgenic protoplasts 24 hpi with TMV. Shown are two cutaway sections of rendered volumes of cells through VRCs along the z axis. The study demonstrates that infection in BY-CPT42W cells does not result in normal VRCs that contain MP and replicase. (a and a′) Nontransgenic BY-2 cell line. (b and b′) BY-CP cell line. (c and c′) BY-CPT42W cell line. Immunostaining color code: green, MP; blue, CP; red, replicase. Colocalization color code: white, MP-CP. N, nucleus. (Scale bars, 1 μm.)
To test the quality of the colocalization analysis, we performed a control assay in which CP was localized in infected protoplasts by using two anti-CP antibodies; one antibody was produced in mice and the other produced in rabbit. Each antibody was labeled with different fluorophores on secondary antibodies. As expected, the results of this study showed extensive colocalization between the fluorophores (data not shown). In another study, we found no interference in the detection of MP in the experimental protocols in which the anti-MP antibody was added after the addition of the primary and secondary antibodies for detection of CP and replicase. Thus, when infected protoplasts were incubated with anti-MP and the anti-CP antibodies simultaneously, followed by two secondary labeled antibodies, we obtained the same result as with the standard labeling protocol (data not shown).
VRCs and ER. To investigate the spatial relationship between the VRC and the ER, we used a transgenic BY-2 cell line that expresses a Ds-Red protein targeted to the lumen of the ER (ER-DsRed) (Fig. 4b). The cell line that expresses ER-DsRed was infected with TMV, and aliquots were harvested at 16 and 24 hpi. Viral proteins were localized by antibody reactions concurrent with detection of ER-DsRed (Fig. 3). Three-dimensional reconstructions and background subtraction were performed as previously described. We generated two colocalization channels, white showing colocalization between the ER (red in the reconstruction) and the CP (blue in the reconstruction), and yellow showing colocalization between ER and MP (green in the reconstruction).
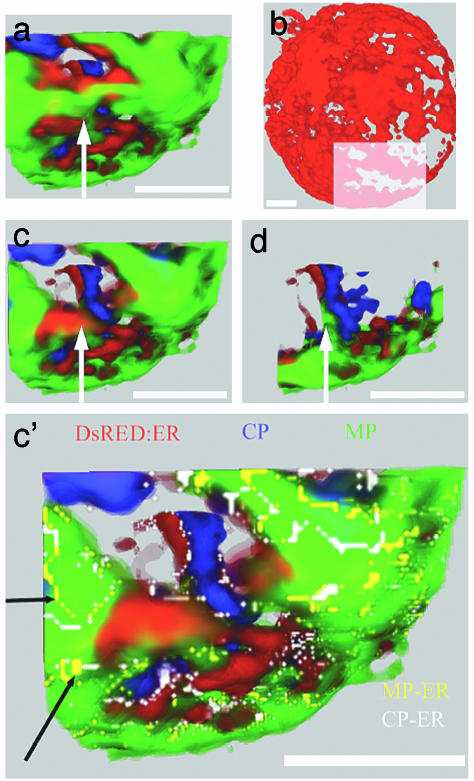
Fig. 4.
ER-DsRed BY-2 protoplasts infected with TMV at 24 hpi after immunostaining to localize CP and MP. Data collected from one region of an infected cell (shown in b) were enlarged to identify the precise manner of colocalization of MP and CP within a single VRC. Color code: green, MP; blue, CP; red, ER-DsRed. Colocalization color code: yellow, MP-ER; white, CP-ER. (b) ER channel only. Shown are a detailed view of the region delineated in b and three cutaway sections of rendered volume (a, c, d, and c′) throughout the VRC along the z axis. (a, b, and c) Fluorophore channels. (c′) Fluorophore and colocalization channels. The black arrows identify the colocalization MP and ER; white arrows indicate the same relative positions in the sectioned volume. Three-dimensional reconstructions are shown in Movies 5 and 6. (Scale bars, 5 μm.)
Fig. 3.
Three-dimensional reconstruction of ER-DsRed protoplasts infected with TMV, after immunostaining to identify the location of viral proteins in positional relationship to the ER. Frontal cell cortex was removed in the visualization to observe the interior of the cell. The study shows strong localization of viral proteins with ER and that localization of MP changes during the infection cycles. Immunostaining color code: green, MP; blue, CP; red, ER-DsRed. Colocalization color code: yellow, MP-ER; white, CP-ER. (a) Fluorophore channels, 16 hpi. (b) Colocalization channels, 16 hpi. (c) Fluorophores channels, 24 hpi. (d) Colocalization channels, 24 hpi. N, nucleus. Three-dimensional reconstructions are shown in Movies 5 and 6. (Scale bars, 5 μm.)
In the majority of cells examined, VRCs were closely associated with and surrounding the ER strands. In mid-stage infection (16 hpi), MP shows strong colocalization with the peripheral ER (yellow channel) and in cytoplasm strands (Fig. 3 a and b). At 16 hpi, CP was colocalized with the ER (Fig. 3b, white channel) and surrounding the face of the cortical MP bodies that faces toward the interior of the cell (Fig. 3a; movies of the 3D reconstructions are available as Movies 5 and 6, which are published as supporting information on the PNAS web site). At late stages of infection (24 hpi), the MP was located near the cell surface and colocalized with the network of cortical ER (Fig. 3 c and d). At this stage, CP was primarily observed in cytoplasmatic strands and proximal to ER adjacent to the nucleus; CP shows substantial colocalization with perinuclear ER (Fig. 3 c and d).
In more detailed views of volume-rendered VCRs at 24 hpi (Fig. 4) it was clear that MP is on the surface of strands of cortical ER. Furthermore, CP accumulates at or near the surface of the large inclusion bodies that contains MP, as well as in the cytoplasm. The white arrow in Fig. 4 a, c, and d points to the same X and Y coordinates in the different cutaway (z axis) sections of one area of the infected cell (displayed in Fig. 4b). Starting from Fig. 4a, one can visualize MP that is adjacent to ER (Fig. 4c; movies of the 3D reconstructions are available as Movies 5 and 6). On the opposite side of the ER, MP is also detected (Fig. 4d), suggesting that the body containing MP surrounds the ER. In Fig. 4d, CP is localized in a strand of cytoplasm and is appressed to the ER. When the colocalization analysis was applied, we observed that MP and ER are colocalized and visualized as a very thin wire-like network immersed in the MP inclusion bodies (Fig. 4c′, black arrows, yellow channel). The white channel reveals the presence of very low amounts (only visible in the colocalization channel) of CP inside the MP bodies and in close association with the ER (Fig. 4c′).
Regulatory Role of CP. Transgenic plants that produce TMV CP interfere with disassembly of TMV and thereby confer resistance (see ref. 16 for review). Plants expressing a mutant CP in which the amino acid at position 42 was mutated from threonine to tryptophan (CPT42W) exhibit very high levels of CP-mediated resistance. The CPT42W mutation interferes with the production of MP and, thus, affects virus cell-to-cell movement. Bendahmane et al. (17) proposed a positive regulatory effect on the production of MP by wild-type CP and a negative effect of MP production by CPT42W. To gain a more complete understanding of the mechanisms of CP-mediated resistance, we analyzed the effects of CP and mutant CP in transgenic BY-2 cell lines on the formation of the VRC. BY-2 cells and transgenic BY-2 cell lines expressing CP (BY-CP) and CPT42W (BY-CPT42W) were infected with TMV RNA, and the distribution of viral proteins was examined at 10 and 24 hpi.
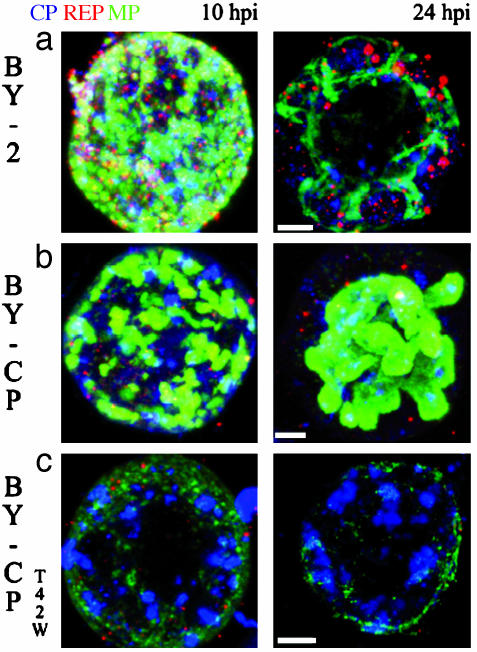
There were considerable differences in accumulation of viral protein in cell lines that contain CP or CPT42W (Fig. 5). First, the large VRCs appeared more rapidly in cells that produced CP than in cells that did not express the CP or the mutant CP. Second, cell lines that contains CPT42W accumulated fewer bodies containing MP, and the small amount of MP present in these cells was localized near the plasma membrane (Fig. 5c). Third, the VRCs produced in the BY-CP line were much larger than in the other cell lines.
Fig. 5.
Maximum-intensity projection of TMV-infected protoplasts at 10 and 24 hpi. In the 24-hpi projections, the frontal cell cortex was removed to observe the interior of the cell. The study shows that accumulation of MP is significantly enhanced in BY-CP protoplasts compared with BY-2; the amount of MP in infected BY-CPT42W protoplasts is much reduced compared with the other cell lines. Immunostaining color code: green, MP; blue, CP; red, replicase. (a) BY-2. (b) BY-CP. (c) BY-CPT42W cell line. (Scale bars, 5 μm.)
Additional differences were detected when a more detailed analysis of the VRCs in all three lines was carried out (Fig. 6). The VRC-like structures located around the nucleus in the BY-CPT42W cells contain only CP; in contrast, VRC-like structures located near the periphery of the cell contain very little MP. The size of the bodies that contain CP was comparable with the other cell lines.
The VRCs in the BY-CP line contain each viral protein; the most abundant, MP, was mixed with replicase (yellow spots in Fig. 6 b and b′). CP colocalized with the MP at the interface between bodies that contain primarily CP and those that contain MP (Fig. 6 b and b′, white channel). The viral proteins in the VRC present in the nontransgenic line show a large amount of colocalization of MP and CP, but there is not a clear indication of replicase in the MP bodies. These results, taken together, suggest that CP enhances the formation of VRCs, and that CPT42W may interfere with the formation of the VRCs.
Another difference between the cell lines was that the replicase bodies (red color in the visualizations) scattered throughout the cell in the nontransgenic line (Fig. 5a) are smaller in size in the BY-CP line and appear to be localized mostly in MP bodies (Fig. 5b). The fact that the number and size of replicase-containing bodies that are not in contact with the VRCs are greater in the BY-2 cell line may indicate a mechanism to control the concentration of the active replicase in such bodies.
Discussion
The application of 3D microscopy to our studies provided substantial spatial and temporal resolution of the cellular processes during TMV infection and coat protein-mediated resistance. It is, however, difficult to present the information in 2D format without loss of detail. Interpretations of the data, described here, are based on the results of many independent experiments and observations of a large number of different cells. The reader is invited to view the 3D images rotations (Movies 1–6) for additional information.
VRCs. Our study focused on the accumulation of TMV proteins during the early and middle stages, during which VRCs are formed and large amounts of virus are produced. In the early stages of the replication cycle, VRCs are formed on ER membranes and, in comparison with late stages of infection, contain relatively large amounts of MP and small amount of replicase. During progression of the cycle, the relative amount of MP decreases while CP accumulation increases, presumably as virions. Throughout the infection cycle, a significant proportion of replicase is not in close proximity with the VRCs. In earlier work (14), we showed that replicase can accumulate in sites that lack vRNA and presumably do not participate in virus replication; Hagiwara et al. (21) demonstrated that there is a pool of replicase that does not cofractionate with replicase activity. These observations may indicate that there exists a mechanism to regulate the amount of replicase that participates in virus replication. This mechanism may limit the amount of replication and further negative impacts on the host cell. Restrepo et al. (22) suggested that there may be a mechanism that controls the participation of NIa and NIb in potyvirus replication.
Most or all of the VRCs are assembled on, and eventually surround, strands or bodies of ER. Earlier studies indicate that the MP is an integral membrane protein (12) that exposes of the N and C termini to the cytoplasm. We applied an analytical program to identify the colocalization of MP with replicase and CP: although the accuracy of the colocalization is limited by the detection methods (i.e., fluorescent-tagged antibodies), the results give greater 3D spatial resolution to the VRCs than previous studies. As the VRCs grow, replicase-rich bodies are found within the MP-rich body or on the periphery of the VRCs, and colocalization of replicase and MP is apparent (Fig. 2). In contrast, the large bodies that contains coat protein (presumably as virions) are generally observed on the surface of the VRCs, and colocalization with MP occurs at the interface of the two types of bodies. This finding is consistent with electron microscopy studies showing MP adjacent to large aggregates of virus particles (23).
The “X-bodies” described by Ivanowski 100 years ago (5), also referred to as viroplasm (24, 25), have features in common with VRCs. Viroplasm contains aggregates of tubules and a ribosome-rich matrix that contains TMV replicase (6, 7). Replicative intermediates of TMV are found on polyribosomes and are presumably associated with the viroplasm (26). Studies of cells infected with brome mosaic virus demonstrated that replicase proteins (1a and 2a) colocalize with the MP (3a) of this virus in electron-dense cytoplasmic inclusions similar to TMV viroplasm/VRCs infection (27). Like the TMV MP, protein 3a is membrane associated.
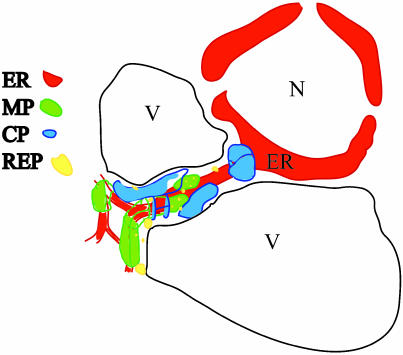
Most, if not all, enzymatically active viral replicase is isolated with membrane-rich complexes; these structures may be organizationally similar to the VRCs that we describe here. We propose a model of VRCs that are anchored to the ER by the MP and replicase in which replication of viral RNA occurs. Fig. 7 presents a schematic visualization of the structural composition and localization of VRCs. MP and replicase can independently bind to ER (10, 14), but it is not known whether binding is at common sites or at different sites that are then brought together during assembly of the VRCs. Overexpression of MP in absence of virus infection causes accumulation of large aggregates of ER (11); we assume that this represents aggregation of existing membranes rather than synthesis of new membranes (28). Electron microscopy studies showed that viroplasms are not membrane enclosed. Virus accumulates to high levels at the periphery of the bodies and may indicate that virion assembly occurs here.
Fig. 7.
Model of VRC structure at late stage of TMV infection. In the middle-to-late stages of infection, VRCs comprising MP and replicase are associated with and surround the cytoplasmic and cortical ER (14). Virus assembly and accumulation is adjacent to VRCs and can accumulate in large aggregates (22) near the nucleus. N, nucleus; V, vacuole; ER, endoplasmic reticulum.
Recently, we showed that structures similar or identical to VRCs are transferred to and through plasmodesmata: those that transmit to adjacent cells are referred to as virus movement complexes (VMCs) (S. Kawakami and R.N.B., unpublished data). The differences, if any exist, between VMCs and VRCs are not known.
The Role of CP in VRC Formation. Our studies revealed significant differences in accumulation of viral proteins after infection by TMV-RNA in nontransgenic and transgenic BY-2 cell lines that contain CP or mutant CPT42W. For example, VRCs appeared more rapidly and were larger in size in cells that produce CP than in nontransgenic cells or cells that contains CPT42W. In VRCs produced in nontransgenic cells and in the transgenic BY-CP line, virus infection resulted in abundant interface colocalization of CP and MP. In cell lines that produce mutant CPT42W, the VRC-like bodies were rich in CP and contain little or no MP; this result is in agreement with a previous studies showing that CPT42W restricts the production of MP (17). The cellular distribution of CP and replicase was not significantly altered in this cell line, although the accumulation of the replicase may be different from in nontransgenic cells.
The CP may have a direct or indirect regulatory role in the establishment of VRCs; it is well known that CP is not required for replication of TMV RNA. However, because infection by TMV that lacks CP produce VRC-like bodies, we consider it likely that a role of CP is to regulate production of subgenomic (Sg) mRNAs that encode MP [and perhaps CP (17)] or translation of mRNAs.
In the case of alfalfa mosaic virus, CP is required for replication of vRNAs and plays an essential role in the asymmetric synthesis of plus-strand RNA; CP–RNA interactions are critical for CP activity (29–31). TMV CP may play a positive but additive role in regulating transcription and on translation pathways of vRNAs; although the mechanisms that enhance the production of MP are not yet known. CPT42W, which restricts production of MP mRNA, does not encapsidate vRNA (18), and self-assembly is altered. We suggest that interactions between CP, CPT42W, and vRNAs are altered and, thus, reduce transcription of Sg mRNAs.
As a consequence, cells that contains CPT42W lack functional VRCs. This restricts the efficiency of virus replication and the formation of virus movement complexes and results in restriction of cell–cell spread of infection. This explains the high level of plant protection provided by CPT42W (18).
The observation that CP has a role in the formation of the VRC should not be surprising because a number of viral proteins are known to have multiple functions. However, until recently the function described for TMV CP was to provide structural protection to the RNA in the virion. The role of CP in other functions, e.g., in regulation production of one or more viral proteins, is consistent with multifunctionality of CP molecules.
Supplementary Material
Acknowledgments
We thank Drs. T. Woodford-Thomas and M. Fujiki for kindly providing the anti-MP antibody coupled with Alexa 488 and the ER-DsRed BY-2 cell line, respectively. This work was supported by National Institutes of Health Grant AI27161.
Abbreviations: CP, capsid protein; hpi, h postinfection; MP, movement protein; TMV, tobacco mosaic virus; VRC, virus replication complex; vRNA, viral RNA.
References
- 1.Osman, T. A. & Buck, K. W. (1996) J. Virol. 70, 6227–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noueiry, A. O. & Ahlquist, P. (2003) Annu. Rev. Phytopathol. 41, 77–98. [DOI] [PubMed] [Google Scholar]
- 3.Restrepo-Hartwig, M. A. & Ahlquist, P. (1996) J. Virol. 70, 8908–8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaad, M. C., Jensen, P. E. & Carrington, J. C. (1997) EMBO J. 16, 4049–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanowski, D. (1903) Z. Pflanzenkr. Pflanzenschutz 13, 1–41. [Google Scholar]
- 6.Hills, G. J., Plaskitt, K. A., Young, N. D., Dunigan, D. D., Watts, J. W., Wilson, T. M. & Zaitlin, M. (1987) Virology 161, 488–496. [DOI] [PubMed] [Google Scholar]
- 7.Saito, T., Hosokawa, D., Meshi, T. & Okada, Y. (1987) Virology 160, 477–481. [DOI] [PubMed] [Google Scholar]
- 8.Martelli, G. P. & Russo, M. (1977) Adv. Virus Res. 21, 175–266. [DOI] [PubMed] [Google Scholar]
- 9.Esau, K. & Cronshaw, J. (1967) J. Cell Biol. 33, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.dos Reis Figueira, A., Golem, S., Goregaoker, S. P. & Culver, J. N. (2002) Virology 301, 81–89. [DOI] [PubMed] [Google Scholar]
- 11.Reichel, C. & Beachy, R. N. (1998) Proc. Natl. Acad. Sci. USA 95, 11169–11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brill, L. M., Nunn, R. S., Kahn, T. W., Yeager, M. & Beachy, R. N. (2000) Proc. Natl. Acad. Sci. USA 97, 7112–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinlein, M., Padgett, H. S., Gens, J. S., Pickard, B. G., Casper, S. J., Epel, B. L. & Beachy, R. N. (1998) Plant Cell 10, 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mas, P. & Beachy, R. N. (1999) J. Cell Biol. 147, 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahn, T. W., Lapidot, M., Heinlein, M., Reichel, C., Cooper, B., Gafny, R. & Beachy, R. N. (1998) Plant J. 15, 15–25. [DOI] [PubMed] [Google Scholar]
- 16.Beachy, R. N. (1999) Philos. Trans. R. Soc. London B 354, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bendahmane, M., Szecsi, J., Chen, I., Berg, R. H. & Beachy, R. N. (2002) Proc. Natl. Acad. Sci. USA 99, 3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendahmane, M., Fitchen, J. H., Zhang, G. & Beachy, R. N. (1997) J. Virol. 71, 7942–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holt, C. A., Hodgson, R. A., Coker, F. A., Beachy, R. N. & Nelson, R. S. (1990) Mol. Plant–Microbe Interact. 3, 417–423. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe, Y., Ohno, T. & Okada, Y. (1982) Virology 120, 478–480. [DOI] [PubMed] [Google Scholar]
- 21.Hagiwara, Y., Komoda, K., Yamanaka, T., Tamai, A., Meshi, T., Funada, R., Tsuchiya, T., Naito, S. & Ishikawa, M. (2003) EMBO J. 22, 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Restrepo, M. A., Freed, D. D. & Carrington, J. C. (1990) Plant Cell 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meshi, T., Hosokawa, D., Kawagishi, M., Watanabe, Y. & Okada, Y. (1992) Virology 187, 809–813. [DOI] [PubMed] [Google Scholar]
- 24.Matthews, R. (1981) Plant Virology (Academic, New York).
- 25.Buck, K. W. (1999) Philos. Trans. R. Soc. London B 354, 613–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beachy, R. N. & Zaitlin, M. (1975) Virology 63, 84–97. [DOI] [PubMed] [Google Scholar]
- 27.Dohi, K., Mori, M., Furusawa, I., Mise, K. & Okuno, T. (2001) Arch. Virol. 146, 1607–1615. [DOI] [PubMed] [Google Scholar]
- 28.Carette, J. E., Stuiver, M., Van Lent, J., Wellink, J. & Van Kammen, A. (2000) J. Virol. 74, 6556–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenllado, F. & Bol, J. F. (2000) Virology 268, 29–40. [DOI] [PubMed] [Google Scholar]
- 30.Choi, J., Kim, B. S., Zhao, X. & Loesch-Fries, S. (2003) Virology 305, 44–49. [DOI] [PubMed] [Google Scholar]
- 31.Bol, J. F. (1999) J. Gen. Virol. 80, 1089–1102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.